Abstract
Cancer metastasis is a frequent manifestation of malignant melanoma progression. Successful invasion into distant organs by tumor cells must include attachment to microvessel endothelial cells, and degradation of basement membranes and extracellular matrix (ECM). Heparan sulfate proteoglycans (HSPG) are essential and ubiquitous macromolecules associated with the cell surface and ECM of a wide range of cells and tissues. Heparanase (HPSE-1) is an ECM degradative enzyme, which degrades the heparan sulfate (HS) chains of HSPG at specific intrachain sites. To investigate effects of changes in heparanase gene expression in metastatic melanoma cells, we constructed adenoviral vectors containing the full-length human HPSE-1 cDNA in both sense (Ad-S/hep) and antisense orientations (Ad-AS/hep). We found increased HPSE-1 expression and activity in melanoma cell lines following Ad-S/hep infection by Western blot analyses and specific HPSE-1 activity assay. Conversely, HPSE-1 content was significantly inhibited following infection with Ad-AS/Hep. Importantly, HPSE-1 modulation by these adenoviral constructs correlated with invasive cellular properties in vitro and in vivo. Our results suggest that HPSE-1 not only contributes to the invasive phenotype of melanoma cells, but also that the Ad-AS/hep-mediated inhibition of its enzymatic activity can be efficacious in the prevention and treatment of melanoma metastasis.
Keywords: Malignant melanoma, heparanase, heparan sulfate, invasion and metastasis, anti-sense gene delivery
Introduction
Mechanisms responsible for melanoma progression to highly aggressive metastatic disease are not fully understood. However, it is known that proteolytic enzymes play important roles due to their involvement in extracellular matrix (ECM) disassembly, which allows tumor cells to invade into distant organs. Heparan sulfate proteoglycans (HSPG) [1] are now recognized as cell surface/ECM active biologic modulators [2], and their degradation at the level of heparan sulfate glycosaminoglycan chains (HS) has significant regulatory consequences in cancer metastasis. Elevated levels of heparanase (HPSE-1) are known to be associated with brain-metastatic melanoma [3–7]. HPSE-1 is an endo-β-d-glucuronidase [8] involved in the degradation of cell surface/ECM HSPG of a wide range of normal and neoplastic tissues [9–13] and a molecular determinant of metastatic events. The enzymatic activity of HPSE-1 is characterized by specific intrachain HS cleavage of glycosidic bonds with a hydrolase (but not eliminase) type of action, therefore facilitating the release of several protein modulators of cell function, including migration, adhesion, inflammation, angiogenesis, embryogenesis, and metastatic invasion [8–17]. Secondly, HPSE-1 levels have been found to be increased in sera and urine of human patients with metastatic cancers [8,18]. Thirdly, HPSE-1 activity correlates with the metastatic potential of tumor cells in animal models, resulting in increased mortality [15]. Finally, enhanced HPSE-1 mRNA levels correlate with reduced post-operative survival in cancer patients [10,15].
HPSE-1 from various mammalian sources has been cloned as a single gene family [9–12], representing the dominant HS-degradative enzyme. Importantly, HPSE-1 regulation has been shown to change metastatic properties of tumors [10,19,20]. For example, an upregulation of the enzyme has been shown to increase the metastatic properties of tumor cells [10,19]. Conversely, downregulating HPSE-1 by antisense strategies [19,20] demonstrated a reduction in the invasive properties of neoplastic cells.
Evidence has also demonstrated that HPSE-1 is a potential target for antimetastasis drugs because of its critical roles in angiogenic and invasive processes: treatment of experimental animals with HPSE-1 inhibitors considerably reduced the incidence of tumor invasion and angiogenesis in animal models [16,21]. Extensive available animal and clinical data suggest that HPSE-1 may play an important role in the progression of a variety of human tumors. Nevertheless, the possible role(s) of HPSE-1 in tumor progression at the molecular and cellular levels is poorly understood.
These observations led us to hypothesize that inhibition of HPSE-1 expression could inhibit tumor cell invasion by metastatic melanoma cells. Here we demonstrate that an adenoviral vector expressing the full-length human heparanase gene in an antisense orientation specifically inhibits HPSE-1 expression as well as its invasiveness, leading to a reduction of invasive capabilities by metastatic melanoma cells in vitro and in vivo. Our results further support the fact that inhibiting HPSE-1 can change the invasive properties of melanoma cells like others have found in lung or breast carcinoma cell lines.
Experimental Procedures
Materials
HS from bovine kidney was acquired from Sigma Chemical Co. (St. Louis, MO). Dulbecco's modified Eagle's medium (DMEM), Ham's F-12 nutrient medium, and trypsin-ethylenediaminetetraacetic acid (EDTA) were purchased from Gibco (Grand Island, New York, NY), and fetal bovine serum (FBS) was from Hyclone Laboratories (Logan, UT). Human recombinant HPSE-1 was kindly provided by Dr. Edward McKenzie [22]. The polyclonal antibodies to human HPSE-1 were generously provided by Dr. Laurie A. Dempsey (Mayo Clinic, Rochester, MN) and Dr. Robert L. Heinrikson (Pharmacia-Upjohn, Inc., Kalamazoo, MI).
Transwell cell culture chambers were purchased from Corning Incorporated Life Sciences (Acton, MA), whereas Matrigel was obtained from BD Biosciences Discovery Labware (Bedford, MA) and fibronectin was from ICN Biochemical (Irvine, CA). All other chemicals used were of reagent-grade or better.
Cells and Tissue Culture Conditions
Early-passage melanoma cells with varying metastatic abilities, both of murine (B16B15b line [23]) and human origin (70W line [24,25]) were maintained as monolayer cultures in a 1:1 (vol/vol) mixture of DMEM/F-12 supplemented with 10% (vol/vol) FBS and 5 mM sodium butyrate (B16B15b) or 10% (vol/vol) FBS (70W). B16B15b and 70W metastatic melanoma cells (with the lung being the primary colonization site) were chosen as a source of HPSE-1 because they are highly invasive and produce HPSE-1 at elevated levels versus their respective parental counterparts [3,4]. Cells were maintained at 37°C in a humidified 5% CO2/95% air (vol/vol) atmosphere and passaged using 2 mM EDTA (B16B15b) or trypsin-EDTA (70W) before reaching confluence. The transformed embryonic kidney cell line 293 was grown in DMEM/F-12 and supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 µg/ml). The 293 cells were used for the production of adenoviral vectors [19].
