Abstract
Bacterial strains belonging to Burkholderia cepacia can be human opportunistic pathogens, plant pathogens, and plant growth promoting and have remarkable catabolic activity. B. cepacia consists of several genomovars comprising what is now known as the B. cepacia complex. Here we report the quorum-sensing system of a genomovar I onion rot type strain ATCC 25416. Quorum sensing is a cell-density-dependent regulatory response which involves the production of N-acyl homoserine lactone (HSL) signal molecules. The cep locus has been inactivated in the chromosome, and it has been shown that CepI is responsible for the biosynthesis of an N-hexanoyl HSL (C6-HSL) and an N-octanoyl HSL (C8-HSL) and that the cep locus regulates protease production as well as onion pathogenicity via the expression of a secreted polygalacturonase. A cep-lacZ-based sensor plasmid has been constructed and used to demonstrate that CepR responded to C6-HSL with only 15% of the molar efficiency of C8-HSL, that a cepR knockout mutant synthesized 70% less HSLs, and that CepR responded best towards long-chain HSLs. In addition, we also report the cloning and characterization of the stationary-phase sigma factor gene rpoS of B. cepacia ATCC 25416. It was established that quorum sensing in B. cepacia has a negative effect on rpoS expression as determined by using an rpoS-lacZ transcriptional fusion; on the other hand, rpoS-null mutants displayed no difference in the accumulation of HSL signal molecules.
Burkholderia cepacia was first described as a potent phytopathogen responsible for the bacterial rot of onions (5). Bacterial strains of B. cepacia are recognized as major opportunistic pathogens in patients with fibrocystic lung disease (20). B. cepacia strains can degrade complex herbicides and pesticides (10) and also behave as plant growth-promoting rhizobacteria by suppressing soil-borne plant pathogens (3, 6, 35). B. cepacia consists of groups or subpopulations termed genomovars (49), the term genomovar refers to a group of strains with phenotypic similarity but genotypic uniqueness. The group of genomovars in this case is called the B. cepacia complex, which comprises at least nine genomic species originally referred to as genomovars I to IX (7, 48, 49). Taxonomic studies have shown that this is a very heterogeneous group of genotypically distinct strains that show a low level of DNA hybridization.
Quorum sensing is a mechanism for regulating gene expression in response to changes in cell density of a bacterial population (15). In gram-negative bacteria, N-acyl homoserine lactone (HSL) autoinducers appear to be the most commonly used signaling molecules; in most cases they are produced by an autoinducer synthase protein belonging to the LuxI family. A transcriptional activator belonging to the LuxR family forms a complex with the cognate autoinducer at high threshold levels to induce transcriptional activation of target genes. The autoinducers produced by different bacterial species differ in the length and structure of the acyl chain, and they are believed to be readily diffusible across the cell envelope into the growth medium where they accumulate. Accumulation continues until the cell density, and consequently also the HSL concentration, reaches a quorum, thereby activating the LuxR type protein, which then elicits the desired cellular response. Quorum sensing has been implicated in the regulation of biofilm formation, plasmid transfer, and motility and in several virulence factors (53).
Another important regulatory component involved in regulation in response to high cell density is the stationary-phase sigma factor RpoS (25, 30). RpoS is an alternative sigma factor which directs transcription of a large number of genes involved in adaptation to nutrient-limiting conditions and several environmental stresses as well as inducing the production of virulence factors (30, 47). In Escherichia coli and Pseudomonas spp., RpoS levels are induced as bacterial cultures enter the stationary phase (14, 26, 27), and it has also been reported that quorum sensing and RpoS cross-regulate their gene expression in Pseudomonas aeruginosa (28, 51).
The quorum-sensing system of a B. cepacia genomovar III cystic fibrosis respiratory isolate has been identified and characterized and consists of cepI and cepR genes. This bacterial isolate, designated K56-2, synthesizes an N-octanoyl HSL (C8-HSL) and an N-hexanoyl HSL (C6-HSL) (29, 32). The CepI/R quorum-sensing system of B. cepacia K56-2 has been implicated in the negative regulation of the siderophore ornibactin and in the positive regulation of a secreted protease. Similarly, the CepI/R quorum-sensing system from another B. cepacia genomovar III cystic fibrosis respiratory isolate, designated H111, has been recently identified and characterized and shown to be involved in regulating biofilm formation and swarming motility (23). In addition, quorum-sensing systems are present and conserved among the heterogeneous B. cepacia complex (19). The stationary-phase sigma factor RpoS, on the other hand, has to our knowledge not been reported in any bacterial strains belonging to the B. cepacia complex.
In this study we report the identification and characterization of the quorum-sensing system of the onion pathogen B. cepacia type strain ATCC 25416 which belongs to genomovar I. This strain has been isolated from a rotten onion, and it has a genome of 8.1 Mb composed of four circular replicons of 3.65 Mb, 3.17 Mb, 1.07 Mb and 200 Kbp (39). We have constructed a cepR-PcepI-lacZ reporter plasmid and shown (i) that a cepR knockout mutant results in a 70% decrease in HSL production, CepR responded to C6-HSL with only 15% the molar efficiency of C8-HSL, and CepR responds best to long chain HSL autoinducers and (ii) that cepI and cepR knockout mutants are attenuated in onion pathogenicity. The rpoS gene of B. cepacia ATCC 25416 has also been identified and characterized. B. cepacia RpoS did not display very high identity in its primary structure to RpoS belonging to the γ-Proteobacteria. RpoS was not involved in HSL production, and quorum sensing had a negative effect on rpoS expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The plasmids used in this study are listed in Table 1. B. cepacia ATCC 25416 (or LMG 1222) is an isolate from a rotten onion, and it was routinely grown in Luria-Bertani (LB) broth (40), M9 minimal medium (40), or M9GP (18) at 30°C. Chromobacterium violaceum CVO26 is a double mini-Tn5 mutant derived from ATCC 31532. This mutant is nonpigmented, and production of the purple pigment can be induced by providing exogenous HSL inducer molecules (34). The E. coli strains used in this study included HB101 (40), DH5α (21), and HB101::Tn5 (33) and were grown in LB medium (36) at 37°C. Antibiotics were added as required at the following final concentrations: tetracycline, 10 (for E. coli), 20 (for C. violaceum), or 300 (for B. cepacia) μg/ml; gentamicin, 10 (for E. coli) or 300 (for B. cepacia) μg/ml; ampicillin, 100 μg/ml (for E. coli); kanamycin, 50 (for E. coli) or 300 (for B. cepacia) μg/ml; and streptomycin, 100 μg/ml (for C. violaceum).
