Abstract
Prostate-specific antigen (PSA) is a serine protease that is widely used as a surrogate marker in the early diagnosis and management of prostate cancer. The physiological relevance of tissue PSA levels and their role in prostate tumor growth and metastasis are not known. Free-PSA (f-PSA) was purified to homogeneity from human seminal plasma by column chromatography, eliminating hk2 and all known PSA complexes and retaining its protease activity. Confluent mono-layers of prostate cancer cell lines, PC-3M and LNCaP, were treated with f-PSA in a series of in vitro experiments to determine the changes in expression of various genes that are known to regulate tumor growth and metastasis. Gene array, quantitative polymerase chain reaction (QPCR), and enzyme-linked immunosorbent assay (ELISA) results show significant changes in the expression of various cancer-related genes in PC-3M and LNCaP cells treated with f-PSA. In a gene array analysis of PC-3M cells treated with 10 µM f-PSA, 136 genes were upregulated and 137 genes were downregulated. In LNCaP cells treated with an identical concentration of f-PSA, a total of 793 genes was regulated. QPCR analysis reveals that the genes for urokinase-type plasminogen activator (uPA), VEGF, and Pim-1 oncogene, known to promote tumor growth, were significantly downregulated, whereas IFN-γ, known to be a tumor-suppressor gene, was significantly upregulated in f-PSA-treated PC-3M cells. The effect of f-PSA on VEGF and IFN-γ gene expression and on protein release in PC-3M cells was distinctly dose-dependent. In vivo studies showed a significant reduction (P = .03) in tumor load when f-PSA was administered in the tumor vicinity of PC-3M tumor-bearing BALB/c nude mice. Our data support the hypothesis that f-PSA plays a significant role in prostate tumor growth by regulating various proangiogenic and antiangiogenic growth factors.
Keywords: Prostate cancer, prostate-specific antigen, f-MRI, gene array, angiogenic growth factors
Introduction
Prostate cancer is the most frequently diagnosed malignancy in men in North America and Northwestern Europe, with an estimated 230,110 new cases and 29,900 deaths occurring in the United States during the year 2004 [1]. In prostate cancer, the identification of high-risk patients and the prediction of tumor aggressiveness have great importance for prognosis. Prostate-specific antigen (PSA), a member of the kallikrein protein family, is widely used as a surrogate marker in the early diagnosis and management of prostate cancer [2]. PSA is a serine protease with chymotrypsin-like activity [3] and is largely found in prostatic tissue and seminal plasma. In prostate cancer, excess PSA spills into circulation where it exists as a mixture of various molecular forms, including free-PSA (f-PSA) and several PSA-protease inhibitor complexes [4]. At present, there is no consensus as to whether PSA has any role in prostate tumor growth and metastasis. The only well-known physiological function of PSA is to liquefy seminal clot [5]. PSA and/or its gene expression has been detected at low concentrations in the endometrium [6], normal breast tissue, breast milk [7], breast tumors, female serum [8], renal cell carcinoma [9], and ovarian cancer [10]. The PSA gene is known to be regulated by androgen and progestin [11].
Angiogenesis is a tightly regulated process modulated by the dynamic interplay between angiogenic stimulators and inhibitors, which control endothelial cell proliferation, migration, and invasion [12,13]. Tumor neovascularization contributes to cancer progression, facilitating its growth and metastasis. The initiation of secretion of angiogenic proteins by tumor cells is important in vascular hyperpermeability, protease secretion, capillary basement membrane breakdown, and endothelial cell migration and proliferation [12].
Several reports in the literature suggest that PSA has antiangiogenic activity [14,15]. PSA is downregulated in prostate cancer tissue compared to hyperplastic tissue [16,17]. Lower tissue PSA level is often associated with a more aggressive form of prostate cancer [18]. PSA has been shown to reduce melanoma lesions in lungs of mice [14]. Transfection of PSA c-DNA into PC-3 prostate carcinoma cells prolongs their doubling time, and reduces their tumorigenicity and metastasis in nude mice [19]. Furthermore, it is reported that breast cancer patients with high levels of PSA in the tumor tissue had a better prognosis than patients whose tumor tissue had lower PSA levels [20]. One study has shown that “the PSA bounce” phenomenon is not associated with a higher rate of disease progression [21]. High expression of PSA in prostate cancer cells is often associated with low microvessel density [22]. PSA has been shown to cleave plasminogen to produce the antiangiogenic protein “angiostatin,” which is known to suppress angiogenesis [23]. More recently, Fortier et al. [15] reported that recombinant PSA has antiangiogenic activity both in in vitro and in vivo. PSA also has been shown to stimulate reactive oxygen species (ROS) generation in the prostate cancer cell lines, PC-3, Du145, and LNCaP [24]. Although the intact PSA molecule is necessary for stimulating ROS, its proteolytic activity is not required [18]. ROS has been linked to the activation of transcriptional factors as well as signal transduction in apoptosis [25]. In a recent study, we have shown that LNCaP cells that are known to express PSA have lower invasive potential in a cell invasion assay compared to non-PSA-expressing cell lines DU-145 and PC3 [26].
Several studies have used microarray technology as a “gene discovery tool” to identify genetic markers that discriminate between normal and cancerous tissues. In other investigations, gene expression profiles of thousands of genes in normal and prostate tumor tissues were used in hierarchical clustering analysis [27,28]. Dhanasekaran et al. [28] could distinguish normal prostate, benign prostatic hyperplasia (BPH), localized prostate cancer, and metastatic prostate cancer samples with spotted microarrays using hierarchical clustering analysis. Luo et al. [29] also were able to differentiate 16 prostate cancer tissue specimens from nine BPH specimens on the basis of differences in gene expression profiles using cDNA microarrays.
In the present study, we examined the effect of enzymatically active f-PSA in modulating gene expression in prostate cancer cell lines using gene array analysis and real-time quantitative polymerase chain reaction (QPCR). We also documented, for the first time, that f-PSA administered to nude mice inoculated with prostate cancer PC-3M cells effectively reduces overall tumor growth. Gene array analysis of control and PSA-treated tumor tissues also revealed significant differences in expression of genes that are known to be involved in tumor growth and metastasis.
Materials and Methods
Purification and Characterization of Enzymatically Active f-PSA
f-PSA was purified from human seminal plasma by a simple two-step column chromatography procedure [30–32]. The Institutional Review Board at Roswell Park Cancer Institute approved the procurement and use of human seminal plasma. Initially, all known forms of PSA, including f-PSA and various PSA complexes, were isolated from the seminal plasma by thiophilic interaction chromatography and, subsequently, f-PSA was separated from other PSA complexes by “molecular size” chromatography. Briefly, seminal plasma dialyzed against buffer containing 25 mM Hepes and 1 M sodium sulfate, pH 7.0, was applied to a column packed with T-gel. The column was washed with the buffer and the bound proteins were eluted with 25 mM Hepes buffer, pH 7.0, containing no sodium sulfate. The T-gel column eluates containing some semino proteins and all forms of PSA (f-PSA and complexed PSA) were pooled, concentrated, and applied to a column packed with Ultrogel AcA-54 having a fractionation range of 5000 to 70,000 kDa. The column was equilibrated with 10 mM sodium acetate buffer containing 0.15 M sodium chloride, pH 5.6. PSA was monitored in various column eluates ranging in molecular weight between 25 and 40 kDa by Western blot analysis. The column eluates containing f-PSA were concentrated, filter-sterilized, and stored at -70°C. Details of the f-PSA purification procedure are described elsewhere [31,32].
The final quantitation of PSA was based on double-determined enzyme-linked immunosorbent assay (ELISA) using polyclonal and monoclonal anti-PSA antibodies. The f-PSA was characterized for purity by SDS-PAGE/Western blot analysis using anti-PSA and anti-PSA complex monoclonal antibodies, and by 2-D gel electrophoresis with silver and antibody staining. Purified f-PSA was assayed for enzymatic activity using a fluorogenic substrate (Mu-His-Ser-Ser-Lys-Leu-Gln-AFC; Calbiochem, San Diego, CA). The assay is based on the hydrolysis of this fluorogenic substrate and is specific for PSA enzymatic activity [33]. Fluorescence was monitored using an Aminco-Bowman spectrophotofluorometer (American Instrument Co., Silver Spring, MD).
Prostate Cancer Cell Lines
PC-3M cells were obtained from Dr. Isaiah Fidler (The University of Texas M. D. Anderson Cancer Center, Houston, TX). PC-3M is a subline of PC3 isolated from a liver metastasis that originally formed a solid tumor in a nude mouse [34]. PC-3M cells are androgen-independent and do not produce any PSA in the culture medium [35]. The LNCaP cell line was initially established from a metastatic lesion of human prostatic adenocarcinoma [36]. LNCaP cells are androgen-sensitive but their growth is independent of androgen. LNCaP cells are known to produce low levels of PSA in the culture medium [36]. Both cell lines were maintained in RPMI 1640 medium containing 10% fetal bovine serum at 37°C and 5% CO2.