Construction of Recombinant Adenovirus Containing the Human Heparanase Gene
An adenovirus expression vector kit (Takara Biomedicals, Inc., Tokyo, Japan) was used to generate recombinant adenovirus for the expression of human HPSE-1 gene in both sense and antisense orientations. Replication-deficient E1-deleted and E3-deleted recombinant adenovirus serotype 5 (Ad5) was used as the viral backbone. Plasmid DNA containing the cloned HPSE-1 gene and synthetic oligonucleotides Hep-5′ (complementary to the 5′ end of the gene) and Hep-3′ (complementary to the 3′ end of the gene) were used to produce polymerase chain reaction (PCR) products encompassing the HPSE-1 sequence (1632 bp). PCR products were treated with T4 DNA polymerase to generate uniformed blunt ends required for the ligation reaction into pAxCAwt cosmid provided by the Takara kit. After cloning the HPSE-1 gene into the cosmid vector, availability of the recombinant cosmids containing the target gene in sense (5′-3′ under the CAG promoter of the vector) and antisense (3′–5′) orientations was confirmed by restriction analysis. Cosmid DNA were produced in large quantities and, after gradient purification, were used for cotransfection with adenovirus genomic DNA-terminal protein complex (DNA-TPC provided by the kit) into 293 cells. Following the kit protocol, recombinant adenoviruses expressing human HPSE-1 in both sense (Ad-S/hep) and antisense (Ad-AS/hep) orientations were generated. Integrity of these recombinant viruses was confirmed by PCR and restriction analysis. The E1-deleted and E3-deleted replication-deficient adenovirus pAd5-Blue was used as the control vector. Viral titer was quantified by determination of 50% infectivity on tissue culture (TCID50) in 293 cells.
Adenoviral Infection
70W and B16B15b cells were plated on 100-mm dishes (106 cells/dish). Twenty-four hours later, cells were washed twice with phosphate-buffered saline (PBS) containing 2 mM EDTA, and infected with virus diluted in serum-free DMEM/F-12. To maximize cell viability and protein expression, cells were infected with viral vectors at a multiplicity of infection (MOI) of 30 to 50. Plates were then incubated at 37°C for 1 hour, gently rocking every 10 minutes. Infection was terminated by adding a culture medium containing penicillin (100 U/ml) and streptomycin (100 µg/ml). Cells were then incubated at 37°C for an appropriate period of time. Immunofluorescent control experiments were performed to confirm the cell infectivity of our adenoviral vectors.
Reverse Transcription (RT) PCR Analysis
Total RNA was isolated from cells using the RNeasy kit (Qiagen, Inc., Valencia, CA). Briefly, RT was performed at 42°C for 1 hour, and PCR was performed using specific primers (final volume, 25 µl). The following specific primers were used: HPSE-1 sense (HPSE-S: 5′-TTC GAT CCC AAG AAG GAA TCA AC-3′) and HPSE-1 antisense (HPSE-AS: 5′- TAC ATG GCA TCA CTA C-3′); for the sequence just after the inserted fragment in the vector (Ad5 pax: 5′-ATC GAT TCT AGA CTA GTT TAA TTA ATT T-3′); control glyceraldehyde-3-phosphate dehydrogenase, sense (5′-TGA AGG TCG GAG TCA ACG GAT TTG GT-3′) and antisense (5′-CAT GTG GGC CAT GAG GTC CAC CAC-3′). The amplification reaction for HPSE-1-, Ad-S/hep-, and Ad-AS/hep-specific constructs involved 35 cycles of denaturation at 95°C for 45 seconds, annealing at 52°C for 1 minute, and 72°C for 1 minute. The amplification for GAPDH reaction involved 35 cycles of denaturation at 94°C for 30 seconds and annealing at 68°C for 3 minutes in the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). PCR products were subsequently resolved on 1% agarose gels and visualized by ethidium bromide staining.
Western Blot Analysis
Cells were plated (106 cells/plate) on 100-mm dishes and allowed to attach and grow for 24 hours. Cells were subsequently infected with Ad-S/hep, Ad-AS/hep, or pAd5-Blue control vector and incubated for 36 to 48 hours. They were then released and centrifuged (300 rpm for 5 minutes) and resuspended in lysis buffer [TBS (pH 7.4) containing Triton-X 100 (0.5%), leupeptin (10 µg/ml), pepstatin (10 µg/ml), and PMSF (0.2 mM)] at a density of 108 cells/ml. Cells were subsequently vortexed and kept on ice for 10 minutes followed by centrifugation for 10 minutes at 13,000 rpm at 4°C. The supernatant was collected and protein concentration was determined by bicinchoninic acid (BCA) assay. Protein samples (60–90 µg) were then incubated at 100°C for 10 minutes with Laemmli sample buffer [26] and separated on 10% Criterion gels (Tris-HCl; Bio-Rad Laboratories Inc., Hercules, CA). Gels were transferred to a PVDF membrane (Pierce Endogen, Inc., Rockford, IL) and incubated overnight (18 hours) in a blocking reagent [3% (wt/vol) nonfat dry milk, 0.5% (wt/vol) bovine serum albumin (BSA), 0.3% (vol/vol) Tween-20 in PBS, pH 7.5]. HPSE-1 protein was labeled using anti-HPSE-1 polyclonal antibody (PAb; 1:5000 dilution; Mayo Clinic) in 3% (wt/vol) nonfat dry milk and 0.5% (wt/vol) BSA, washed for 1 hour with six to eight changes of 0.5% IGE-PAL (CA-630; Sigma Chemical Co.) in Tris buffer (20 mM Tris, 150 mM NaCl, pH 7.4), and then incubated with horseradish peroxidase (HRP)-antirabbit immunoglobulin G (IgG; 1:5000 dilution; Accurate Scientific, Westbury, NY). It was subsequently washed and developed using the Supersignal west femto maximum sensitivity substrate (Pierce Endogen, Inc.). Dual-Color Precision Plus Protein Standards (Bio-Rad Laboratories Inc.) were used as molecular weight markers. Labeling was detected and quantified using a Versadoc imaging system (Quantity One; Bio-Rad Laboratories Inc.).