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pBluescript KS | Apr, Co1E1 replicon | Stratagene |
| pUC18 | Apr, Co1E1 replicon | 54 |
| pMP190 | Cmr, IncQ, promoter probe vector | 44 |
| pUC4K | Apr, pBR322 replicon | Amersham Pharmacia |
| pMP77 | Cmr, IncQ, promoter probe vector | 44 |
| pRK2013 | Kmr, Tra+ Mob+, Co1E1 replicon | 12 |
| pPH1J1 | Gmr IncP1 | 1 |
| pSUP2021 | Apr Cmr Tcr Kmr pBR325 replicon, broad-host-range vector | 43 |
| pLAFR3 | Tcr, broad-host-range cloning vector, IncP1 | 45 |
| pQF50 | Apr, broad-host-range vector, pRO1600 replicon | 11 |
| pLIR5 | Tcr, pLAFR3 containing cepIR locus in a 9-kbp EcoRI fragment | This study |
| pBIR | Apr, pBluescript KS containing cep1R locus in a 9-kbp EcoRI fragment | This study |
| pCQS1 | Tcr, pLAFR3 containing B. cepacia DNA | This study |
| pCQS2 | Tcr, pLAFR3 containing B. cepacia DNA | This study |
| pCQS3 | Tcr, pLAFR3 containing B. cepacia DNA | This study |
| pLIR::Tn53 | Tcr, Kmr, pLIR5 with a Tn5 insertion in cepR | This study |
| pLCIKm | Tcr, Kmr, pLIR5 with a Kmr cassette in cepI | This study |
| pSCR1 | Apr, pQF50 containing PcepI-lacZ and cepR | This study |
| pMPIR | Cmr, pMP77 containing the cepIR locus in a 4.5-kbp PstI fragment | This study |
| pRPC-1 | Apr, pUC18 containing a 300-bp PCR fragment of rpoS gene | This study |
| pCOSRPOS-2C | Tcr, pLAFR3 containing B. cepacia DNA | This study |
| pRBS-2 | Apr, pBluescript KS containing a 5-kbp SmaI fragment from pCOSRPOS-2C | This study |
| pRBS-3 | Apr, pBluescript KS containing a 1-kbp PstI fragment from pCOSRPOS-2C | This study |
| pRBS-3Km | Apr Kmr, pRBS-3 with a Kmr cassette in the rpoS gene | This study |
| pCOSRPOS-2CKm | Tcr Apr Kmr, pCOSRPOS-2C with a Kmr cassette in the rpoS gene | This study |
| pRPR2 | Cmr, PrpoS-lacZ fusion in pMP190 promoter probe vector | This study |
Recombinant DNA techniques.
Digestion with restriction enzymes, agarose gel electrophoresis, purification of DNA fragments, ligation with T4 DNA ligase, end filling with Klenow fragment of DNA polymerase, Southern hybridization, and transformation of E. coli were performed as described previously (40). Analytical amounts of plasmids were isolated as described previously (4), whereas preparative amounts were purified with Qiagen columns. Total DNA from B. cepacia was isolated by the sarcosyl-pronase lysis method (2). Triparental matings from E. coli to B. cepacia were performed with the helper strain E. coli(pRK2013) (12). A genomic cosmid library of B. cepacia ATCC 25416 was constructed by using a Gigapack III XL packaging extract kit (Stratagene, La Jolla, Calif.) and following the instructions provided by the supplier. Briefly, genomic DNA was partially digested with EcoRI and ligated into pLAFR3. Concatemers were then packaged into lambda phages and used to infect E. coli HB101 cells.
Cloning of quorum-sensing genes of B. cepacia ATCC 25416.
About 4 × 109 cells (each) of E. coli HB101 harboring the B. cepacia ATCC 25416 cosmid library and E. coli(pRK2103) and 2 × 108 cells of C. violaceum CVO26 were mixed. The suspension was applied to a 0.45-μm-pore-size membrane filter (Millipore Corp.) on an LB plate. After overnight incubation at 30°C, the cells were resuspended and spread on LB plates containing ampicillin (100 μg/ml), kanamycin (100 μg/ml), streptomycin (100 μg/ml), and tetracycline (20 μg/ml). Strain CVO26 is naturally resistant to ampicillin and streptomycin and resistant to kanamycin due to the mini-Tn5 present on the chromosome. Tetracycline will select transconjugants that have received the pLAFR3-based cosmid clone. These plates were incubated for 48 h at 30°C, and transconjugants that turned purple were further assayed. Three cosmids (pCQS1 to pCQS3) from the cosmid library could restore purple pigmentation, and they only shared a 9-kb EcoRI common insert. Further subcloning experiments confirmed that the 9-kb EcoRI fragment cloned in pLAFR3 (creating pLIR5) could restore pigmentation on strain CVO26. Thin-layer chromatographic analysis with extracts from spent supernatants of strain CVO26(pLIR5) showed that C6- and C8-HSL were now synthesized.
Transposon Tn5 mutagenesis.
Transposon Tn5 insertions within recombinant plasmid pLIR5 were obtained as described previously (33), with E. coli HB101::Tn5 as the source of the transposon. E. coli HB101 cells containing Tn5 insertions within plasmid pLIR5 were identified by purifying plasmid DNA from HB101::Tn5(pLIR5), using it to transform E. coli DH5α, and selecting for plasmids having tetracycline and kanamycin resistance. These recombinant plasmids in DH5α were delivered by triparental conjugation to C. violaceum CVO26 as described above. Transconjugants of strain CVO26(pLIR5::Tn5) lacking the expression of violacein (i.e., those which remained white) were purified, and the position of Tn5 was mapped within the 9-kb EcoRI insert. Three insertions were located in the cepR gene (data not shown).