Treatment of Prostate Cancer Cells with f-PSA
PC-3M and LNCaP cells were grown in six-well tissue culture plates at 37°C in a humidified atmosphere with 5% CO2. When the cells reached 80% confluency, the culture medium was removed and different concentrations of f-PSA (0–10 µM) in the growth medium were added to duplicate wells for 48 hours. Control cells were treated with growth medium alone. The culture supernates were collected and used for determination of levels of different cytokines by ELISA. RNA was extracted from f-PSA-treated and control cells (3 x 106) for the gene expression studies. All samples were frozen at -70°C until processed.
RNA Extraction and Real-Time QPCR
Cytoplasmic RNA was extracted by the acid guanidinium thiocyanate-phenol-chloroform (AGPC) method of Chomczynski and Sacchi [37]. The RNA pellet was dissolved in DEPC water, quantified, and checked for its integrity using agarose gel electrophoresis and stored at -70°C. Real-time QPCR [38] was performed with the GeneAmp 5700 sequence detection system using SYBR green reagents according to the manufacturer's instructions (Applied Bio-systems, Foster City, CA). Equal amounts of RNA from cells treated with f-PSA at different concentrations and controls were reverse-transcribed into first-strand cDNA and used as PCR templates in reactions to obtain the threshold cycle (Ct). Ct was normalized using the known Ct from the housekeeping gene (β-actin) to obtain ΔCt. To compare the relative levels of gene expression of VEGF, uPA, Pim-1 oncogene, and IFN-γ, ΔΔCt values were calculated using gene expression from the untreated control cells. The ΔΔCt values were expressed as the real-fold increase in gene expression.
ELISA
VEGF and IFN-γ protein levels in the culture supernates were quantitated by highly sensitive and specific ELISA kits from R&D Systems (Minneapolis, MN) according to the manufacturer's protocol. The assay performance and inter-assay and intra-assay variations were within the limits defined by the manufacturer.
Gene Array
The genes selected for the “cancer array” included several cytokine genes, apoptotic genes, cell cycle regulators, oncogenes, etc. Cloned cDNA were printed in triplicate using a MicroGridII TAS arrayer and MicroSpot 2500 split pins (Apogent Discoveries, Hudson, NH). From PSA-treated cells, total RNA was extracted and labeled with Cy5 and the untreated reference total RNA with Cy3. For each reverse transcription reaction, 2.5 µg of RNA was mixed with 2µl random primers (Invitrogen, Carlsbad, CA) in a total volume of 10 µl, heated to 70°C for 5 minutes, and cooled to 42°C. To this sample was added an equal volume of reaction mix (4 µl of 5x first-strand buffer, 2 µl of 10x dNTP mix, 2 µl of DTT, 1 µl of deionized H2O, and 1 µl of Powerscript reverse transcriptase) according to the manufacturer's instructions. After 1-hour incubation at 42°C, the RNA-cDNA complex was denatured by incubating at 70°C for 5 minutes. The mixture was cooled to 37°C and incubated for 15 minutes with 0.2 µl of RNase H (10 U/µl). The resultant amino-modified cDNA was purified, precipitated, and fluorescently labeled.
Prior to hybridization, the two separate probes were resuspended in 10 µl of dH2O, combined, and mixed with 2 µl of human Cot1 (20 µg/µl; Invitrogen) and 2 µl of poly A (20 µg/µl; Sigma, St. Louis, MO). The probe mixture was denatured at 95°C for 5 minutes, placed on ice for 1 minute, and prepared for hybridization by addition of 110 µl of preheated (65°C) SlideHyb no. 3 buffer (Ambion, Austin, TX). After a 5-minute incubation at 65°C, the probe solution was placed on the array in an assembled GeneTAC hybridization station module (Genomic Solutions, Inc., Ann Arbor, MI). The slides were incubated overnight at 55°C for 16 to 18 hours with occasional pulsation of the hybridization solution. After hybridization, the slides were spun dry and scanned immediately on an Affymetrix 428 scanner to generate high-resolution images for both Cy3 and Cy5 channels. Two hybridizations for each RNA sample were performed, switching the dyes in the second hybridization to account for possible dye bias.
The hybridized slides were scanned using an Axon GenePix 4200A Scanner. The background-corrected signal for each cDNA spot is the mean signal (of all the pixels in the region) minus the mean local background. The ratios were then normalized on the log scale across the entire slide. For each clone on the slide, the expression ratio is the mean of all of its replicates on the log scale. The results from the two slides that make up the dye flip were then averaged on the log scale, and this became the final expression ratio of that clone generated along with a spreadsheet detailing expression ratios of all normalized spots.
Inoculation of PC-3M Cells Into Nude Mice
Male athymic BALB/c nude mice, 5 weeks old, were used in the study. For prostate tumor cell inoculation, nearly confluent PC-3M cells were harvested with 0.01% trypsin. The cells were suspended in sterile PBS at a concentration of 1 x 107 cells/ml. The cell suspension (100 µl) was injected subcutaneously into the right shoulder region of the mouse.
Treatment of Nude Mice with f-PSA
From the second day of tumor cell inoculation, mice were randomly divided into two groups of six mice each. One group received 50 µM of f-PSA (100 µl) injected subcutaneously in the vicinity of tumor cell inoculation three times a week. The dose of f-PSA was selected to mimic the actual level of PSA, the human prostate tissue, which is in the range of 50 to 150 µM [39]. The second group served as a control and received only physiological saline solution. Tumor volumes were measured with a vernier caliper three times a week for a total of 4 weeks. The small animal functional magnetic resonance imaging (f-MRI) facility at the Roswell Park Cancer Institute was used several times to carry out MRI during period of tumor development in control and in PSA treated mice.
Results
In Vitro Studies
Microarray gene expression analysis Microarray gene expression analysis used to study the effect of f-PSA treatment on prostate cancer cell lines, PC-3M and LNCaP, was performed at Roswell Park Cancer Institute Microarray and Genomics Core Facility. Gene expression in PC-3M and LNCaP cells, grown in six-well tissue culture plates and treated with 10 µM f-PSA for 48 hours, was compared with the untreated controls. Various genes were grouped according to the role they play in tumor progression and/or metastasis.
Tables 1 and 2 show some of the genes involved in angiogenesis, tumor growth, and metastasis that were significantly regulated in PC-3M and LNCaP cells in response to f-PSA treatment. In PC-3M cells, 237 genes were modulated to approximately two-fold. Out of these 237 genes, 136 genes were upregulated (two-fold to seven-fold) and 137 genes were downregulated (range 2-fold to 30-fold). With regard to LNCaP cells, the effect of PSA treatment was much greater than with PC-3M cells. In LNCaP cells, PSA treatment modulated as many as 793 genes by more than two-fold, out of which 433 genes were downregulated (range 2-fold to 131-fold) and 360 genes were upregulated (range 2-fold to 128-fold). Some of the genes such as Epha2, uPA, uPAR, and Pim-1 oncogene are known to be highly expressed in prostate cancer [28,40–42]. In PC-3M cells, uPA, uPAR, EphA2, and Pim-1 oncogene were downregulated by 12.4-, 2.8-, 2.2-, and 8.4-fold, respectively, in response to f-PSA treatment. The same genes were downregulated in LNCaP cells by 131-, 77-, 25-, and 3.6-fold, respectively. In PC-3M cells, maximum downregulation was observed for the cysteine-rich, angiogenic inducer 61 (CYR61) gene [43], whereas in LNCaP cells, it was the uPA gene that was downregulated the most (131-fold).
Table 1.