HPSE-1 Activity Assays
HPSE-1 activity was determined by degradation of FITC-labeled heparan sulfate (FITC-HS) using high-speed gel permeation column chromatography (HPLC) as previously described [9,27]. Briefly, cell lysates (80 µg cell lysate/µg of HS) were prepared as described above and incubated with FITC-HS at 37°C for 18 hours in 100 µl of sodium acetate (100 mM, pH 4.2). Reaction was terminated by heating samples at 100°C for 15 minutes. HS products yielded by HPSE-1 reaction were analyzed by size exclusion chromatography performed on a HP1090 liquid chromatograph with a photodiode array detector (Agilent Technologies, Inc., Wilmington, DE) and a HP1046 programmable fluorescence detector (Agilent Technologies, Inc.). Samples (25 µl) were injected on a TSK-GEL 3000SWXL column (5 µm, 7.8 mm x 30 cm) with a TSK-GEL SWXL guard column (5 µm, 6.0 mm x 4.0 cm) from Tosoh Bioscience (Montgomeryville, PA) at ambient temperature (25°C). Samples were isocratically eluted using Tris-HCl (25 mM), NaCl (150 mM), pH 7.5, at a flow rate of 0.5 ml/min with a sample splitter placed between the column and detectors to maintain a column pressure of less than 70 bar. Fluorescence was monitored with excitation at 490 nm and emission at 520 nm, then data were processed and peaks were integrated using HP ChemStation software (Agilent Technologies, Inc.). HPSE-1 activity was determined by measuring the decrease in fluorescence intensity in the first half area of the intact FITC-HS peak chromatogram. The retention times were calculated from the highest peak of the chromatograms.
Alternatively, a commercial heparan-degrading enzyme assay kit (Takara Mirus Biomedical, Inc., Madison, WI) was used to determine HPSE-1 activity in melanoma cells [27]. Indicated amounts of cell lysate were incubated with biotinylated HS at 37°C for 45 minutes and HPSE-1 activity was determined by an enzyme-linked immunosorbent assay (ELISA)-type assay. Color was developed using the substrate supplied in the kit and plates were read at 450 nm using a microplate reader (EL 808; Bio-Tek Instruments, Inc., Winooski, VT).
Chemoinvasion Assays
Invasive properties of melanoma cells were assayed by Boyden's chambers as previously described [3]. Briefly, cell invasion was assayed using Transwell cell culture chambers (12 µm pore size, 12 mm diameter) coated with Matrigel [diluted as 38 µg/ml in cold DMEM/F-12 with penicillin/streptomycin (P/S), 100 µl final coating volume], which was applied to the upper filter surface and allowed to dry before use. Cells were plated on day 1 and, after 24 hours, were infected with Ad-S/hep, Ad-AS/hep, or the control vector pAd5-Blue as described above. Twenty-four hours later, cells were released from the culture plate and added to the upper chamber of invasion plates (1.2 x 105 cells/chamber) in serum-free DMEM/F-12. Lower chambers contained n-formyl-l-methyl-l-leucine-l-phenylalanine (5 nM) in DMEM/F-12 with 10% (vol/vol) FBS and fibronectin (1 µg/ml) as chemoattractants. Cells were incubated in invasive chambers for 48 to 72 hours at 37°C in a humidified 5% CO2/95% air (vol/vol) atmosphere. At the end of the experiment, Transwell membranes were lifted from the wells carefully without touching the bottom. Noninvasive cells were wiped from the upper chamber with a cotton swab and invasive cells were visualized with Diff-Quick Stain Kit (IMEB, Inc., San Marcos, CA) according to the manufacturer's instructions. Briefly, membranes were dipped in solution A (methanol) for 2 minutes and allowed to air dry. Membranes were then transferred in solution B (Eosin), dipped 25 times, transferred in solution C (Azure), and then dipped 25 more times. After rinsing with deionized water, membranes were inverted and allowed to dry. By using this kit, the nuclei of cells are stained blue and cell cytoplasm is stained pink. The number of invasive cells was obtained by counting the nuclei under the microscope.
In Vivo Tumorigenic Assays
Human malignant melanoma 70W cells were infected with Ad-S/hep, Ad-AS/hep, and control pAd5-Blue at an MOI of 50. Cells were harvested 36 hours after infection, and resuspended in DMEM/F-12 at a density of 5.0 x 106 cells/ml. Six-week-old female athymic nu/nu mice were injected intravenously (tail vein) with 200 µl of cell suspension (106 cells) through a 27-gauge needle. Mice were euthanized and examined for tumor formation as previously described [25]. Metastatic behavior per animal group (number of lung nodules and lungs wet weight; N = 15) was then determined. Additionally, lung tumor tissues were saved in formalin, and random samples were taken and stained with hematoxylin and eosin and analyzed under microscopy.
Immunohistochemical Analysis
Immunohistochemical staining was performed using a Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, CA) according to the instructions provided by the manufacturer. Formalin-fixed, paraffin-embedded tissue sections from in vivo tumorigenic assays were mounted on silanized slides and deparaffinized. Endogenous peroxidase was blocked by incubating the sections in 3.0% H2O2 without pretreatment. After the blocking of nonspecific reactivity with rabbit serum for 30 minutes at room temperature (25°C), sections were incubated at 4°C for 60 minutes with the antihuman HPSE-1 rabbit PAb raised against human HPSE-1 (Pharmacia-Upjohn, Inc.). Following rinsing, slides were incubated with biotinylated antirabbit IgG and then with Vectastain ABC reagent. Peroxidase activity was determined using nova red and counterstained with hematoxylin. As a negative control, sections were subjected to normal serum blocking with omission of the primary antibody.
Results
Detection of HPSE-1 mRNA Expression in Sense or Antisense Orientation
To examine the significance of human HPSE-1 in metastatic melanoma cell invasion, we constructed replication-defective recombinant adenoviruses expressing sense and antisense RNA to a full-length human HPSE-1 sequence, Ad-S/hep, and Ad-AS/hep, respectively. Both sense and antisense transcripts of HPSE-1 could be detected when HPSE-S and HPSE-AS primers were used in 70W cells, but not in the B16B15b cells because primers chosen against human heparanase did not amplify the murine heparanase. Strand-specific primer pairs (HPSE-S and Ad5pax, or HPSE-AS and Ad5pax) detected the expression of the sense and antisense constructs in B16B15b and 70W melanoma cells with Ad-S/Hep or Ad-AS/hep treatment, respectively (Figure 1).
Figure 1.
Expression of human heparanase sense and antisense transcripts in melanoma cells. B16B15b and 70W cell lines were infected with pAd5-Blue (lanes 1 and 4), Ad-S/hep (lanes 2 and 5), or Ad-AS/hep vector (lanes 3 and 6) and subjected to RT-PCR analysis 36 hours after infection. Two primer pairs, HPSE-S/Ad5 pax and HPSE-AS/Ad5pax, were designed to, respectively, detect sense and antisense transcripts of the human heparanase gene. Both sense and antisense transcripts of HPSE-1 could be detected when HPSE-S and HPSE-AS primers were used in 70W cells, but not in the B16B15b cells because the primers chosen against human heparanase gene did not amplify murine heparanase. Primers and predicted products are indicated at the side of panel.