Construction of a B. cepacia ATCC 25416 cepI and cepR knockout mutants.
Plasmid pLCIKm was constructed as follows. A 3.5-kb HindIII DNA fragment from pSUP2021containing the Kmr gene of transposon Tn5 was cloned into the corresponding site in cepI in plasmid pLIR5. Plasmid pLIR::Tn53 (which contain a Tn5 insertion in the cepR gene harbored in pLAFR3) and plasmid pLCIKm (which contains a kanamycin resistance gene cloned in the cepI gene) were homogenized with the corresponding target regions of the genome of B. cepacia ATCC 25416 by a marker-exchange procedure (9). Plasmid pPH1JI was used as the incoming IncP1-incompatible plasmid, and selections were made on LB plates containing kanamycin and gentamicin. This generated two genomic mutants, designated B. cepacia 25416-I and B. cepacia 25416-R, which harbored a Kmr cassette in the cepI gene and a Tn5 insertion in the cepR gene, respectively. The fidelity of each marker-exchange event was confirmed by Southern analysis (data not shown).
Construction of a B. cepacia CepR-based HSL-detecting plasmid.
The cep genes of B. cepacia ATCC 25416 were used to construct a plasmid detecting HSL molecules. A BamHI-HindIII DNA fragment of 3.5 kb from pBIR containing the whole cepR gene and the cepI promoter together with the first 120 bp of cepI were inserted into the corresponding sites of the vector pQF50 (11), yielding pSCR1. Plasmid pQF50 contains a promoterless lacZ gene, and in plasmid pSCR1, the transcription of the lacZ gene is under the control of the cepI promoter. The β-galactosidase activity was determined as described previously (34, 43).
Purification, detection, and visualization of autoinducer molecules (HSLs).
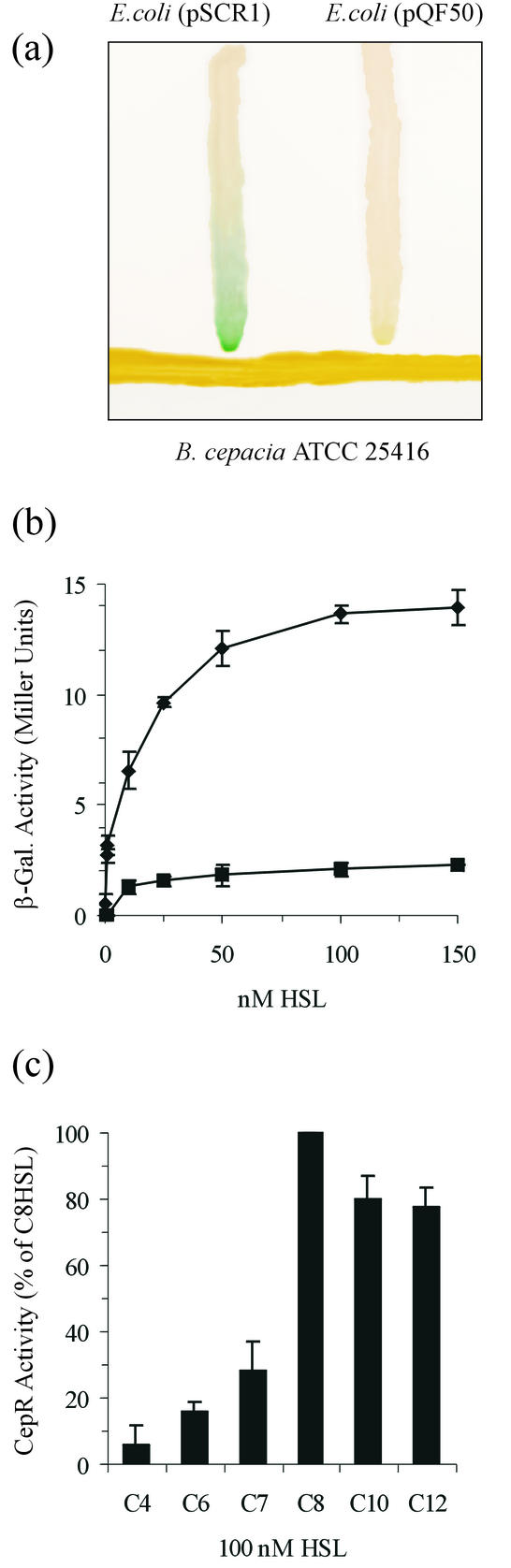
The purification, detection, and visualization of HSL inducer molecules from culture supernatants were performed essentially as described previously (25, 34). Synthetic HSLs (C4-HSL, C6-HSL, C7-HSL, C8-HSL, C10-HSL, and C12-HSL) were purchased from Fluka Chemie AG (Buchs, Switzerland). For quantification of CepR activity, overnight E. coli DH5α(pSCR1) cultures were normalized to an optical density at 600 nm (OD600) of 0.1 in a volume of 20 ml of LB containing the desired HSL at the desired concentration. Cultures were then grown with agitation at 37°C for 6 h, and β-galactosidase activities were determined. The presence of HSL was also detected on solid media by growing E. coli(pSCR1) on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and ampicillin media in close proximity to the tester strain. The presence of HSL was observed when E. coli(pSCR1) turned blue (Fig. 1a).
FIG. 1.