PSA Downregulates Cancer Gene Expression in PC-3M and LNCaP Cells*.
| Gene ID | Gene Name | PC-3M (Fold) | LNCaP (Fold) |
| Apoptosis | |||
| AA931820 | BCL2-like 1 | 2.25 | NC† |
| pBcl-xl | bcl-xl | 2.31 | NC |
| W45688 | Caspase 6, apoptosis-related cysteine protease | 3.05 | 5.1 |
| AA457705 | Immediate early response 3 | 5.17 | 14.7 |
| H74007 | Serum/glucocorticoid-regulated kinase | 3 | NC |
| Angiogenesis/growth factor | |||
| AA478543 | A kinase (PRKA) anchor protein (gravin) 12 | 3.62 | 48.9 |
| AA446120 | Adrenomedullin | 5.32 | 2.1 |
| AA857163 | Amphiregulin (schwannoma-derived growth factor) | 2.49 | 34.8 |
| AA678160 | CD-55 | 1.87 | 42.7 |
| T62636 | Chemokine (C-X-C motif), receptor 4 (fusin) | 2.99 | 3.4 |
| AA101875 | Chondroitin sulfate proteoglycan 2 (versican) | 2.11 | 4.5 |
| AA598794 | Connective tissue growth factor | 8.66 | 11.5 |
| AA777187 | Cysteine-rich, angiogenic inducer, 61 | 30.54 | 2.7 |
| R09561 | Decay-accelerating factor for complement (CD55) | 1.72 | 58.9 |
| R45640 | Diphtheria toxin receptor (heparin-binding epidermal growth factor-like growth factor) | 2.71 | 2.9 |
| R63623 | Dual-specificity tyrosine phosphorylation-regulated kinase 2 | 2.28 | 9.7 |
| H84481 | EphA2 | 2.17 | 25.7 |
| H13623 | Epidermal growth factor receptor pathway substrate 8 | 3.36 | NC |
| N47214 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | NC | 36.8 |
| H96643 | FOS-like antigen-1 | 5.58 | 21.4 |
| W46900 | GRO1 oncogene (melanoma growth-stimulating activity, α) | 3.06 | NC |
| AA911832 | Homo sapiens cDNA: FLJ22182 fis, clone HRC00953 | 2.05 | 27.5 |
| T62547 | Insulin-like growth factor 2 receptor | 2.1 | NC |
| AA598601 | Insulin-like growth factor-binding protein 3 | 3.77 | NC |
| AA479795 | Interferon-stimulated gene (20 kDa) | 2.27 | NC |
| W47101 | Interleukin 1β | 8.66 | 2.2 |
| N69322 | Matrix metalloproteinase 13 (collagenase 3) | 2.17 | 5.9 |
| AA284669 | Plasminogen activator, urokinase | 12.49 | 131.0 |
| AA454879 | Plasminogen activator, urokinase receptor | 2.86 | 77.4 |
| AA453759 | Sprouty (Drosophila) homolog 2 | 3.57 | 8.1 |
| AA001219 | STAT-induced STAT inhibitor 3 | 4.21 | 2.3 |
| AA464532 | Thrombospondin 1 | 3.35 | NC |
| AA399473 | Tissue factor pathway inhibitor 2 | 8.41 | 6.0 |
| AA233738 | Transforming growth factor, β2 | 3.94 | 12.1 |
| W92764 | Tumor necrosis factor, α-induced protein 6 | 2.17 | 34.2 |
| AA430504 | Ubiquitin carrier protein E2-C | 3.3 | NC |
| AA099568 | Uridine phosphorylase | 5.45 | 12.4 |
| R45059 | Vascular endothelial growth factor | 1.9 | 2.13 |
| AA630120 | Vascular endothelial growth factor B | NC | 2.7 |
| H07899 | Vascular endothelial growth factor C | NC | 26.7 |
| Cell cycle | |||
| AA598974 | Cell division cycle 2, G1 to S and G2 to M | 3.42 | NC |
| AA995402 | Colony-stimulating factor 2 (granulocyte-macrophage) | 4.84 | NC |
| AA079775 | c-src tyrosine kinase | 2.32 | 4.4 |
| AA608568 | Cyclin A2 | 2.34 | NC |
| AA774665 | Cyclin B2 | 2.22 | NC |
| AA444049 | Dual-specificity phosphatase 4 | 4.56 | 5.6 |
| AA460152 | Serum-inducible kinase | 13.18 | NC |
| AA504348 | Topoisomerase (DNA) II α (170 kDa) | 3.32 | 2.1 |
| R80790 | Ubiquitin carrier protein E2-C | 3.44 | NC |
| AA457034 | v-myb avian myeloblastosis viral oncogene homolog-like 2 | 2.27 | NC |
| Cell adhesion and cell junction | |||
| AA418564 | Cadherin 12, type 2 (N-cadherin 2) | 2.12 | 5.3 |
| AI336940 | CD6 antigen | 3.08 | NC |
| AA485668 | Integrin, β4 | 1.91 | 80.4 |
| AA490238 | Mitogen-inducible 2 | 3.75 | NC |
| Signal transduction | |||
| AA521232 | HSPC022 protein | 2.12 | 20.6 |
| AA128153 | Interleukin 1 receptor-like 1 | 2.96 | NC |
| AA890663 | p21/Cdc42/Rac1-activated kinase 1 | 2.2 | 17.4 |
| AA447730 | Pim-1 oncogene | 8.35 | 3.6 |
| AI375353 | Serum/glucocorticoid-regulated kinase | 3.46 | 3.8 |
| Transcription | |||
| AA496678 | B-cell CLL/lymphoma 3 | 3.98 | NC |
| W84868 | Cytochrome P450, subfamily IVA, polypeptide 11 | 4 | NC |
| H82442 | Inhibitor of DNA binding 2, helix-loop-helix protein | 2.26 | NC |
| H77597 | Metallothionein 1H | 6.65 | NC |
| AA496628 | Nonmetastatic cells 2, protein (NM23B) expressed | 2.37 | NC |
PC-3M and LNCaP cells were treated with 10 µM f-PSA for 48 hours.
NC = no significant change.
Table 2.
PSA Upregulates Cancer Gene Expression in PC-3M and LNCaP Cells*.
| Gene ID | Gene Name | PC-3M (Fold) | LNCaP (Fold) |
| Apoptosis | |||
| W72310 | Fas-activated serine/threonine kinase | 2.07 | NC† |
| R72244 | 2–5 Oligoadenylate synthetase 2 | 2.3 | 15.5 |
| Angiogenesis/growth factor | |||
| AA148548 | Fatty acid binding protein 3 mammary-derived growth inhibitor | 2.66 | NC |
| AA490996 | Interferon γ-inducible protein 16 | 2.13 | NC |
| AA630800 | Interferon γ-inducible protein 30 | 2.48 | NC |
| AA489640 | Interferon-induced protein, tetratricopeptide repeats 1 | 2.82 | NC |
| AA291389 | Interferon-stimulated transcription factor 3 (48 kDa) | 2.23 | NC |
| AA487020 | Isoprenylcysteine carboxyl methyltransferase | 2.44 | NC |
| AA283030 | Methionine aminopeptidase; eIF-2-associated p67 | 2.07 | NC |
| AA443284 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog) | 2.46 | NC |
| AA490213 | Transducer of ERBB2, 1 | 1.92 | NC |
| AA486088 | Transducer of ERBB2, 2 | 1.71 | 2.1 |
| R36467 | Transforming growth factor, β 1 | 1.91 | 3.4 |
| Cell cycle | |||
| AI360334 | Carbamoyl-phosphate synthetase 2 aspartate transcarbamylase | 2.4 | NC |
| AA029997 | Collagen, type IV, α 5 | 2.48 | NC |
| AA504844 | DKFZp434J1813 protein | 2.02 | NC |
| AA478479 | Heat shock protein (hsp 110 family) | 2.01 | NC |
| AA456636 | RAN, member RAS oncogene family | 2.33 | 2.3 |
| AA487492 | Retinoblastoma-binding protein 2 | 2.47 | NC |
| N66132 | rTS β protein | 2.1 | NC |
| AA419177 | Solute carrier family 7 (cationic amino acid transporter, member 5) | 2.7 | NC |
| AA425853 | Splicing factor proline/glutamine rich | 2.26 | NC |
| AA447515 | Mad4 homolog | 4.37 | NC |
| AA456636 | RAN, member RAS oncogene family | 2.33 | 2.3 |
| AA487492 | Retinoblastoma-binding protein 2 | 2.47 | NC |
| N66132 | rTS β protein | 2.1 | NC |
| AA419177 | Solute carrier family 7 cationic amino acid transporter, member 5) | 2.7 | NC |
| AA425853 | Splicing factor proline/glutamine-rich | 2.26 | NC |
| Signal transduction | |||
| AA676453 | CD37 antigen | 2.45 | NC |
| AI345015 | γ-Glutamyltransferase 2 | 2.03 | NC |
| H58953 | Nuclear factor (erythroid-derived 2), 45 kDa | 2.42 | NC |
| AA485731 | Phosphoinositide-3-kinase (p85 β) | 2.04 | NC |
| AA291742 | Promyelocytic leukemia | 2.12 | NC |
| AA057378 | RAB32, member of RAS oncogene family | 2.57 | NC |
| AA916836 | Small inducible cytokine subfamily A (Cys-Cys), member 23 | 2.46 | NC |
| Transcription | |||
| H11660 | p53-induced protein | 1.96 | 15.3 |
| R98936 | Membrane metalloendopeptidase (neutral endopeptidase, enkephalinase, CALLA, CD10) | 2.33 | 18.1 |
| T53431 | Nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 2.19 | 15.3 |
PC-3M and LNCaP cells were treated with 10 µM f-PSA for 48 hours.
NC = no significant change.
Other genes downregulated in PC-3M cells by f-PSA treatment include the antiapoptotic gene Bcl-xl (2.3-fold); the proangiogenic genes TGF-β2 (2.9-fold) and VEGF (1.9-fold); the cell cycle genes cyclin A2 (2.34-fold) and C-src tyrosine kinase (2.3-fold); the cell adhesion gene N-cadherin (2.1-fold); and the transcription gene metalothionin-1H (6.6-fold).