Modulation of HPSE-1 Protein Levels Following Ad-S/Ad-AS/hep Infection in Melanoma Cells
Western blot analyses were performed to examine the effects of Ad-S/hep and Ad-AS/hep infection on HPSE-1 protein levels in B16B15b and 70W cells (Figure 2). Because HPSE-1 is a low-abundance protein, 60 to 90 µg of total cell lysate had to be electrophoresed in order to detect adequate HPSE-1 levels. HPSE-1 protein expression increased with Ad-S/hep and decreased with Ad-AS/hep-treated melanoma cells compared to treatment with control vector pAd5-Blue in both B16B15b (Figure 2A) and 70W (Figure 2B) cells. HPSE-1 is a heterodimer consisting of a 50-kDa and a 8-kDa band processed from a proenzyme precursor (65 kDa) by proteolytic cleavage, and presence of the 8-kDa band is essential for its enzymatic activity [11,18,28]. We detected the active form of HPSE-1 in the cells; however, it migrated slightly slower than its predicted molecular weight (50 kDa). This is consistent with the recent findings by Simizu et al. [29], and possibly due to posttranslational modification in the glycosylation of the enzyme. We also detected a weak band representing the 65-kDa inactive precursor in B16B15b cells. Furthermore, we found a two-fold increase in HPSE-1 protein level using the Ad-S/hep vector and a 30% decrease with the Ad-AS/hep in the B16B15b cell line (Figure 2A). In 70W cells, we observed a 30% increase or decrease in HPSE-1 protein by densitometric analysis following Ad-S/hep or Ad-AS/hep treatment, respectively (Figure 2B).
Figure 2.
Western blot analysis in melanoma cells for HPSE-1 protein expression. Cells (B16B15b and 70W) were infected with pAd5-Blue (lanes 1 and 4), Ad-S/hep (lanes 2 and 5), or Ad-AS/hep (lanes 3 and 6) (see Experimental Procedures section) and analyzed for HPSE-1 expression after 48 hours. Equal amounts (60–90 µg) of protein were loaded on gels and HPSE-1 protein levels were detected by a rabbit polyclonal antibody and HRP-antirabbit IgG followed by use of the Supersignal west femto maximum sensitivity substrate. Bands were visualized on a Versadoc imaging system. Quantification was performed using Quantity One software program (Bio-Rad Laboratories Inc.). (A) Murine B16B15b. (B) Human 70W.
HPSE-1 Activity Is Upregulated in Ad-S/hep-Treated Melanoma Cells
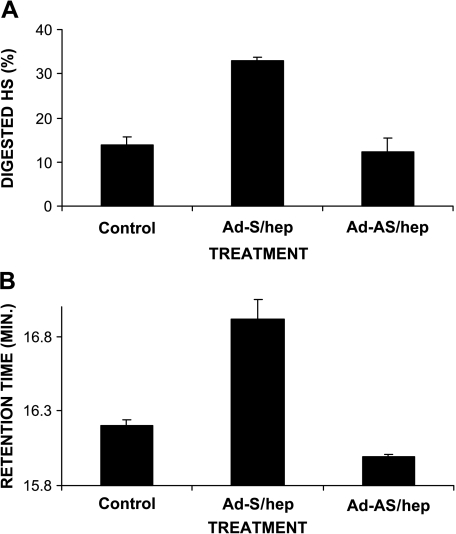
We used a specific HPSE-1 activity assay on fluorescein isothiocyanate (FITC)-labeled HS in 70W cells. This assay detects HPLC profile shifts based on the size of HS fragments [9,27]. Because HPSE-1 cleaves the HS into discrete fragments, we detected a profile shift when compared to FITC-HS alone, which was used as negative control [9,27] (data not shown). We observed a decrease in fragment size in Ad-S/hep-treated 70W cells compared to mock-infected cells. We did not observe a significant difference between Ad-AS/hep- and mock-treated cells because this represents a small shift compared to the total profile area (Figure 3A). However, we observed a significant difference in their respective HPLC retention times, which detect changes in the size of predominant HS fragments (P = .02). This analysis is more sensitive to small changes in HS fragment size and showed less activity with Ad-AS/hep-treated cell lysates (Figure 3B). The Ad-S/hep-treated cell lysates showed a higher retention time compared to Ad-AS/hep or mock treatment, suggesting that increased digestion of HS resulted from an augmented HPSE-1 activity following Ad-S/hep treatment in the 70W cell line. We also performed the ELISA-type assay and found similar changes of HPSE-1 activity in response to pAd5-Blue, Ad-S/hep, or Ad-AS/hep treatments (Table 1).
Figure 3.
HPSE-1 activity increases with Ad-S/hep treatment in 70W cells compared to mock-treated cells. Cells were infected with Ad-S/hep, Ad-AS/hep, or none, and analyzed for HPSE-1 activity after 48 hours. Equal amounts (100 µg) of cell lysates were incubated with FITC-HS for 18 hours (see Experimental Procedures section) and run on HPLC to analyze HPSE-1 activity. (A) Percentage of HS digested with different treatments. (B) Retention times calculated with different treatments. Bars represent the mean with standard deviation of triplicate determinations.
Table 1.
Modulation of HPSE-1 Activity with Ad-S/hep or Ad-AS/hep Treatment.
| Treatment (MOI 50) | A450 (nm)* |
| Pad5-Blue | 0.65 ± 0.05 |
| Ad-S/hep | 0.21 ± 0.03 |
| Ad-AS/hep | 0.73 ± 0.06 |
The higher the OD value, the lower the HPSE-1 activity. The A450 starting point of no enzyme degradation was 0.79.
Secondly, we measured HPSE-1 activity in B16B15b and 70W cells coinfected with Ad-S/hep and Ad-AS/hep, or pAd5-Blue vectors using biotinylated HS and an ELISA kit. We detected a dose-dependent inhibition of HPSE-1 activity with a higher dose of Ad-AS/hep vector (Table 2).
Table 2.