(a) Activation of bacterial sensor strain in cross-streak experiments. The sensor strain E. coli DH5α(pSCR1) was cross streaked on LB agar plates (containing ampicillin and X-Gal) with B. cepacia ATCC 25416 used as a tester strain. (b) HSL bioassay with E. coli DH5α(pSCR1). DH5α(pSCR1) was grown in the presence of variousconcentrations of either C6-HSL (▪) or C8-HSL (♦), and β-galactosidase activities were determined after 6 h. The values were determined with LB medium, the means of triplicate experiments are given, and the standard deviations are shown. (c) CepR-HSL response. E. coli DH5α(pSCR1) was grown for 6 h in the presence of a 100 nM concentration of either C4-, C6-, C8-, C10-, or C12-HSL, and β-galactosidase activities were determined. The values were determined with LB medium, the means of triplicate experiments are given, and the standard deviations are shown. The values are expressed as percentages of the activity determined with C8-HSL.
Cloning of the rpoS gene of B. cepacia ATCC 25416.
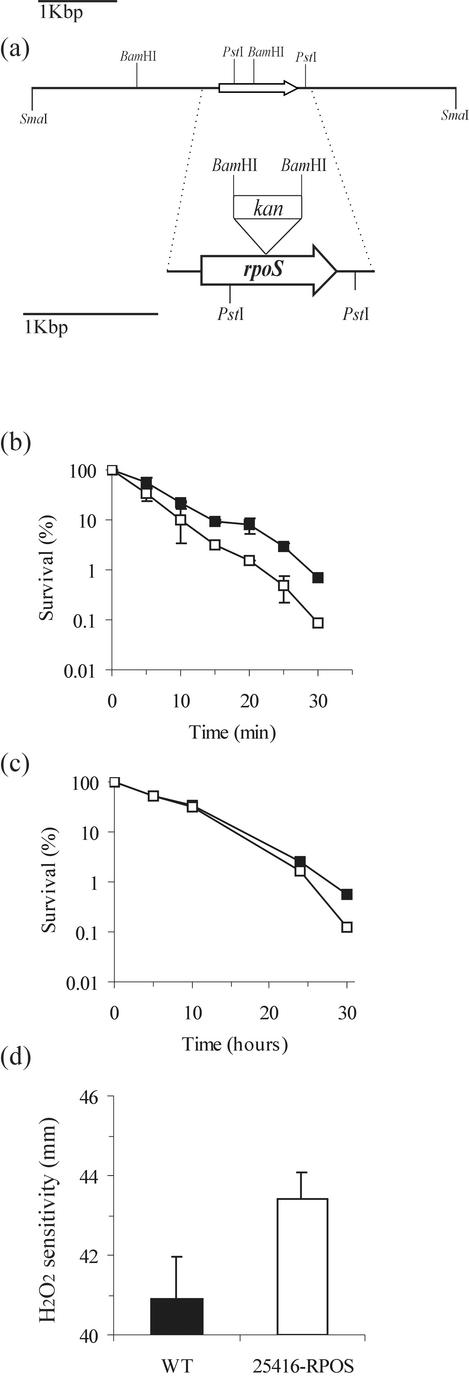
The rpoS DNA sequences from cloned and characterized Pseudomonas spp. (25) were compared with the genome sequence of B. cepacia genomovar III clinical isolate strain J2315 (www.sanger.ac.uk/Projects/B_cepacia/). The putative rpoS of B. cepacia J2315 was localized in contig Bcep1047b02.g1c at positions 10602 to 11690. Two synthetic primers were designed from conserved regions (rpoS-P3, 5′-CTGCTCGACCTGATCGA-3′, and rpos-P6, 5′-AGGCTGCTCGCGGGATC-3′) and used in a PCR amplification reaction. This resulted in the amplification of a 300-bp fragment, which was cloned in pUC18, yielding pRPC-1. This fragment was then used as a probe against the cosmid library of B. cepacia ATCC 25416. A cosmid designated pCOSRPOS-2C was identified, and the rpoS gene was localized within a 5.5-kb SmaI fragment and in part in a 1-kb PstI fragment (see Fig. 4). These were cloned in the corresponding sites in pBluescript KS, yielding pRBS-2 and pRBS-3, respectively.
FIG. 4.
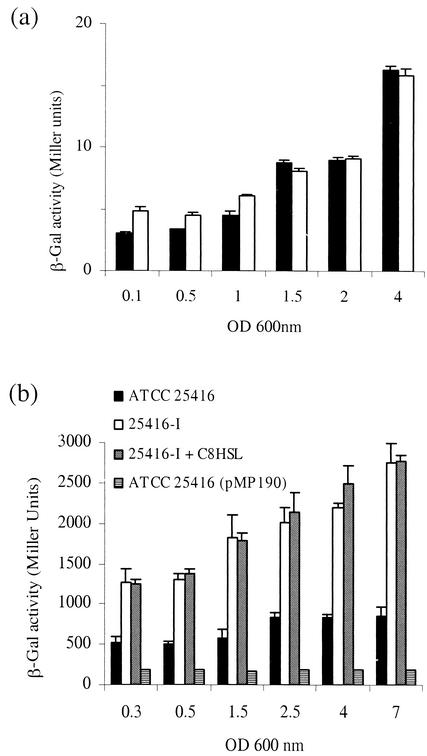
(a) Production of HSLs at different growth stages from B. cepacia ATCC 25416 (filled bars) and from B. cepacia ATCC 25416-RPOS (open bars). Values were determined as described in Materials and Methods. (b) rpoS promoter activity at different growth stages measured in B. cepacia ATCC 25416(pRPR2) (filled bars), B. cepacia 25416-I(pRPR2) (open bars), B. cepacia 25416-I(pRPR2) plus 100 nM C8-HSL (shaded bars), and B. cepacia ATCC 25416(pMP190) (striped bars). Values were determined as described in Materials and Methods.
Construction of a B. cepacia ATCC 25416 rpoS knockout mutant.