As shown in Table 2, more than 30 genes were upregulated in PC-3M cells to nearly two-fold in response to f-PSA treatment. These genes are involved in angiogenesis, apoptosis, and cell cycle. We observed upregulation of the apoptotic gene, Fas-activated serine kinase (2.1-fold), and the cell cycle gene Mad4 homolog (4.4-fold). In addition, IFN-γ induced protein-16 gene (2.1-fold) and IFN γ-induced protein-30 gene (2.5-fold) were also upregulated in PC-3M cells treated with f-PSA. Interferon-related genes are known to be involved in tumor suppression [44]. In LNCaP cells, the following genes were upregulated: 2–5 oligoadenylate synthetase 2 (15.5-fold), TGF-β1 (3.4-fold), p-53-induced protein (15.4-fold), and CD 10 (18-fold).
Our preliminary data on gene array analysis document that PSA can significantly modulate the expression of numerous genes that are known to be involved in apoptosis, cell cycle, angiogenesis, cell growth, etc. Because there is no literature on genes that are affected by PSA, it is premature to speculate on the role of these genes, affected by PSA, in prostate tumor growth beside those genes that are known to be involved in prostate cancer or cancer growth at large.
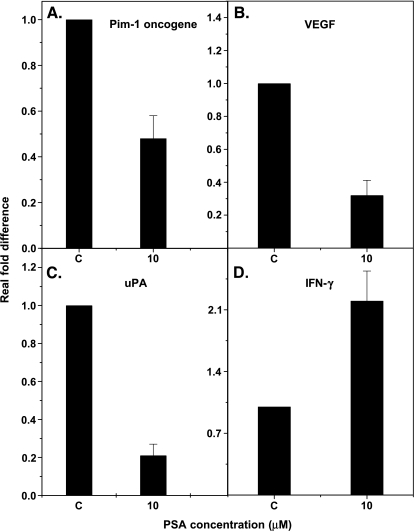
Real-time QPCR To further validate our findings with gene array analysis, we selected a panel of genes that are known for their involvement in tumor growth and metastasis, and carried out gene expression studies using QPCR. These include VEGF, uPA, and Pim-1 oncogene, which are known to promote tumor growth, and the IFN-γ gene, which is known to suppress tumor growth. We examined the relative expression of these genes in PC-3M cells treated with 10 µM f-PSA. For these studies, RNA was reverse-transcribed and cDNA-amplified by QPCR using primers specific for the genes of interest and the housekeeping gene, β-actin. Experiments were repeated three times and the results are shown in Figure 1. Treatment of PC-3M cells with 10 µM PSA for 48 hours significantly downregulates uPA, VEGF, and Pim-1 gene expression and upregulates IFN-γ gene expression. The gene expression of uPA, VEGF, and Pim-1 was downregulated by 80% (P < .001), 65% (P < .001), and 52% (P = .01), respectively. However, IFN-γ gene expression was upregulated by 102% (P < .001) (Figure 1).
Figure 1.
Gene expression levels of Pim-1 oncogene, uPA, VEGF, and IFN-γ in PC 3M cells treated with f-PSA in vitro. The cells were treated with 10 µM F-PSA for 48 hours. RNA was extracted and reverse-transcribed. The c-DNA was amplified by real-time QPCR using specific primers. The results shown are the average of three experiments. (A) Pim-1 oncogene; (B) VEGF; (C) uPA; and (D) IFN-γ.
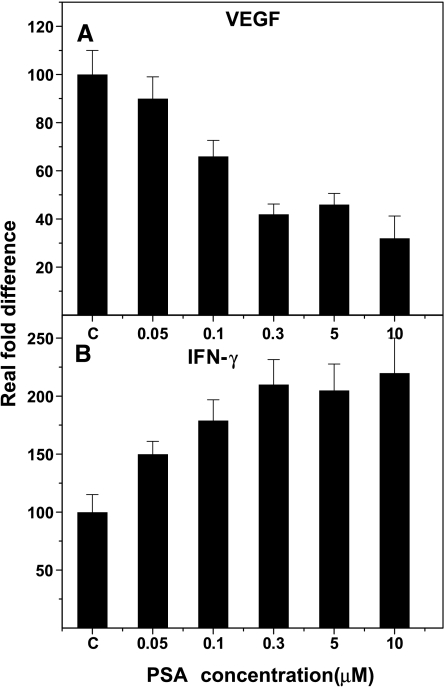
Effect of different concentrations of f-PSA on VEGF and IFN-γ gene expression and production in PC-3M cells Because there is no literature to provide guidelines for the optimal dose of f-PSA for a maximal biologic response, we were compelled to investigate the effect of a range of f-PSA concentrations on production and gene expression of some of the genes that regulate tumor growth. We selected two cytokines—VEGF, a proangiogenic factor, and IFN-γ, an antiangiogenic factor—and studied the effect of a range of f-PSA concentrations on gene expression and production of these two cytokines. PC-3M cells were treated for 48 hours with f-PSA at concentrations ranging from 0 to 10 µM. Culture supernates were collected from treated and control cells to determine the amounts of VEGF and IFN-γ released into the culture medium, and total RNA was extracted from the cells. For gene expression studies, RNA was reverse-transcribed and cDNA-amplified by QPCR using primers specific for VEGF and IFN-γ and the housekeeping gene, β-actin. The experiment was repeated three times and results are shown in Figure 2. A distinct dose-response curve was seen with VEGF gene expression in the range of 0.05 to 10 µM f-PSA. Concentrations in the range of 0.006 to 0.05M did not show any significant effect on VEGF gene expression. However, at concentrations of 0.1 to 5 µM, f-PSA treatment resulted in a significant inhibition of VEGF gene expression (Figure 2A; P < .001). In our experiments, maximal downregulation of VEGF gene expression in PC-3M cells was seen at 10 µM f-PSA (Figure 2A). On the contrary, IFN-γ gene expression was upregulated in PC-3M cells treated with f-PSA, again in a dose-dependent manner. The maximal enhancement of IFN-γ gene expression was seen at 10 µM f-PSA (P < .001; Figure 2B).
Figure 2.
Dose-response effects of f-PSA treatment of PC-3M cells on gene expression of VEGF and IFN-γ. Cells were treated for 48 hours with f-PSA concentrations as shown. RNA was extracted and reverse-transcribed. Real-time QPCR using specific primers amplified the c-DNA for VEGF (A) and IFN-γ (B). The experiment was repeated three times.
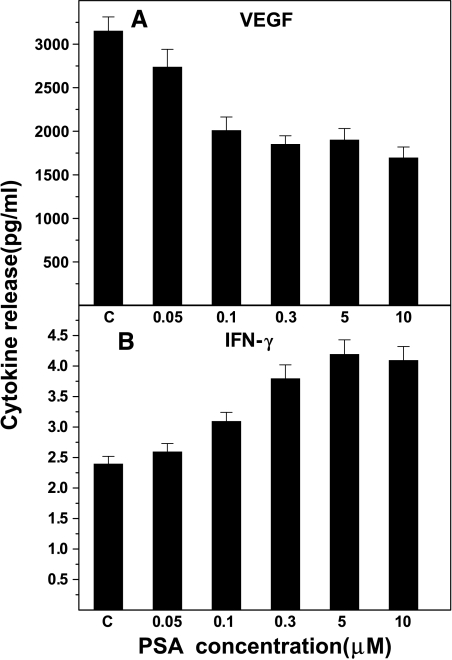
Protein levels of VEGF and IFN-γ were determined in the culture supernates using ELISA kits. The experiment was repeated three times with essentially identical results. The results are shown in Figure 3. The culture supernates from untreated control PC-3M cells contained an average of 3155 pg/ml VEGF protein compared to 2014 pg/ml from cells treated with 0.1 µM f-PSA. This is a significant reduction (36.2%; P < .001). Treating PC-3M cells with higher concentrations of f-PSA (up to 10 µM) did not result in further reduction in the amount of VEGF released into the culture medium (Figure 3A). The culture supernates from control cells had an average of 2.4 pg/ml IFN-γ protein. This constitutive expression of IFN-γ was not affected when cells were treated with f-PSA in the range of 0.006 to 0.025 µM. However, when cells were treated with 0.1 µM f-PSA, an increase of 66.7% in IFN-γ synthesis was observed (P < .001; Figure 3B). At a higher concentration of f-PSA (5 µM), IFN-γ protein release was increased by 175% (P < .001). No further increases in IFN-γ protein release were seen when f-PSA concentrations higher than 5 µM were used (Figure 3B).
Figure 3.
Dose-response effects of f-PSA treatment of PC-3M cells on secretion of VEGF and IFN-γ proteins. Cells were treated with indicated concentrations of f-PSA for 48 hours. The culture medium was collected and quantitated for VEGF (A) and IFN-γ (B) protein by sandwich ELISA. The results shown are the average of three experiments.