HPSE-1 Activity Decreases with Ad-AS/hep Treatment in a Dose-Dependent Manner.
| Cell Line | Treatment (MOI) | A450 (nm)* | ||
| pAd5-Blue | Ad-S/hep | Ad-AS/hep | ||
| B16B15b | - | - | - | 1.1 ± 0.19 |
| 125 | - | - | 1.0 ± 0.14 | |
| 100 | 25 | - | 0.87 ± 0.01 | |
| 50 | 25 | 50 | 0.92 ± 0.03 | |
| - | 25 | 100 | 1.2 ± 0.10 | |
| 70W | - | - | - | 0.90 ± 0.33 |
| 125 | - | - | 0.83 ± 0.04 | |
| 100 | 25 | - | 0.59 ± 0.01 | |
| 50 | 25 | 50 | 0.72 ± 0.02 | |
| - | 25 | 100 | 0.87 ± 0.16 | |
The higher the OD value, the lower is the HPSE-1 activity. The A450 starting point of no enzyme degradation was 1.7.
Next, we used suramin, an HPSE-1 antagonist, to inhibit the activity of Ad-S/hep-infected cells (Figure 4). This suggests that: 1) increased activity (Figure 4A) and retention time (Figure 4B) in the assay are due to an upregulation of HPSE-1 in the Ad-S/hep-infected cells; and 2) this activity can be further modulated by suramin (500 µM) as an HPSE-1 inhibitor (Figure 4).
Figure 4.
HPSE-1 activity is inhibited by suramin in Ad-S/hep-treated 70W cells. Cells were infected with Ad-S/hep or pAd5-Blue as control and analyzed for HPSE-1 activity after 48 hours. Equal amounts (80 µg of protein/µg of HS) of cell lysates were incubated with FITC-HS for 18 hours (see Experimental Procedures section) with or without suramin (500 µM) and run on HPLC to analyze HPSE-1 activity. (A) The percentage of HS digested with different treatments. (B) Retention times calculated with different treatments. Bars represent the mean with standard deviation of triplicate determinations.
Inhibition of HPSE-1-Induced Invasion by Ad-AS/hep Infection of Melanoma Cells
To investigate HPSE-1 mechanisms in metastatic cell invasion, chemoinvasion assays were performed using Matrigel-coated Transwell chamber systems. B16B15b and 70W cells, when infected with Ad-S/hep, possessed augmented invasive properties compared with pAd5-Blue-infected cells, whereas Ad-AS/hep-treated cells showed a significant inhibition of invasion (Figure 5). These results suggest that the antisense adenoviral construct selectively inhibited HPSE-1-induced invasive properties in B16B15b and 70W cells.
Figure 5.
Ad-AS/hep treatment inhibits HPSE-1-mediated invasion of melanoma cells. Metastatic melanoma cells (B16B15b and 70W) infected with pAd5-Blue, Ad-S/hep, or Ad-AS/hep were placed in invasion chambers (12 µm diameter pore size) for 72 hours at 37°C in a humidified 5% CO2/95% air (vol/vol) atmosphere. Noninvasive cells were wiped from the upper chamber with a cotton swab and invasive cells were detected with the Diff-Quick Stain Kit. Bars represent the mean with standard deviation of triplicate determinations (*P < .01, **P < .1). Student's t test was used as statistical method.
Inhibition of Tumor Formation with Ad-AS/hep-Treated 70W Cells
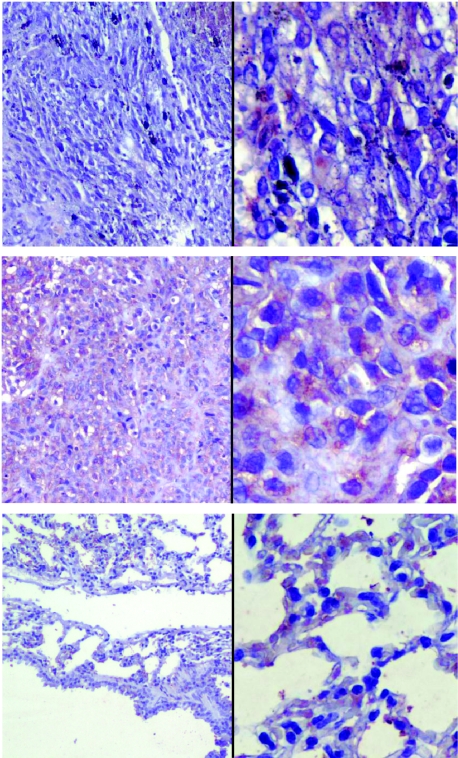
To evaluate HPSE-1 mechanism in metastatic cell invasion, human malignant melanoma 70W cells were infected with Ad-S/hep, Ad-AS/hep, or pAd5-Blue vectors and injected in 6-week-old female athymic nude (nu/nu) mice. Mice injected with pAd5-Blue- and Ad-S/hep-treated 70W cells developed tumors, but none of the Ad-AS/hep-treated cells was capable of extensive tumor formation (Figure 6, Table 3). During quantitation of tumor formation, we found microscopic tumors in one of the mice treated with Ad-AS/hep, suggesting that the antisense adenoviral construct selectively inhibited HPSE-1-induced invasive properties in 70W cells. Because control cells already expressed relatively high levels of HPSE-1, we were unable to see a significant difference between our control and Ad-S/hep groups; however, we found a significant difference between the control and Ad-AS/hep groups. Furthermore, formalin-fixed tissues from each group were analyzed for immunohistochemistry (Figure 7): lung tumor tissue from the Ad-S/hep group expressed HPSE-1 more consistently and with an increased immunohistochemical intensity than in the control group. HPSE-1 staining was also observed in the Ad-AS/hep group but was mainly due to the presence of the enzyme in the endothelium [30] and macrophages [13] of lung tissues (Figure 7).
Figure 6.
Ad-AS/hep treatment inhibits tumor formation in athymic nude (nu/nu) mice. Human malignant melanoma 70W cells were infected with pAd5-Blue control vector (A), Ad-S/hep (B), or Ad-AS/hep (C) and were injected intravenously in 6-week-old female athymic nu/nu mice. At the end of the experimental period, mice were euthanized and examined for tumor formation, as described [25]. Quantification of lung tumor formation and statistical analyses were performed on a number of experimental animals and shown in Table 3.
Table 3.
Ad-AS/hep Treatment Inhibits Tumor Formation in Nude (nu/nu) Mice.
| Treatment | Number of Lung Tumor Nodules | Lung Weight (mg, mean ± SD) | |
| Median | Range | ||
| pAd5-Blue | 16 | 0–32* | 571 ± 154 |
| Ad-S/hep | 12 | 0–44† | 453 ± 210 |
| Ad-AS/hep | 0 | 0–4‡ | 366 ± 75 |
Eighty percent of mice had lung tumors.