The kanamycin-resistant gene from pUC4K as a BamHI fragment was cloned in the corresponding site in the pRBS-3, creating pRBS-3Km and resulting in an interruption of the rpoS gene of B. cepacia ATCC 25416 (see Fig. 4). This rpoS-interrupted locus in pRBS-3Km was transferred by homologous recombination to the rpoS gene harbored in pCOSRPOS-2C in the following way: pRBS-3Km was used to transform DH5α(pCOSRPOS-2C), the resulting DH5α(pCOSRPOS-2C)(pRBS-3Km) was grown overnight, and the culture was used in a triparental conjugation into Pseudomonas putida WCS358 (16) with E. coli(pRK2013) as a helper. After appropriate selection, pCOSRPOS-2CKm was selected. Transfer of the Kmr cassette by double-crossover homologous recombination from pRBS-3Km to pCOSRPOS-2 was verified by restriction and Southern analysis. The plasmid pCOSRPOS-2CKm was then used in a marker-exchange technique, as described above, in order to introduce site-specific insertion mutations into the rpoS gene of B. cepacia ATCC 25416. The fidelity of the marker-exchange event in the B. cepacia rpoS::Kmr mutant was confirmed by Southern analysis (data not shown). This mutant was designated B. cepacia 25416-RPOS.
Exoenzyme assays, siderophore production, in vitro maceration of onion tissue, and stress response assays.
Proteolytic, lipolytic, and chitinolytic activity were determined on the appropriate indicator plates (23) and also determined spectrophotometrically (29). Polygalacturonase activity was determined as previously described (18). In order to determine the ability to macerate onion tissue, overnight LB cultures were normalized at an OD600 of 1, and 100 μl of culture was inoculated onto the surface of onion tissue prepared as described previously (52). Clean, disease-free onions (Allium cepa) were wiped with 90% (vol/vol) alcohol before being cut aseptically into slices. Onion slices were placed into sterile petri dishes, and nicks of approximately 2 mm were made in the tissue surface. Plates were incubated at 30°C, and readings were taken after 48 h of incubation. The measurement of cell viability and ability to survive heat stress and osmotic stress and sensitivity to hydrogen peroxide were determined as described previously (25).
DNA sequence determination and analysis.
The DNA sequence of the cepI/R locus was determined by using pBIR as template. The DNA sequence of the rpoS gene was determined by using plasmids pRBS-2 and pRBS-3 as templates. Nucleotide sequences were determined by the dideoxy chain-termination method (41) by using [35S]dATPαS for labeling and 7-deaza-dGTP (Pharmacia) instead of dGTP.
Nucleotide accession number.
The sequence of cepI-cepR locus and the rpoS gene have been deposited in the GenBank/EMBL/DDBJ database under accession numbers AJ422183 and AJ457984, respectively.
RESULTS
The quorum-sensing system of B. cepacia ATCC 25416.
The quorum-sensing systems of two B. cepacia clinically isolated strains (genomovar III) have been associated with roles in siderophore production and ability to form biofilms (23, 29). In addition, it has been reported that many strains belonging to the B. cepacia complex produce HSL molecules with an aliphatic chain composed of C6 and C8 (19). The quorum-sensing system of B. cepacia genomovar I ATCC 25416 produces both C8- and C6-HSL, and cepI and cepR genes have been previously cloned by PCR amplification (19). In this study, we have cloned and characterized the complete genomic locus coding for CepIR in B. cepacia ATCC 25416 (accession number AJ422183). In order to study this quorum-sensing system, we constructed a sensor plasmid pSCR1 which is made of components of the cep quorum-sensing system of strain ATCC 25416. This sensor plasmid contains the promoter of cepI transcriptionally fused to a promoterless lacZ reporter gene together with the cepR gene and intergenic region in plasmid pQF50. It was observed that on solid media containing X-Gal, pSCR1 harbored in E. coli can conveniently be used to detect the production of HSL when streaked in close proximity to a tester strain (Fig. 1a).
Since B. cepacia ATCC 25416, like many other strains of the B. cepacia complex (19), produced C6- and C8-HSL molecules, we tested the activity of these two molecules by using plasmid pSCR1. CepR responded to C6-HSL with only 15% of the molar efficiency of C8-HSL; the plasmid detected the latter molecule at a concentration of less than 10 nM and reached its highest β-galactosidase activity when a 100 nM concentration of the molecule was present. Higher concentrations did not result in higher reporter enzyme activity (Fig. 1b). The C6-HSL, on the other hand, did not result in a good response with CepR (Fig. 1b), even at very high concentrations as high as 1,000 nM (data not shown).
It was of interest to determine the response of the CepR-based sensor plasmid with respect to different HSL molecules. This was determined by measuring the β-galactosidase activities of E. coli(pSCR1) exposed to 100 nM concentrations of various HSL molecules. It was decided to use 100 nM since it was the minimum concentration of C8-HSL (the major molecule produced by B. cepacia) necessary to achieve a good response (see Fig. 3a). As expected, the constructed biosensor responded best to C8-HSL when compared to C4-, C6-, C7-, C10-, and C12-HSL. In Fig. 1c, the percentage of the CepR response to C8-HSL is presented, and as shown, CepR displayed a good response to long-chain signal molecules and very poor one to short-chain signal molecules.
FIG.3.
(a) Map of the 5.5-kbp SmaI DNA fragment from B. cepacia ATCC 25416 isolated in this study. Shown are several enzyme restriction sites and the location of the rpoS gene within this fragment. Also shown is the position where a Kmr-containing BamHI fragment derived from pUC4K was cloned in the corresponding site of the rpoS gene to create pLCIKm. (b, c, and d) Effect of rpoS on stress responses of B. cepacia ATCC 25416. (b) Response to heat shock (50°C). Viability is expressed as a percentage of the number of CFU at time zero. (c) Response to osmotic shock (2 M NaCl). Viability is expressed as a percentage of the number of CFU at time zero. (d) Effect of rpoS mutation on oxidative stress. The sensitivity to H2O2 was measured on cells grown for 16 h in LB at 30°C. The zones of inhibition were measured in millimeters. Filled symbols, WT strain; open symbols, B. cepacia 25416-RPOS strain.