Our data demonstrate that in PC-3M cells treated with f-PSA, the release of VEGF protein, a potent proangiogenic factor, is downregulated, whereas the release of IFN-γ protein, a well-known antiangiogenic factor, is upregulated.
In Vivo Studies
Effect of f-PSA treatment on growth of PC-3M tumors in nude mice The results of in vitro studies, both gene array and QPCR, strongly support the notion that PSA significantly modulates the gene expression and production of many growth factors that are known to be involved in angiogenesis, tumor growth, and metastasis. However, it is not known if exogenously administered PSA can significantly modulate prostate tumor growth in vivo. In our studies, we observed that PSA administered to nude mice inoculated with PC-3M prostate tumor cells indeed can significantly suppress tumor growth. Five-week-old athymic BALB/c nude mice were inoculated subcutaneously with 1 x 106 PC-3M cells in the neck region. After 48 hours of tumor cell inoculation, a group of animals was treated with 150 µg of f-PSA in PBS, administered subcutaneously within the vicinity of tumor cell inoculation. PSA was administered on alternate days for a total of 4 weeks. In our preliminary studies, we were able to detect circulating levels of PSA up to 48 hours postadministration when it was given either subcutaneously or intravenously by tail vein (data not shown). The dose of PSA was selected to attain a PSA level in the tumor vicinity comparable to physiological PSA levels known to be present in the human prostate gland (50–150 µM) [39]. Furthermore, there are no data available with regard to PSA dose, frequency, and duration of administration in any experimental situation. Tumors began to grow in control animals at the inoculated site by the second week, whereas no palpable tumors were seen in animals treated with f-PSA by this time period. Tumor growth was followed for 4 weeks until termination of the experiment. Tumor volumes in f-PSA-treated mice were monitored periodically either by measuring with a caliper or f-MRI, and were found to be significantly lower than untreated control animals. At termination of the experiments after 4 weeks, tumors were removed, weighed, and snap-frozen for RNA extraction.
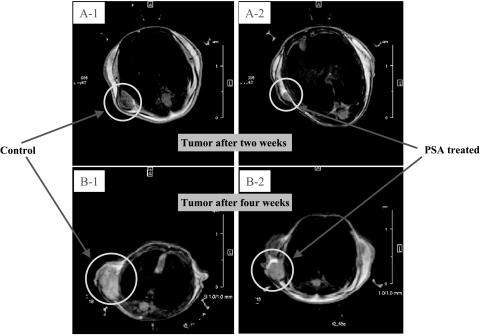
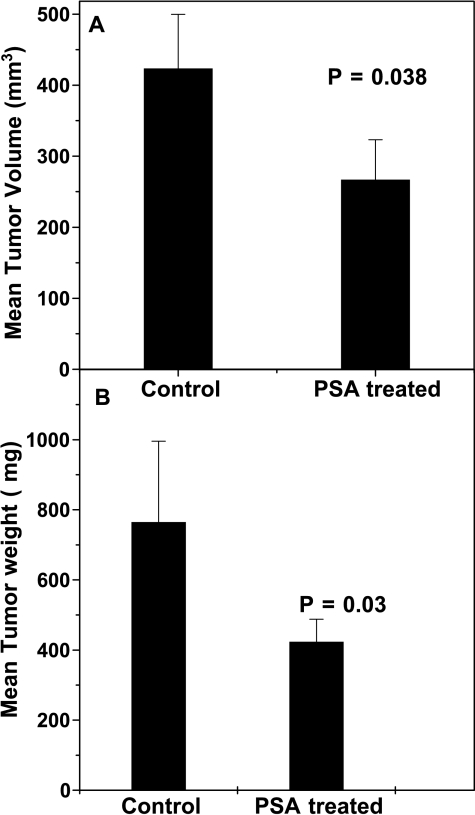
Administration of f-PSA to PC-3M tumor-bearing mice produced a significant reduction in the extent of tumor growth (Figure 4). The mean tumor weight of the treated mice was 420 ± 37.8 mg, whereas that of the untreated control animals was 767 ± 69 mg (P = .03; Figure 5A). Mean tumor volumes measured using f-MRI [45] also were significantly lower in f-PSA-treated mice (Figure 5B). The mean tumor volume of the treated animals by the end of 4 weeks was 267.2 ± 24 mm3, whereas that of control animals was 423.9 ± 38 mm3 (P = .029). The suppression of the tumor growth as observed may be, in part, due to the changes in the expression of various growth factors in response to PSA treatment.
Figure 4.
Effect of f-PSA treatment on growth of PC-3M tumor in nude mice. Five-week-old male athymic BALB/c nude mice were injected subcutaneously with 1 x 106 PC-3M tumor cells in the neck region. Two days later, 150 µg of f-PSA was administered subcutaneously within the tumor vicinity and on alternate days for a total of 4 weeks. Control animals were treated with PBS injections. There were five animals each in control and treatment groups. Tumor growth was monitored by f-MRI. Tumor growth—Control [A-1, at 2 weeks (mean tumor volume = 58 mm3); B-1, at 4 weeks (mean tumor volume = 424 mm3)]; Tumor growth—PSA-treated [A-2, at 2 weeks (mean tumor volume = 13 mm3); B-2, at 4 weeks (mean tumor volume = 267 mm3)].
Figure 5.
Effect of f-PSA treatment of PC-3M tumor-bearing nude mice on tumor volume and weight. BALB/c nude mice bearing PC-3M tumors, control and f-PSA-treated, were sacrificed at the end of 4 weeks. Both tumor volume and tumor weights were measured for each animal. There were five animals in each group. Average tumor volume (A); and average tumor weight (B). Both tumor volume and tumor weight were significantly reduced in PSA-treated animals.
Gene array analysis of PC-3M tumors in nude mice treated with PSA Gene array profiling of the effect of f-PSA treatment on growth of PC-3M tumors in nude mice was also carried out. Table 3 shows the effect of f-PSA treatment on the expression of various genes in prostate tumor tissues when PC-3M tumor cells were implanted in nude mice. Of the downregulated genes, uPA (2.1-fold) and platelet-derived growth factor receptor gene (2.3-fold) are known to have important role(s) in prostate cancer progression [46]. In addition, others such as MMP-7, S100 calcium-binding protein A9 gene, TNF-α-induced protein 6, and IL-7 receptor genes were also downregulated to a minimum of two-fold. Among the upregulated genes, chondrotin sulfate proteoglycan 2 and phospholipase C gamma-1 genes are known to be involved in the inhibition of tumor progression (Table 3).
Table 3.
PSA Modulates Gene Expression in Human Prostate Tumor Tissue in Nude Mice*.
| Gene ID | Name | Fold Downregulated |
| AA489331 | Adenosine deaminase, RNA-specific, B1 | 2.17 |
| AA862465 | α2-Glycoprotein 1, zinc | 2.27 |
| AA055979 | Integrin, α 7 | 2.00 |
| AA485865 | Interleukin 7 receptor | 2.04 |
| AA031514 | Matrix metalloproteinase 7 (matrilysin, uterine) | 2.22 |
| AA284669 | Plasminogen activator, urokinase | 2.13 |
| R56211 | Platelet-derived growth factor receptor, β polypeptide | 2.30 |
| AA402883 | Progestagen-associated endometrial protein | 2.59 |
| R59165 | Protein phosphatase 2, regulatory subunit B (B56), α isoform | 2.22 |
| R45941 | Protein tyrosine phosphatase, receptor type, N | 4.19 |
| AA004638 | Ribosomal protein L4 | 2.94 |
| AA864554 | S100 calcium-binding protein A9 (calgranulin B) | 2.29 |
| W92764 | Tumor necrosis factor, a-induced protein 6 | 2.17 |
| AA426227 | Uridine monophosphate synthetase | 2.04 |
| R62813 | v-myc avian myelocytomatosis viral oncogene homolog 1 | 2.17 |
| AA521434 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | 1.77 |
| AI347538 | BCL2-interacting killer (apoptosis-inducing) | 1.82 |
| AA101875 | Chondroitin sulfate proteoglycan 2 (versican) | 2.62 |
| AA450009 | Endothelin receptor type A | 2.15 |
| AA872402 | Eukaryotic translation initiation factor 4B | 1.78 |
| R62612 | Fibronectin 1 | 1.84 |
| N26285 | Fibronectin 1 | 1.78 |
| AA482119 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 2.88 |
| R52796 | Interleukin 13 receptor, α2 | 2.81 |
| T71686 | KIAA 1254 protein | 2.19 |
| AA463946 | Mitochondrial carrier homolog 2 | 1.89 |
| AA682423 | Monoamine oxidase B | 1.95 |
| R76690 | Phospholipase C, γ1 (formerly subtype 148) | 2.07 |
| H95960 | Secreted protein, acidic, cysteine-rich (osteonectin) | 1.79 |
| AA281635 | Suppression of tumorigenicity 16 (melanoma differentiation) | 1.76 |
Athymic BALB/c nude mice were treated with 50 µM f-PSA (100µl) in the tumor vicinity three times a week for a total of 4 weeks.