Eighty percent of mice had lung tumors.
Twenty percent of mice had lung tumors (P = .04). Student's t test was used as statistical method.
Figure 7.
Ad-S/hep/Ad-AS/hep-mediated HPSE-1 expression in nude (nu/nu) mice. Animals were injected with pAd5-Blue-, Ad-S/hep-, or Ad-AS/hep-infected 70W cells and formalin-fixed lung tumor tissues were analyzed by immunohistochemistry. Intense staining of HPSE-1 was found in lung tissues of animals treated with Ad-S/hep-infected 70W cells (C and D) compared to pAd5-Blue (A and B) or Ad-AS/hep (E and F) treatment. HPSE-1 staining observed in the Ad-AS/hep is due to the presence of the enzyme in the endothelium [30] and macrophages [13] in lung tissues. Digital images were produced on an Axioplan microscope with advanced spot imaging program at x10 (A, C, and E) and x40 (B, D, and F) objectives using identical conditions for all photographs and antibodies used; x40 objectives show the different intensities of HPSE-1 staining by various treatments.
Discussion
Invasion and metastasis are characteristic features of malignant tumors and are among the greatest impediments to curing cancer. Inhibition of tumor invasion is an attractive approach for the treatment of highly malignant tumors [31,32]. Substantial evidence accumulated over the last three decades indicates that HSPG act to inhibit invasion by promoting tight cell-cell and cell-ECM interactions [1,33,34]. Degradation of HS weakens cell-cell and cell-matrix adhesion in melanoma and other tumor cells [35–37]. HSPG can also act both as reservoirs of growth factors [15,38] and as coreceptors for ligand binding and subsequent intracellular signaling. These heparin-binding factors are involved in growth, invasion, angiogenesis, and tumor progression [1,15,33]. HS-degradative enzymes, such as heparitinases, heparinases from Flavobacterium heparinum, or endoglucosaminidases, cleave HS to disaccharides and tetrasaccharides, which are too short for growth factor and ECM ligand binding [39–41]. However, HPSE-1 is an endo-β-d-glucuronidase that cleaves HS at specific intrachain sites, resulting in fragments of appreciable size (10–20 sugar units) [8,15,30,42]. This confirms the notion that HPSE-1, in degrading HSPG and releasing HS-bound angiogenic/growth factors, may aid the modulation of growth factor activities in metastasis [27,43]. Thus, HPSE-1 is an attractive target for the development of novel antimetastatic drugs because of the considerable evidence implicating this enzyme in tumor cell invasion [15].
In the present study, we have demonstrated that overexpression of human HPSE-1 enhanced tumor cell invasiveness in vitro and in vivo, and that this invasive ability was significantly reduced by inhibiting HPSE-1 expression using an adenovirus-mediated antisense gene delivery strategy. HPSE-1 expression, as well as its biologic activity, were enhanced in metastatic melanoma cell lines following Ad-S/hep treatment. Conversely, treatment with Ad-AS/hep resulted in decreased HPSE-1 expression as well as cell invasiveness in vitro and in vivo. For example, in our experiments designed to analyze the effects of Ad-S/hep, we found that there was an upregulation of protein expression in both B16B15b and 70W melanoma cell lines. Conversely, we observed the opposite using Ad-AS/hep (Figure 2). The reduction in HPSE-1 protein level was consistent over multiple experiments. The inability to completely inhibit HPSE-1 protein expression was possibly due to slow protein turnover rate. However, the reduction results in significant biologic effects in in vitro invasion assays or in in vivo tumorigenic assays, despite this small change in the protein level.
HPSE-1 activity was inhibited in a dose-dependent manner when cells were coinfected with Ad-S/hep and different doses of Ad-AS/hep (Table 2). Furthermore, we detected an increase in HPSE-1 activity with Ad-S/hep infection (Figure 3, Table 1). There was a small reduction of activity with Ad-AS/hep treatment compared to mock-treated cells (Figure 3A), which significantly decreased when HPLC retention times were calculated from these experiments (P = .02) (Figure 3B).
We were also able to inhibit the increased activity of Ad-S/hep with suramin, a known HPSE-1 inhibitor, therefore relating this augmented cell invasiveness to HPSE-1 (Figure 4). The poor sensitivity of HPSE-1 activity assays may reflect a lack of sensitivity at low HPSE-1 levels. Alternatively, differences between HPSE-1 activity and biologic assays can be due to lysis of the cells (HPSE-1 activity assays) versus using intact cells in in vitro (invasion assays) and in vivo (tumorigenic assays) analyses. In fact, we observed both a significant biologic effect in chemoinvasion assays and a reduction in both B16B15b and 70W cell-invasive properties (Figure 5). Of equal importance, mice injected with pAd5-Blue- and Ad-S/hep-treated 70W cells showed extensive tumor formation, whereas none of the mice injected with Ad-AS/hep-treated 70W cells presented any evidence of macroscopic malignancy (Figure 6, Table 3). Because control cells already expressed high levels of HPSE-1, we were unable to detect a significant difference between control and Ad-S/hep groups; however, this was found between the control and Ad-AS/hep groups (P = .04). Secondly, immunohistochemical analyses on the animal tissue from each group revealed more consistent and intense HPSE-1 staining in Ad-S/hep-infected cells (Figure 7, C and D) compared to using pAd5-Blue (Figure 7, A and B) or Ad-AS/hep vectors (Figure 7, E and F). These findings indicate that even a small reduction in HPSE-1 activity can lead to a great difference in its biologic function and may also suggest that the antisense-mediated gene delivery strategy can be a very effective way in preventing tumor formation and its metastatic potential.
Many chemotherapeutic agents are available to target HPSE-1 to suppress metastasis [44–47]. However, these agents have their limitations in affecting malignant cells as well as normal cells, thus affecting the normal growth and functions of vital organs. Secondly, delivering these agents to specific sites can be difficult. Finally, interactions between the antimetastatic agents and biologic molecules like HSPG can interfere with normal biologic processes. Accordingly, to specifically inhibit the metastatic potential, which is a frequent malignant phenotype of many tumor types, a more specific therapeutic approach, such as the antisense-mediated RNA expression, needs to be considered. Antisense RNA can bind to the mRNA expressing the gene of interest and can block protein expression by the target cells.