The cepI and cepR genes were insertionally inactivated independently, creating two genomic mutants, 25416-I and 25416-R, respectively. The 25416-I mutant no longer produced the C6- and C8-HSL (data not shown). The 25416-R synthesizes approximately 30% of the signal molecules produced by the wild-type parent strain (data not shown). Since the detecting CepR-based plasmid is not specific to C6-HSL, this value refers mainly to C8-HSL. Furthermore, on the basis of the β-galactosidase activity obtained, it was estimated that a culture of B. cepacia ATCC 25416 at an OD600 of 1 contains an approximately 100 nM concentration of C8-HSL signal molecules. Both 25416-I and 25416-R could be complemented for HSL production when plasmid pMPIR, containing the cepI and cepR genes, was introduced by triparental conjugation (data not shown).
CepIR regulates onion pathogenicity in B. cepacia ATCC 25416.
It was observed that 25416-I and 25416-R displayed similar lipolyitc and chitinolytic activities and siderophore production when compared to the wild-type parent strain, but the proteolytic activity, on the other hand, was significantly lower in the two mutants. B. cepacia ATCC 25416 was originally isolated as a pathogen responsible for the rot of onions (5), and it was therefore of interest to check if quorum sensing was related to rot. Experiments with in vitro maceration of the onion A. cepa resulted in both 25416-I and 25416-R mutants having attenuated maceration compared to the wild type; introduction in trans of pMPIR carrying the cepI/R locus resulted in more-severe onion pathogenicity than in the wild type (Fig. 2). The attenuated maceration in the onion could be attributed to the decrease in production of extracellular enzymes involved in the onion maceration. One such example could be polygalacturonase, which has been identified and characterized in this strain and implicated in onion disease development (18). We determined polygalacturonase activity in spent culture supernatants of ATCC 25416 and mutant 25416-I. The enzyme activity in inducing conditions was reduced 40% in 25416-I, whereas in the complemented mutant, 25416-I(pMPIR), the activity was 140% of the wild-type levels (Table 2). Quorum sensing was therefore implicated in onion pathogenicity in this strain, and this is at least in part due to the hydrolytic secreted enzyme polygalacturonase. Similar attenuation and complementation levels of enzyme activity were obtained with 25416-R (data not shown).
FIG. 2.
Role of quorum sensing in onion rot. One hundred microliters of a culture of the indicated strain with an OD600 of 1 was inoculated on the right half of a sterile onion as described in the text. This picture was taken after 48 h of incubation at 30°C.
TABLE 2.
Polygalacturonase activity from spent culture supernatants of B. cepacia ATCC 2516 and 25416-I
| Strain | Polygalacturonase activity (U/mg)a | % of ATCC 25416 polygalacturonase activity |
|---|---|---|
| ATCC 25416 | 2,300 | 100 |
| 25416-I | 1,380 | 60 |
| 25416-I(pMPIR) | 3,220 | 140 |
Cultures were grown for 16 h in M9GP medium and had similar OD650s (18). For growth media and enzyme activity, see Materials and Methods. The mean values of triplicate experiments are given, and standard deviations were ±5%.
Stationary-phase rpoS gene of B. cepacia ATCC 25416.
The nucleotide sequence of the putative rpoS gene of strain ATCC 25416 was determined and consists of 1,089 nucleotides (Fig. 3a) (accession number AJ457984). The RpoS protein consists of 362 amino acids with a molecular mass of 41 kDa. Interestingly, it displayed the highest amino acid identity with other putative RpoS proteins of the Burkholderia genus (98% with B. cepacia genomovar III J2315, >88% with Burkholderia fungorum, and 93% with Burkholderia pseudomallei and Burkholderia mallei) and 74% identity with the experimentally determined RpoS of Ralstonia solanacearum, whereas it displayed approximately 50% identity with the sigma factors of γ-Proteobacteria (e.g., P. aeruginosa, E. coli, and Vibrio cholerae). In order to investigate the physiological role that rpoS played in B. cepacia, we insertionally inactivated rpoS in strain ATCC 25416, yielding an rpoS::Kmr knockout mutant called B. cepacia 25416-RPOS. We tested the response of 25416-RPOS to various environmental conditions, as the RpoS sigma factor is known to confer cross-protection against several stresses in other gram-negative bacteria (17, 38, 47). We tested the resistance of the parent strain and 25416-RPOS against heat shock, increased osmolarity, and hydrogen peroxide. Strain 25416-RPOS was more sensitive to heat shock and oxidative stress than the wild-type strain; however, there was no significant difference when cells were exposed to an increase in osmotic pressure (Fig. 3b, c, and d)
Do rpoS and quorum sensing cross-regulate each other in B. cepacia?
It was of interest to investigate whether RpoS and quorum sensing cross-regulate each other since the genes regulated by these two systems are maximally expressed at the stationary phase.
In order to determine whether rpoS influenced the accumulation of HSL signal molecules, we quantified HSL production in 25416-RPOS and compared it to the values obtained with the wild type strain. Signal molecules were extracted from spent supernatants of the wild-type and mutant strains at different growth stages, and the extracts were assayed with E. coli(pSCR1). Figure 4a depicts the results showing that the B. cepacia mutant lacking RpoS synthesizes at different growth stages approximately the same amount of signal molecules produced by the wild-type parent strain, demonstrating that the absence of rpoS does not influence the production of HSL molecules.
In order to determine whether the quorum-sensing system is regulating rpoS expression, the rpoS promoter was cloned as a 2-kb BamHI fragment from pCOSRPOS-2C in the BglII site of β-galactosidase promoter probe vector pMP190, creating pRPR2. rpoS promoter activity was then assayed at different growth stages in the 25416-I mutant and compared to the promoter activity obtained with the wild-type strain. As depicted in Fig. 4b, rpoS expression in the wild-type strain was relatively constant during the early, exponential, and stationary phases. It was then determined that in 25416-I, rpoS expression increased two- to threefold in all growth stages; this increase in activity was retained even when a 100 or 1,000 nM concentration of C8-HSL was added to the growth media (Fig. 4b). A similar increase in rpoS promoter activity was also observed in the cepR mutant 25416-R (data not shown). In view of the fact that rpoS expression increased in the quorum-sensing mutants, the resistance against heat shock and hydrogen peroxide was determined in mutant 25416-I. It was observed, however, that the resistance to the two stresses was comparable to what was observed with the wild-type strain (data not shown).