Discussion
For the past 20 years, PSA has been an important biomarker for the early diagnosis and management of prostate cancer [2]. Despite the importance of PSA as a surrogate marker for early diagnosis of prostate cancer, relatively little is known about its biological/physiological function(s). There are several observations that suggest that PSA may have tumor-promoting and/or tumor-suppressing activities. PSA is a serine protease that, through its proteolytic activity, can release free bioactive IGF-1 previously bound to IGFPB-3. [47]. IGF-1, being a mitogen, may constitute a risk factor for prostate carcinoma. However, there are several recent reports suggesting that circulating free IGF-1, total IGF-1, and IGFPB-3 do not predict the future risk to development of prostate cancer [48,49]. On the contrary, there are several reports suggesting that PSA may act as an antiangiogenic molecule or as an inducer of apoptosis [50]. Lower tissue PSA levels are associated with a more aggressive form of prostate cancer [18]. Transfection of PSA c-DNA in PC-3 prostate carcinoma cells prolongs their doubling time, reduces their tumorigenicity and metastasis in nude mice, and induces apoptosis [19]. PSA is also able to convert Lysplasminogen to biologically active "angiostatin" such as fragments that are known to inhibit angiogenesis [23]. More recently, PSA purified from seminal plasma and recombinant PSA protein produced in the Pichia pastoris expression system were shown to inhibit angiogenesis both in vitro and in vivo [15]. In this report, we provide further evidence that f-PSA has antiangiogenic/antitumor activities both in vitro and in vivo. Treatment of prostate tumor cells with f-PSA resulted in significant modulation of several genes that are known to be involved in angiogenesis, tumor growth, and metastasis.
We performed gene array profiling of genes that are modulated in human prostate cancer cells, PC-3M and LNCaP, that vary significantly in their metastatic potential and in PC-3M tumors growing in nude mice treated with f-PSA. In both studies, treatment with f-PSA resulted in a significant modulation of the expression of various cancer-related genes. The response of prostate tumor cells to f-PSA treatment varied significantly. Treatment of PC-3M (highly metastatic) and LNCaP (poorly metastatic) cells with f-PSA resulted in both upregulation and downregulation of 273 and 793 cancer-related genes, respectively (Tables 1 and 2). Although the expression of cancer genes in LNCaP cells is influenced to a greater magnitude compared to PC-3M cells, it is premature to speculate the relevance of such changes. It is also known that LNCaP cells produce low levels of PSA in the culture medium (30–100 ng/ml); the concentration of PSA in prostate tissue is significantly higher. Consequently, low levels of PSA produced by LNCaP cells may not be of any consequence.
The modulation of gene expression in PC-3M cells treated with f-PSA varied between 2-fold and 30-fold. Among the downregulated genes, VEGF [51], uPA [52], uPAR [53], Pim-1 oncogene [28], and EphA2 [40] have already been implicated in prostate cancer progression. In addition, Bcl-x [54], MMP13 [55], and CYR61 [43] genes, also known to promote tumor growth, were downregulated in response to f-PSA treatment. On the other hand, the Mad4 [56] gene and several interferon-related genes that are known to have antitumor properties were significantly upregulated in PC-3M cells treated with f-PSA (Table 2).
Treatment of LNCaP cells with f-PSA modulated the expression of as many as 793 genes in the range of 2-fold to 131-fold, wherein 433 genes were downregulated and 360 genes were upregulated. In the group of downregulated genes, uPA showed the greatest suppression (131-fold). VEGF-C gene, a well-known tumor promoter, was downregulated 26-fold (Table 1). In addition, the MMP13 gene that has protumor activity was downregulated in LNCaP cells. Other genes such as EphA2, uPAR, and Pim-1 oncogene were also significantly downregulated in LNCaP cells (Table 1).
Several cancers overexpress tyrosine kinases [57] and these kinases have been exploited for cancer diagnosis and therapy. In prostate cancer, tyrosine kinases are known to be overexpressed [58]. EphA2 is a transmembrane receptor tyrosine kinase of the Eph family [59]. EphA2 is also overexpressed in adult epithelial malignancies of the colon and esophagus [60]. It has been reported that EphA2 is overexpressed in metastatic prostate cancer [40]. One of the important molecules involved in cancer metastasis is uPA. An elevated concentration of uPA is a strong indicator of poor prognosis. It is known that uPA binds to its receptor, uPAR, on the cell surface, resulting in conversion of plasminogen to plasmin. Plasmin then digests fibrin, which subsequently allows endothelial tube formation. uPAR gene expression was inhibited in LNCaP cells as well as in PC-3M cells. PSA treatment also downregulated CYR61 gene in PC-3M cells. CYR61 is a prototypical member of the CCN family. This protein functions as extracellular matrix (ECM)-associated signaling molecule. It is transcriptionally activated in endothelial cells in response to bFGF or VEGF, and can interact with various cellular integrins [61]. Purified Cyr61 protein has been reported to mediate cell adhesion, stimulate chemotaxis, augment growth factor-induced DNA synthesis, foster cell survival, and enhance angiogenesis in vivo [61].
Because there is no literature available regarding the dosage, duration, and frequency of PSA treatment for optimal response(s), we investigated the effect of PSA dosage on VEGF and IFN-γ gene and protein expression in PC-3M cells. VEGF gene expression was maximally suppressed (>60%) at 10 µM f-PSA (Figure 2A). With regard to IFN-γ gene, maximal upregulation was observed at concentrations of 5 µM f-PSA (Figure 2B). VEGF protein release was downregulated with increasing concentrations of f-PSA. At 0.3 µM f-PSA, VEGF protein release was suppressed by over 40% compared to untreated control cells (Figure 3A). Similarly, the release of IFN-γ protein was maximal at f-PSA concentration of 0.3 µM (Figure 3B). INF-γ is a pleiotropic cytokine that plays a central role in innate and adaptive immunity [62]. IFN-γ acts in various ways on host and tumor cells to favor tumor regression. IFN-γ is known to have a significant role in tumor surveillance [63]. In our in vivo study, BALB/c nude mice bearing PC-3M tumors were treated with f-PSA and monitored by f-MRI. Results as shown in Figure 4 clearly document that f-PSA treatment results in significant reduction in the overall growth of prostate tumor.
A widely accepted mechanism of action for the antiangiogenic activity of PSA is due to its protease activity. There are reports that antiangiogenic drugs such as TNP470 and thalidomide enhance PSA production by prostate cancer cells in vitro [64]. The antiangiogenic activity of PSA could explain some of the growth characteristics of prostate cancer such as the association of low microvascular density and a slow proliferation rate in the early stages of prostate cancer [22]. Blocking the serine protease activity of PSA with an inhibitor such as α-chymotrypsin effectively blocks its antiangiogenic effects [14]. The finding that PSA can cleave plasminogen to yield the antiangiogenic fragment “angiostatin” supports the above premise [23]. The f-PSA used in this study was 99.9% pure and was enzymatically active. The possibility of contamination by another kallikrein family protein such as hK2 was ruled out. This study shows that, in addition to endothelial cell-specific inhibition of angiogenesis, PSA can induce changes at the transcriptional and translational levels, which, in turn, regulate the production of proangiogenic and antiangiogenic factors. Serine proteases have the ability to induce apoptosis in cells [65], but a study with enzymatically inactive, N-1 variant, recombinant PSA, in which the first codon is removed, shows that its antiangiogenic activity is retained. Because both full-length PSA (enzymatically active) and the N-1 variant (enzymatically inactive) have antiangiogenic activity, Fortier et al. [15] suggest that PSA can exert its antitumor activity by mechanisms in addition to its protease activity.
Another potential mechanism for the antiangiogenic activity of PSA may be due to its action with cell surface receptors. PSA has extensive homology with γ-nerve growth factor (56%), epidermal growth factor-binding protein (53%), and α-nerve growth factor (51%) [66]. Thus, it may interact with the natural cell surface receptors for these growth factors, potentially exerting antiangiogenic effects by mechanisms yet to be determined. Anti-PSA antibodies abrogate the effect of PSA on ROS, suggesting that PSA acts at an extracellular site [24]. We have shown that blocking the enzymatic activity of f-PSA with the protease inhibitor, aprotinin, resulted in an incomplete reversal of its ability to regulate the expression of VEGF (data not shown). Our data support the notion that the antiangiogenic activity of PSA may be mediated by more than one mechanism including its enzymatic activity.
Ours are the first observations to document that treatment of prostate cancer cells with f-PSA significantly modulates the expression of various growth factors involved in tumor growth. Such a change may be responsible for the suppression of growth of prostate tumor xenograft in nude mice as observed. Our findings are in general agreement with another report that documents the antiangiogenic effect of PSA in murine melanoma model [14]. In view of our current findings that PSA may have tumor-protective activities in vivo, it is interesting to speculate that increased expression of PSA may also be an adaptive host antitumor response. Further research in this area is warranted.