Several groups have reported that tumor cells transfected with HPSE-1 cDNA acquire a highly metastatic phenotype in vivo [10] (reviewed in Ref. [15]). Conversely, inhibition of HPSE-1 by means of antisense strategy, antimetastatic drugs, or gene silencing has resulted in decreased invasiveness in vitro [20] and in vivo [19,48]. However, in other antisense animal models, tumor cells treated with viral vectors were directly injected into the thoracic cavity [19], which bypasses several steps in the metastatic process that includes adhesion to endothelium and invasion [8–17]. A recent report of HPSE-1 inhibition by siRNA strategy also demonstrated reduced tumor load and increased survival in animals [48]. Consistent with these findings, we have observed a significant inhibition of tumor formation in in vivo experiments where HPSE-1-expressing 70W cells were infected with Ad-AS/hep and subsequently injected in athymic nu/nu mice (Figure 6, Table 3). This suggests that Ad-AS/hep has the potential to suppress the metastatic phenotype of highly invasive tumor cells. Thus, approaches that selectively block the expression of molecules implicated in cellular invasion may be clinically more efficacious in preventing tumor cell dissemination than the ones used to induce tumor cell apoptosis.
The metastatic phenotype is considered decisive for tumor progression and several key molecules involved in these complex biologic events are potential candidates for therapeutic intervention. HPSE-1 should be considered one of these.
Acknowledgements
We express our gratitude to Marian Waguespack of Steven Barker's laboratory (LSU-Baton Rouge) for her expert assistance in HPLC analyses, Andrea Greiter-Wilke for her input in animal experiments, and Daniel Paulsen (LSU-Baton Rouge) for his pathological expertise. We also thank Jason Blust for his editorial help.
Abbreviations
- BCA
bicinchoninic acid
- BSA
bovine serum albumin
- DMEM
Dulbecco's modified Eagle's medium
- ECM
extracellular matrix
- EDTA
ethylenediaminetetraacetic acid
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- FITC-HS
FITC-labeled heparan sulfate
- HPLC
high-speed gel permeation column chromatography
- HPSE-1
heparanase
- HRP
horseradish peroxidase
- HS
heparan sulfate glycosaminoglycan chains
- HSPG
heparan sulfate proteoglycans
- IgG
immunoglobulin G
- MAb
monoclonal antibody
- PAb
polyclonal antibody
- PBS
phosphate-buffered saline
- P/S
penicillin/streptomycin
Footnotes
This work was supported by a National Institutes of Health grant 5R0-1 CA86832 (to D.M.) and by a grant from the Louisiana Governor's Biotechnology Initiative.
References
- 1.Iozzo RV. Heparan sulfate proteoglycans: intricate molecules with intriguing functions. J Clin Invest. 2001;108:165–167. doi: 10.1172/JCI13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 3.Marchetti D, Menter DG, Jin L, Nakajima M, Nicolson GL. Nerve growth factor effects on human and mouse melanoma cell invasion and heparanase production. Int J Cancer. 1993;55:692–699. doi: 10.1002/ijc.2910550430. [DOI] [PubMed] [Google Scholar]
- 4.Marchetti D, McQuillan DJ, Spohn WC, Carson DD, Nicolson GL. Neurotrophin stimulation of human melanoma cell invasion: selected enhancement of heparanase activity and heparanase degradation of specific heparan sulfate subpopulations. Cancer Res. 1996;56:2856–2863. [PubMed] [Google Scholar]
- 5.Marchetti D. Brain-metastasis associated genes. In: Welch DR, editor. Cancer Metastasis-Related Genes. The Netherlands: Kluwer Academic Publishing; 2002. pp. 89–108. [Google Scholar]
- 6.Marchetti D. Heparanase: a target for therapy of brain invasive tumors? Expert Rev Neurotherapeutics. 2002;2:89–93. doi: 10.1586/14737175.2.4.459. [DOI] [PubMed] [Google Scholar]
- 7.Marchetti D, Nicolson GL. Human heparanase: a molecular determinant of brain metastasis. Adv Enzyme Regul. 2001;41:343–360. doi: 10.1016/s0065-2571(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 8.Nakajima M, Irimura T, Nicolson GL. Heparanase and tumor metastasis. J Cell Biochem. 1988;36:157–167. doi: 10.1002/jcb.240360207. [DOI] [PubMed] [Google Scholar]
- 9.Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274:24153–24160. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- 10.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 11.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 12.Kussie PH, Hulmes JD, Ludwig DL, Patel S, Navarro EC, Seddon AP, Giorgio NA, Bohlen P. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun. 1999;261:183–187. doi: 10.1006/bbrc.1999.0962. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki N, Higashi N, Taka T, Nakajima M, Irimura T. Cell surface localization of heparanase on macrophages regulates degradation of extracellular matrix heparan sulfate. J Immunol. 2004;2:3830–3835. doi: 10.4049/jimmunol.172.6.3830. [DOI] [PubMed] [Google Scholar]
- 14.Vlodavsky I, Bar-Shavit R, Ishai-Michaeli R, Bashkin P, Fuks Z. Extracellular sequestration and release of fibroblast growth factor: a regulatory mechanism? Trends Biochem Sci. 1991;16:268–271. doi: 10.1016/0968-0004(91)90102-2. [DOI] [PubMed] [Google Scholar]
- 15.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parish CR, Coombe DR, Jakobsen KB, Bennett FA, Underwood PA. Evidence that sulphated polysaccharides inhibit tumour metastasis by blocking tumour-cell-derived heparanases. Int J Cancer. 1987;40:511–518. doi: 10.1002/ijc.2910400414. [DOI] [PubMed] [Google Scholar]
- 17.Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- 18.Kelly T, Miao HQ, Yang Y, Navarro E, Kussie P, Huang Y, MacLeod V, Casciano J, Joseph L, Zhan F, et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63:8749–8756. [PubMed] [Google Scholar]
- 19.Uno F, Fujiwara T, Takata Y, Ohtani S, Katsuda K, Takaoka M, Ohkawa T, Naomoto Y, Nakajima M, Tanaka N. Antisense-mediated suppression of human heparanase gene expression inhibits pleural dissemination of human cancer cells. Cancer Res. 2001;61:7855–7860. [PubMed] [Google Scholar]