DISCUSSION
In this study we identified and characterized the genetic determinants coding for the quorum-sensing system and for the stationary-phase sigma factor RpoS in B. cepacia genomovar I type strain ATCC 25416. To our knowledge, this is the first report of the rpoS gene in the B. cepacia complex, other genes have been sequenced and are highly identical (83 to 98%, see above). This is only the second rpoS gene characterized from a β-Proteobacteria; interestingly, RpoS proteins belonging to β-Proteobacteria are not so closely related to the RpoS proteins of γ-Proteobacteria (22). These two loci, cepI/R and rpoS, involved in stationary-phase gene expression were inactivated and studied.
B. cepacia ATCC 25416, like other members of the B. cepacia complex, synthesizes C6- and C8-HSL molecules (19). The synthesis of both appears to require the CepI protein reported here, since a cepI knockout mutant did not synthesize either (data not shown). It is possible that the cepI synthase directs the synthesis of C8-HSL and lower traces of C6-HSL by recognizing two different acyl-acyl carrier proteins (37). It was observed that our CepR-based biosensor in E. coli has a stronger response to long-chain HSLs than to short-chain HSLs, and it displays the best response to the C8-HSL, the major molecule synthesized by B. cepacia. It must be stressed, however, that these results will not necessarily reflect what the in vivo situation is in B. cepacia, since it has been reported that for the LuxR homologue CarR of Erwinia carotovora, the protein concentration as well as the bacterial host can influence its specificity towards different HSL molecules (50). However, the fact that the C8-HSL has the strongest response in E. coli is an indication that the results obtained here are likely to also occur in B. cepacia. It is believed that HSL-protein interaction reduces susceptibility to proteolysis as well as allowing multimerization important for activating transcription (31, 50). It is likely that CepR has a binding pocket to fit molecules with a longer acyl chain. Recently, similar experiments were performed by using a cep-gfp-based biosensor showing that CepR from B. cepacia H111 responds best to C8 and other long-chain HSL autoinducers (46). CepR displayed a very low response towards the C6-HSL, questioning the function of this molecule in B. cepacia. However, it cannot be excluded that in the genome of B. cepacia there is another luxR family gene coding for another LuxR family protein, which displays a better response to the C6-HSL autoinducer; in this case, this molecule might have a biological function. The presence of this molecule could slightly affect our results regarding our estimation in E. coli(pSCR1) that a 100 nM concentration of C8-HSL is regarded as the minimum necessary for the E. coli(pSCR1) response (Fig. 1b), since C6-HSL molecules could interfere with CepR activity.
Various extracellular phenotypes were tested in relation to quorum sensing, and most did not display any alteration in the quorum-sensing-deficient mutant, with the exception of protease activity. Proteolytic activity was significantly lower in the 25416-I mutants; this observation was similar to what was observed in two different B. cepacia genomovar III quorum-sensing mutants (23, 29). Interestingly, the two B. cepacia genomovar III quorum-sensing mutants displayed either an increase or decrease in siderophore production, whereas in the genomovar I B. cepacia strain used in this study there was no alteration of siderophore production. The discrepancies can be attributed to the fact that the strains belong to different genomovars, highlighting the fact that there is no common quorum-sensing phenotype found within the B. cepacia complex, with the sole exception of protease activity. Interestingly, polygalacturonase activity was significantly reduced in the quorum-sensing mutants (Table 2), and this was also indirectly observed by the attenuated onion maceration activity (Fig. 2). Polygalacturonase activity in B. cepacia ATCC 25416 has been associated with onion maceration, and the plasmid-borne pehA gene is a virulence factor in this strain (18). The complemented mutants harboring the cep locus in trans resulted in higher polygalacturonase activity and higher onion maceration, thus further indicating the involvement of quorum sensing in the onion pathogenicity of this strain. It was not possible, however, to restore wild-type levels of onion maceration activity of the cepI and cepR mutants by dispersing synthetic C8-HSL on the onion (data not shown).
The stationary-phase sigma factor RpoS has also been implicated in several gram-negative bacteria in the change in gene expression necessary for the adaptation to stationary phase (17, 38, 47), and it has also been shown to be involved in the regulation of virulence factors (8, 24, 42). We have observed that RpoS in B. cepacia is necessary for adaptation to heat and oxidative stress, whereas it was not important for osmotic shock adaptation. It was of interest to study whether RpoS and quorum sensing cross-regulate each other since the gene(s) regulated by these two systems are maximally expressed at the stationary phase. It was determined that a genetic background without RpoS has no influence in the accumulation of the HSL molecules produced by B. cepacia (Fig. 4a); on the other hand, rpoS promoter activity is positively influenced by the absence of the quorum-sensing system (Fig. 4b). This doubling in promoter activity did not reflect an increased resistance to RpoS-regulated stresses; this might be because increasing the promoter activity does not automatically result in an increasing amount of RpoS protein as posttranscriptional regulation of RpoS might occur (22). This increase in rpoS promoter activity in the cepI mutant 25416-I could not be restored to wild-type levels by the addition to the growth media of synthetic C8-HSL (Fig. 4b). The reason for this is not known; in addition, it was observed that rpoS promoter activity was higher in 25416-I already at low cell densities and activity increased as the cells entered the stationary phase (Fig. 4b). In the wild-type strain, however, the expression of rpoS remained relatively constant during bacterial growth; this is in contrast to rpoS regulation in Pseudomonas where transcription dramatically increases at the onset of the stationary phase (23). It is similar, however, to what happens with E. coli since RpoS levels are mainly controlled at the posttranscriptional and posttranslational levels (22). In R. solanacearum it has been reported that RpoS regulates the HSL production by the decrease in solR and solI expression (13). Similarly, in P. aeruginosa, quorum sensing and rpoS cross-regulate each other (28, 51). Future work will determine how quorum sensing regulates rpoS and whether the two systems are involved in the regulation of similar phenotypes of B. cepacia.