Acknowledgements
We are grateful to Richard Mazurchuck for f-MRI, and Norma Novak and Devin McQuaid for the gene array analysis.
Abbreviations
- PSA
prostate-specific antigen
- f-MRI
functional magnetic resonance imaging
- ROS
reactive oxygen species
Footnotes
This work was supported by the Alliance Foundation, Roswell Park Cancer Institute (Buffalo, NY), and the Margaret Duffy and Cameron Troup Memorial Fund for Cancer, Buffalo General Hospital (Buffalo, NY).
References
- 1.ACS, author. Cancer Figures and Facts 2004. Atlanta, GA: American Cancer Society; 2004. [Google Scholar]
- 2.Diamandis EP. Prostate-specific antigen: its usefulness in clinical medicine. Trends Endocrinol Metab. 1998;9:310–316. doi: 10.1016/s1043-2760(98)00082-4. [DOI] [PubMed] [Google Scholar]
- 3.Armbruster DA. Prostate-specific antigen: biochemistry, analytical methods, and clinical application. Clin Chem. 1993;39:181–195. [PubMed] [Google Scholar]
- 4.Zhang WM, Finne P, Leinonen J, Vesalainen S, Nordling S, Stenman UH. Measurement of the complex between prostate-specific antigen and alpha1-protease inhibitor in serum. Clin Chem. 1999;45:814–821. [PubMed] [Google Scholar]
- 5.Lilja H, Oldbring J, Rannevik G, Laurell CB. Seminal vesicle-secreted proteins and their reactions during gelation and liquefaction of human semen. J Clin Invest. 1987;80:281–285. doi: 10.1172/JCI113070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements J, Mukhtar A. Glandular kallikreins and prostate-specific antigen are expressed in the human endometrium. J Clin Endocrinol Metab. 1994;78:1536–1539. doi: 10.1210/jcem.78.6.7515392. [DOI] [PubMed] [Google Scholar]
- 7.Diamandis EP. New diagnostic applications and physiological functions of prostate-specific antigen. Scand J Clin Lab Invest. 1995;(Suppl 221):105–112. doi: 10.3109/00365519509090573. [DOI] [PubMed] [Google Scholar]
- 8.Hautmann S, Huland E, Grupp C, Haese A, Huland H. Super-sensitive prostate-specific antigen (PSA) in serum of women with benign breast disease or breast cancer. Anticancer Res. 2000;20:2151–2154. [PubMed] [Google Scholar]
- 9.Pummer K, Wirnsberger G, Purstner P, Stettner H, Wandschneider G. False positive prostate-specific antigen values in the sera of women with renal cell carcinoma. J Urol. 1992;148:21–23. doi: 10.1016/s0022-5347(17)36497-2. [DOI] [PubMed] [Google Scholar]
- 10.Kucera E, Kainz C, Tempfer C, Zeillinger R, Koelbl H, Sliutz G. Prostate-specific antigen (PSA) in breast and ovarian cancer. Anticancer Res. 1997;17:4735–4737. [PubMed] [Google Scholar]
- 11.Zarghami N, Grass L, Sauter ER, Diamandis EP. Prostate-specific antigen in serum during the menstrual cycle. Clin Chem. 1997;43:1862–1867. [PubMed] [Google Scholar]
- 12.Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339:58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- 13.Folkman J, Shing Y. Angiogenesis. J Biol Chem. 1992;267:10931–10934. [PubMed] [Google Scholar]
- 14.Fortier AH, Nelson BJ, Grella DK, Holaday JW. Antiangiogenic activity of prostate-specific antigen. J Natl Cancer Inst. 1999;91:1635–1640. doi: 10.1093/jnci/91.19.1635. [DOI] [PubMed] [Google Scholar]
- 15.Fortier AH, Holaday JW, Liang H, Dey C, Grella DK, Holland-Linn J, Vu H, Plum SM, Nelson BJ. Recombinant prostate-specific antigen inhibits angiogenesis in vitro and in vivo. Prostate. 2003;56:212–219. doi: 10.1002/pros.10256. [DOI] [PubMed] [Google Scholar]
- 16.Pretlow TG, Pretlow TP, Yang B, Kaetzel CS, Delmoro CM, Kamis SM, Bodner DR, Kursh E, Resnick MI, Bradley EL., Jr Tissue concentrations of prostate-specific antigen in prostatic carcinoma and benign prostatic hyperplasia. Int J Cancer. 1991;49:645–649. doi: 10.1002/ijc.2910490503. [DOI] [PubMed] [Google Scholar]
- 17.Magklara A, Scorilas A, Stephan C, Kristiansen GO, Hauptmann S, Jung K, Diamandis EP. Decreased concentrations of prostate-specific antigen and human glandular kallikrein 2 in malignant versus nonmalignant prostatic tissue. Urology. 2000;56:527–532. doi: 10.1016/s0090-4295(00)00621-x. [DOI] [PubMed] [Google Scholar]
- 18.Stege R, Grande M, Carlstrom K, Tribukait B, Pousette A. Prognostic significance of tissue prostate-specific antigen in endocrinetreated prostate carcinomas. Clin Cancer Res. 2000;6:160–165. [PubMed] [Google Scholar]
- 19.Balbay MD, Juang P, Ilansa N, Williams SD, Fidler IJ, et al. Stable transfection of human prostate cancer cell line PC-3 with prostate-specific antigen induced apoptosis both in vivo and in vitro. Proc Am Assoc Cancer Res. 1999;49:225–228. (Abstract) [Google Scholar]
- 20.Yu H, Levesque MA, Clark GM, Diamandis EP. Prognostic value of prostate-specific antigen for women with breast cancer: a large United States cohort study. Clin Cancer Res. 1998;4:1489–1497. [PubMed] [Google Scholar]
- 21.Rosser CJ, Kuban DA, Levy LB, Chichakli R, Pollack A, Lee AK, Pisters LL. Prostate-specific antigen bounce phenomenon after external beam radiation for clinically localized prostate cancer. J Urol. 2002;168:2001–2005. doi: 10.1016/S0022-5347(05)64282-6. [DOI] [PubMed] [Google Scholar]
- 22.Papadopoulos I, Sivridis E, Giatromanolaki A, Koukourakis MI. Tumor angiogenesis is associated with MUC1 overexpression and loss of prostate-specific antigen expression in prostate cancer. Clin Cancer Res. 2001;7:1533–1538. [PubMed] [Google Scholar]
- 23.Heidtmann HH, Nettelbeck DM, Mingels A, Jager R, Welker HG, Kontermann RE. Generation of angiostatin-like fragments from plasminogen by prostate-specific antigen. Br J Cancer. 1999;81:1269–1273. doi: 10.1038/sj.bjc.6692167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun XY, Donald SP, Phang JM. Testosterone and prostate-specific antigen stimulate generation of reactive oxygen species in prostate cancer cells. Carcinogenesis. 2001;22:1775–1780. doi: 10.1093/carcin/22.11.1775. [DOI] [PubMed] [Google Scholar]
- 25.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 26.Aalinkeel R, Nair MP, Sufrin G, Mahajan SD, Chadha KC, Chawda RP, Schwartz SA. Gene expression of angiogenic factors correlates with metastatic potential of prostate cancer cells. Cancer Res. 2004;64:5311–5321. doi: 10.1158/0008-5472.CAN-2506-2. [DOI] [PubMed] [Google Scholar]
- 27.Bull JH, Ellison G, Patel A, Muir G, Walker M, Underwood M, Khan F, Paskins L. Identification of potential diagnostic markers of prostate cancer and prostatic intraepithelial neoplasia using cDNA microarray. Br J Cancer. 2001;84:1512–1519. doi: 10.1054/bjoc.2001.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 29.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 30.Bindukumar B, Kawinski E, Cherrin C, Gambino ML, Nair PM, Schwartz SA, Chadha KC. Two step procedure for purification of enzymatically active Prostate-specific antigen from seminal plasma. J Chromatogr B. doi: 10.1016/j.jchromb.2004.09.014. in press. [DOI] [PubMed] [Google Scholar]
- 31.Kawinski E, Levine E, Chadha K. Thiophilic interaction chromatography facilitates detection of various molecular complexes of prostate-specific antigen in biological fluids. Prostate. 2002;50:145–153. doi: 10.1002/pros.10042. [DOI] [PubMed] [Google Scholar]
- 32.Chadha KC, Kawinski E, Sulkowski E. Thiophilic interaction chromatography of prostate-specific antigen. J Chromatogr B Biomed Sci Appl. 2001;754:521–525. doi: 10.1016/s0378-4347(00)00622-8. [DOI] [PubMed] [Google Scholar]
- 33.Denmeade SR, Lou W, Lovgren J, Malm J, Lilja H, Isaacs JT. Specific and efficient peptide substrates for assaying the proteolytic activity of prostate-specific antigen. Cancer Res. 1997;57:4924–4930. [PMC free article] [PubMed] [Google Scholar]