- 20.Zhang YL, Fu ZR, Ding GS, Fu H, Wang YH, Wang Q. Expression of heparanase mRNA and its clinical significance in primary hepatocellular carcinoma. Zhonghua Zhong Liu Za Zhi. 2003;25:359–361. [PubMed] [Google Scholar]
- 21.Marchetti D, Reiland J, Erwin B, Roy M. Inhibition of heparanase activity and heparanase-induced angiogenesis by suramin analogues. Int J Cancer. 2003;104:167–174. doi: 10.1002/ijc.10930. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie E, Young K, Hircock M, Bennett J, Bhaman M, Felix R, Turner P, Stamps A, McMillan D, Saville G, et al. Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. Biochem J. 2003;373:423–435. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima M, Irimura T, Di Ferrante D, Di Ferrante N, Nicolson GL. Heparan sulfate degradation: relation to tumor invasive and metastatic properties of mouse B16 melanoma sublines. Science (Washington, DC) 1983;220:611–613. doi: 10.1126/science.6220468. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa M, Dennis JW, Man S, Kerbel RS. Isolation and characterization of spontaneous wheat germ agglutinin-resistant human melanoma mutants displaying remarkably different metastatic profiles in nude mice. Cancer Res. 1988;48:665–670. [PubMed] [Google Scholar]
- 25.Ishikawa M, Fernandez B, Kerbel RS. Highly pigmented human melanoma variant which metastasizes widely in nude mice, including to skin and brain. Cancer Res. 1988;48:4897–4903. [PubMed] [Google Scholar]
- 26.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: functional implications for tumor cell invasion. J Biol Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- 28.Fairbanks MB, Mildner AM, Leone JW, Cavey GS, Mathews WR, Drong RF, Slightom JL, Bienkowski MJ, Smith CW, Bannow CA, et al. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem. 1999;274:29587–29590. doi: 10.1074/jbc.274.42.29587. [DOI] [PubMed] [Google Scholar]
- 29.Simizu S, Ishida K, Wierzba MK, Osada H. Secretion of heparanase protein is regulated by glycosylation in human tumor cell lines. J Biol Chem. 2004;279:2697–2703. doi: 10.1074/jbc.M300541200. [DOI] [PubMed] [Google Scholar]
- 30.Marchetti D. Specific degradation of subendothelial matrix proteoglycans by brain-metastatic melanoma and brain endothelial cell heparanases. J Cell Physiol. 1997;2:334–342. doi: 10.1002/(SICI)1097-4652(199709)172:3<334::AID-JCP7>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 31.Zetter BR. Angiogenesis and tumor metastasis. Annu Rev Med. 1998;49:407–412. doi: 10.1146/annurev.med.49.1.407. [DOI] [PubMed] [Google Scholar]
- 32.Folkman J. Angiogenesis-dependent diseases. Semin Oncol. 2001;28:536–542. doi: 10.1016/s0093-7754(01)90021-1. [DOI] [PubMed] [Google Scholar]
- 33.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 34.Sanderson RD. Heparan sulfate proteoglycans in invasion and metastasis. Semin Cell Dev Biol. 2001;12:89–98. doi: 10.1006/scdb.2000.0241. [DOI] [PubMed] [Google Scholar]
- 35.Dhodapkar MV, Kelly T, Theus A, Athota AB, Barlogie B, Sanderson RD. Elevated levels of shed syndecan-1 correlate with tumour mass and decreased matrix metalloproteinase-9 activity in the serum of patients with multiple myeloma. Br J Haematol. 1997;99:368–371. doi: 10.1046/j.1365-2141.1997.3893203.x. [DOI] [PubMed] [Google Scholar]
- 36.Engbring JA, Hoffman MP, Karmand AJ, Kleinman HK. The B16F10 cell receptor for a metastasis-promoting site on laminin-1 is a heparan sulfate/chondroitin sulfate-containing proteoglycan. Cancer Res. 2002;62:3549–3554. [PubMed] [Google Scholar]
- 37.Ma YQ, Geng JG. Heparan sulfate-like proteoglycans mediate adhesion of human malignant melanoma A375 cells to P-selectin under flow. J Immunol. 2000;165:558–565. doi: 10.4049/jimmunol.165.1.558. [DOI] [PubMed] [Google Scholar]
- 38.Iozzo RV, Cohen IR, Grassel S, Murdoch AD. The biology of perlecan: the multifaceted heparan sulphate proteoglycan of basement membranes and pericellular matrices. Biochem J. 1994;302:625–639. doi: 10.1042/bj3020625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst S, Langer R, Cooney CL, Sasisekharan R. Enzymatic degradation of glycosaminoglycans. Crit Rev Biochem Mol Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 40.Ernst S, Venkataraman G, Winkler S, Godavarti R, Langer R, Cooney CL, Sasisekharan R. Expression in Escherichia coli, purification and characterization of heparinase I from Flavobacterium heparinum. Biochem J. 1996;315:589–597. doi: 10.1042/bj3150589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu D, Shriver Z, Venkataraman G, El Shabrawi Y, Sasisekharan R. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc Natl Acad Sci USA. 2002;99:568–573. doi: 10.1073/pnas.012578299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakajima M, Irimura T, Di Ferrante N, Nicolson GL. Metastatic melanoma cell heparanase. Characterization of heparan sulfate degradation fragments produced by B16 melanoma endoglucuronidase. J Biol Chem. 1984;259:2283–2290. [PubMed] [Google Scholar]
- 43.Marchetti D, Denkins Y, Reiland J, Greiter-Wilke A, Galjour J, Murry B, Roy M, Roy M. Brain-metastatic melanoma: a neurotrophic perspective. Pathol Oncol Res. 2003;9:147–158. doi: 10.1007/BF03033729. [DOI] [PubMed] [Google Scholar]
- 44.Miao HQ, Elkin M, Aingorn E, Ishai-Michaeli R, Stein CA, Vlodavsky I. Inhibition of heparanase activity and tumor metastasis by laminarin sulfate and synthetic phosphorothioate oligodeoxynucleotides. Int J Cancer. 1999;83:424–431. doi: 10.1002/(sici)1097-0215(19991029)83:3<424::aid-ijc20>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 45.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- 46.Dhar S, Gullbo J, Csoka K, Eriksson E, Nilsson K, Nickel P, Larsson R, Nygren P. Antitumour activity of suramin analogues in human tumour cell lines and primary cultures of tumour cells from patients. Eur J Cancer. 2000;36:803–809. doi: 10.1016/s0959-8049(00)00024-1. [DOI] [PubMed] [Google Scholar]
- 47.Meyers MO, Gagliardi AR, Flattmann GJ, Su JL, Wang YZ, Woltering EA. Suramin analogs inhibit human angiogenesis in vitro. J Surg Res. 2000;91:130–134. doi: 10.1006/jsre.2000.5920. [DOI] [PubMed] [Google Scholar]
- 48.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]