Acknowledgments
C.A. is financially supported by an ICGEB (International Centre for Genetic Engineering and Biotechnology) predoctoral fellowship. I.B. is the beneficiary of a CNR (Consiglio Nazionale delle Ricerche) fellowship.
REFERENCES
- 1.Beringer, J. E., J. L. Beynon, A. V. Buchanan-Wollaston, and A. W. B. Jonhston. 1978. Transfer of the drug-resistance transposon Tn5 to Rhizobium. Nature 276:633-634. [Google Scholar]
- 2.Better, M., B. Lewis, D. Corbin, G. Ditta, and D. R. Helinski. 1983. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell 35:479-485. [DOI] [PubMed] [Google Scholar]
- 3.Bevivino, A., S. Tabacchioni, L. Chiarini, M. V. Carusi, M. Del Gallo, and P. Visca. 1994. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology 140:1069-1077. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 5.Burkholder, W. 1950. Sour skin, a bacterial rot of onions bulbs. Phytopathology 40:115-118. [Google Scholar]
- 6.Cartwright, D. K., W. S. Chilton, and D. M. Benson. 1995. Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol. 43:211-216.B. D. M. [Google Scholar]
- 7.Coenye, T., P. Vandamme, J. R. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbell, N., and J. E. Loper. 1995. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbin, D., G. Ditta, and D. R. Helinski. 1982. Clustering of nitrogen fixation (nif) genes in Rhizobium meliloti. J. Bacteriol. 149:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubaras, D. L., C. D. Hershberger, K. Kitano, and A. M. Chakrabarty. 1995. Sequence analysis of a gene cluster involved in metabolism of 2,4,5-trichlorophenoxyacetic acid by Burkholderia cepacia AC1100. Appl. Environ. Microbiol. 61:1279-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flavier, A. B., M. A. Schell, and T. P. Denny. 1998. An RpoS (sigmaS) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol. Microbiol. 28:475-486. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, M., K. Tanaka, H. Takahashi, and A. Amemura. 1994. Transcription of the principal sigma-factor genes, rpoD and rpoS, in Pseudomonas aeruginosa is controlled according to the growth phase. Mol. Microbiol. 13:1071-1077. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 16.Geels, F. P., and B. Schippers. 1983. Reduction in yield depression in high frequency potato cropping soil after seed tuber treatments with antagonist fluorescent Pseudomonas spp. Phytopathol. Z. 108:207-221. [Google Scholar]
- 17.Gerard, F., A. M. Dri, and P. L. Moreau. 1999. Role of Escherichia coli RpoS, LexA and H-NS global regulators in metabolism and survival under aerobic, phosphate-starvation conditions. Microbiology 145:1547-1562. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, C. F., E. A. Pettit, V. A. Valadez, and E. M. Provin. 1997. Mobilization, cloning, and sequence determination of a plasmid-encoded polygalacturonase from a phytopathogenic Burkholderia (Pseudomonas)cepacia. Mol. Plant-Microbe Interact. 10:840-851. [DOI] [PubMed] [Google Scholar]
- 19.Gotschlich, A., B. Huber, O. Geisenberger, A. Togl, A. Steidle, K. Riedel, P. Hill, B. Tummler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 20.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 24.Iriarte, M., I. Stainier, and G. R. Cornelis. 1995. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 63:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojic, M., G. Degrassi, and V. Venturi. 1999. Cloning and characterisation of the rpoS gene from plant growth-promoting Pseudomonas putida WCS358: RpoS is not involved in siderophore and homoserine lactone production. Biochim. Biophys. Acta 1489:413-420. [DOI] [PubMed] [Google Scholar]
- 26.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 29.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 31.Luo, Z. Q., and S. K. Farrand. 1999. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc. Natl. Acad. Sci. USA 96:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magazin, M. D., J. C. Moores, and J. Leong. 1986. Cloning of the gene coding for the outer membrane receptor protein for ferric pseudobactin, a siderophore from a plant growth-promoting Pseudomonas strain. J. Biol. Chem. 261:795-799. [PubMed] [Google Scholar]
- 34.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 35.McLoughlin, T. J., J. P. Quinn, A. Bettermann, and R. Bookland. 1992. Pseudomonas cepacia suppression of sunflower wilt fungus and role of antifungal compounds in controlling the disease. Appl. Environ. Microbiol. 58:1760-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramos-Gonzalez, M. I., and S. Molin. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440 J. Bacteriol. 180:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodley, P. D., U. Romling, and B. Tummler. 1995. A physical genome map of the Burkholderia cepacia type strain. Mol. Microbiol. 17:57-67. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarniguet, A., J. Kraus, M. D. Henkels, A. M. Muehlchen, and J. E. Loper. 1995. The sigma factor sigma s affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc. Natl. Acad. Sci. USA 92:12255-12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 44.Spaink, H. P., R. J. H. Okker, C. A. Wijffelmann, E. Pees, and B. J. J. Lugtemberg. 1987. Promoter in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 45.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steidle, A., K. Sigl, R. Schuhegger, A. Ihring, M. Schmid, S. Gantner, M. Stoffels, K. Riedel, M. Givskov, A. Hartmann, C. Langebartels, and L. Eberl. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67:5761-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 49.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch, M., D. E. Todd, N. A. Whitehead, S. J. McGowan, B. W. Bycroft, and G. P. Salmond. 2000. N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wigley, P., and N. F. Burton. 1999. Genotypic and phenotypic relationships in Burkholderia cepacia isolated from cystic fibrosis patients and the environment. J. Appl. Microbiol. 86:460-468. [DOI] [PubMed] [Google Scholar]
- 53.Williams, P., M. Camara, A. Hardman, S. Swift, D. Milton, V. J. Hope, K. Winzer, B. Middleton, D. I. Pritchard, and B. W. Bycroft. 2000. Quorum sensing and the population-dependent control of virulence. Philos. Trans. R. Soc. Lond. B 355:667-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]