- 34.Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2:1627–1636. [PubMed] [Google Scholar]
- 35.Stephenson RA, Dinney CP, Gohji K, Ordonez NG, Killion JJ, Fidler IJ. Metastatic model for human prostate cancer using orthotopic implantation in nude mice. J Natl Cancer Inst. 1992;84:951–957. doi: 10.1093/jnci/84.12.951. [DOI] [PubMed] [Google Scholar]
- 36.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- 37.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 38.Mahajan SD, Schwartz SA, Nair MP. Immunological assays for chemokine detection in in vitro culture of CNS cells. Biol Proc Online. 2003;5:90–102. doi: 10.1251/bpo50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sensabaugh GF, Blake ET. Seminal plasma protein p30: simplified purification and evidence for identity with prostate-specific antigen. J Urol. 1990;144:1523–1526. doi: 10.1016/s0022-5347(17)39790-2. [DOI] [PubMed] [Google Scholar]
- 40.Walker-Daniels J, Coffman K, Azimi M, Rhim JS, Bostwick DG, Snyder P, Kerns BJ, Waters DJ, Kinch MS. Overexpression of the EphA2 tyrosine kinase in prostate cancer. Prostate. 1999;41:275–280. doi: 10.1002/(sici)1097-0045(19991201)41:4<275::aid-pros8>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 41.Hollas W, Hoosein N, Chung LW, Mazar A, Henkin J, Kariko K, Barnathan ES, Boyd D. Expression of urokinase and its receptor in invasive and non-invasive prostate cancer cell lines. Thromb Haemost. 1992;68:662–666. [PubMed] [Google Scholar]
- 42.Vassalli JD, Pepper MS. Tumour biology. Membrane proteases in focus. Nature. 1994;370:14–15. doi: 10.1038/370014a0. [DOI] [PubMed] [Google Scholar]
- 43.Brigstock DR. Regulation of angiogenesis and endothelial cell function by connective tissue growth factor (CTGF) and cysteine-rich 61 (CYR61) Angiogenesis. 2002;5:153–165. doi: 10.1023/a:1023823803510. [DOI] [PubMed] [Google Scholar]
- 44.Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 45.Wu M, Mazurchuk R, Chaudhary ND, Spernyak J, Veith J, Pera P, Greco W, Hoffman RM, Kobayashi T, Bernacki RJ. High-resolution magnetic resonance imaging of the efficacy of the cytosine analogue 1-[2-C-cyano-2-deoxy-beta-d-arabino-pentofuranosyl]-N(4)-palmitoyl cytosine (CS-682) in a liver-metastasis athymic nude mouse model. Cancer Res. 2003;63:2477–2482. [PubMed] [Google Scholar]
- 46.Ustach CV, Taube ME, Hurst NJ, Jr, Bhagat S, Bonfil RD, Chel ML, Schuger L, Kim HR. A potential oncogenic activity of platelet-derived growth factor D in prostate cancer progression. Cancer Res. 2004;64:1722–1729. doi: 10.1158/0008-5472.can-03-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen P, Peehl DM, Lamson G, Rosenfeld RG. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins in primary cultures of prostate epithelial cells. J Clin Endocrinol Metab. 1991;73:401–407. doi: 10.1210/jcem-73-2-401. [DOI] [PubMed] [Google Scholar]
- 48.Lopez JB, Sahabudin RM, Chin LP. Are plasma insulin-like growth factor I (IGF-I) and IGF-binding protein 3 (IGFBP-3) useful markers of prostate cancer? Int J Biol Markers. 2004;19:164–167. doi: 10.1177/172460080401900213. [DOI] [PubMed] [Google Scholar]
- 49.Janssen JA, Wildhagen MF, Ito K, Blijenberg BG, Van Schaik RH, Roobol MJ, Pols HA, Lamberts SW, Schroder FH. Circulating free insulin-like growth factor (IGF)-I, total IGF-I, and IGF-I, and IGF binding protein-3 levels do not predict the future risk to develop prostate cancer: results of a case-control study involving 201 patients within a population-based screening with a 4-year interval. J Clin Endocrinol Metab. 2004;89:4391–4396. doi: 10.1210/jc.2004-0232. [DOI] [PubMed] [Google Scholar]
- 50.Diamandis EP. Prostate-specific antigen: a cancer fighter and a valuable messenger? Clin Chem. 2000;46:896–900. [PubMed] [Google Scholar]
- 51.Nicholson B, Theodorescu D. Angiogenesis and prostate cancer tumor growth. J Cell Biochem. 2004;91:125–150. doi: 10.1002/jcb.10772. [DOI] [PubMed] [Google Scholar]
- 52.Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. FASEB J. 2003;17:1081–1088. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- 53.Gallicchio MA, Kaun C, Wojta J, Binder B, Bach LA. Urokinase type plasminogen activator receptor is involved in insulin-like growth factor-induced migration of rhabdomyosarcoma cells in vitro. J Cell Physiol. 2003;197:131–138. doi: 10.1002/jcp.10352. [DOI] [PubMed] [Google Scholar]
- 54.Lebedeva IV, Sarkar D, Su ZZ, Kitada S, Dent P, Stein CA, Reed JC, Fisher PB. Bcl-2 and Bcl-x(L) differentially protect human prostate cancer cells from induction of apoptosis by melanoma differentiation associated gene-7, mda-7/IL-24. Oncogene. 2003;22:8758–8773. doi: 10.1038/sj.onc.1206891. [DOI] [PubMed] [Google Scholar]
- 55.Freije JM, Diez-Itza I, Balbin M, Sanchez LM, Blasco R, Tolivia J, Lopez-Otin C. Molecular cloning and expression of collagenase-3, a novel human matrix metalloproteinase produced by breast carcinomas. J Biol Chem. 1994;269:16766–16773. [PubMed] [Google Scholar]
- 56.Hurlin PJ, Queva C, Koskinen PJ, Steingrimsson E, Ayer DE, Copeland NG, Jenkins NA, Eisenman RN. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1996;15:2030. [PMC free article] [PubMed] [Google Scholar]
- 57.Patarca R. Protein phosphorylation and dephosphorylation in physiologic and oncologic processes. Crit Rev Oncogen. 1996;7:343–432. doi: 10.1615/critrevoncog.v7.i5-6.20. [DOI] [PubMed] [Google Scholar]
- 58.Kondapaka BS, Reddy KB. Tyrosine kinase inhibitor as a novel signal transduction and antiproliferative agent: prostate cancer. Mol Cell Endocrinol. 1996;117:53–58. doi: 10.1016/0303-7207(95)03725-x. [DOI] [PubMed] [Google Scholar]
- 59.Lindberg RA, Hunter T. cDNA cloning and characterization of eck, an epithelial cell receptor protein-tyrosine kinase in the eph/elk family of protein kinases. Mol Cell Biol. 1990;10:6316–6324. doi: 10.1128/mcb.10.12.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenberg IM, Goke M, Kanai M, Reinecker HC, Podolsky DK. Epithelial cell kinase-B61: an autocrine loop modulating intestinal epithelial migration and barrier function. Am J Physiol. 1997;273:G824–G832. doi: 10.1152/ajpgi.1997.273.4.G824. [DOI] [PubMed] [Google Scholar]
- 61.Xie D, Yin D, Tong X, Kelly O'J, Mori A, Miller C, Black K, Gui D, Said JW, Koeffler HP. Cyr61 is overexpressed in gliomas and involved in integrin-linked kinase-mediated Akt and beta-catenin-TCF/Lef signaling pathways. Cancer Res. 2004;64:1987–1996. doi: 10.1158/0008-5472.can-03-0666. [DOI] [PubMed] [Google Scholar]
- 62.Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 63.Dighe AS, Richards E, Old LJ, Schreiber RD. Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity. 1994;1:447–456. doi: 10.1016/1074-7613(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 64.Dixon SC, Kruger EA, Bauer KS, Figg WD. Thalidomide upregulates prostate-specific antigen secretion from LNCaP cells. Cancer Chemother Pharmacol Suppl. 1999;43:S78–S84. doi: 10.1007/s002800051103. [DOI] [PubMed] [Google Scholar]
- 65.Torriglia A, Perani P, Brossas JY, Altairac S, Zeggai S, Martin E, Treton J, Courtois Y, Counis MF. A caspase-independent cell clearance program. (2000) The LEI/L-DNase II pathway. Ann NY Acad Sci. 1999;926:192–203. doi: 10.1111/j.1749-6632.2000.tb05612.x. [DOI] [PubMed] [Google Scholar]
- 66.Watt KW, Lee PJ, M'Timkulu T, Chan WP, Loor R. Human prostate-specific antigen: structural and functional similarity with serine proteases. Proc Natl Acad Sci USA. 1986;83:3166–3170. doi: 10.1073/pnas.83.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]