Abstract
In a microarray-based methylation analysis of astrocytomas [World Health Organization (WHO) grade II], we identified a CpG island within the first exon of the protocadherin-γ subfamily A11 (PCDH-γ-A11) gene that showed hypermethylation compared to normal brain tissue. Bisulfite sequencing and combined bisulfite restriction analysis (COBRA) was performed to screen low- and high-grade astrocytomas for the methylation status of this CpG island. Hypermethylation was detected in 30 of 34 (88%) astrocytomas (WHO grades II and III), 20 of 23 (87%) glioblastomas (WHO grade IV), and 8 of 8 (100%) glioma cell lines. There was a highly significant correlation (P = .00028) between PCDH-γ-A11 hypermethylation and decreased transcription as determined by competitive reverse transcription polymerase chain reaction in WHO grades II and III astrocytomas. After treatment of glioma cell lines with a demethylating agent, transcription of PCDH-γ-A11 was restored. In summary, we have identified PCDH-γ-A11 as a new target silenced epigenetically in astrocytic gliomas. The inactivation of this cell-cell contact molecule might be involved in the invasive growth of astrocytoma cells into normal brain parenchyma.
Keywords: Glioma, PCDH-γ-A11, methylation, cRT-PCR, expression
Introduction
Diffuse infiltration of normal brain tissue is a hallmark of astrocytomas and a major obstacle to neurosurgical intervention. Gliomas are the most frequent tumors of the brain and account for 50% to 60% of all intracranial neoplasms [1]. Even though these tumors rarely metastasize outside the brain, tumor cells not removed by surgery and/or chemotherapy tend to infiltrate into surrounding normal brain parenchyma, giving rise to a more aggressive recurrent tumor. Although much is known about genetic alterations in gliomas (e.g., deletions in chromosomes 1p, 9p, 11p, 19q, and 22q, and inactivation of tumor-suppressor genes TP53 and CDKN2A/B) [2], there is relatively little information on epigenetic alterations and gene silencing, especially those implicated in cell adhesion and locomotion.
In a microarray-based methylation analysis, we have identified a CpG island within the first exon of the protocadherin-γ subfamily A11 (PCDH-γ-A11) gene that showed frequent methylation in diffuse astrocytoma World Health Organization (WHO) grade II. Protocadherins constitute the largest subgroup within the cadherin superfamily of calcium-dependent cell-cell adhesion molecules and have been implicated in neural cell-cell interactions. They are abundantly expressed in the central nervous system during embryonic development and in adulthood [3,4]. PCDH-γ-A11 is located in the protocadherin gene cluster of the chromosomal region 5q31. This region contains nearly 60 protocadherin genes organized in three large sequential clusters (α-, β-, and γ-protocadherins), some of which have been associated with specific neuronal connectivity and synaptic junctions in the nervous system [5,6]. Loss of expression of members of the cadherin superfamily (e.g., CDH1, CDH13, CDH11, and protocadherin LKC) has been demonstrated in a number of different cancer entities including colorectal, liver, and lung cancers [7–10], and may be involved in tumor cell invasion and metastasis [11,12]. Candidate gene approaches as well as genomewide methylation analysis by RLGS demonstrated that epigenetic alterations are common in gliomas and other central nervous system tumors [13–16]. This points to the likelihood that DNA methylation may be involved in tumorigenesis and tumor progression in gliomas, and it is therefore of interest to uncover novel gene promoters that are hypermethylated in this tumor entity.
Here we present evidence for de novo methylation and the associated transcriptional silencing of PCDH-γ-A11, a member of the protocadherin-γ subfamily, in 57 astrocytomas and eight glioma cell lines. Methylation status of PCDH-γ-A11 was determined by bisulfite sequencing and COBRA analysis. A competitive reverse transcription polymerase chain reaction (cRT-PCR) protocol was used to determine transcript levels of PCDH-γ-A11 in astrocytomas and glioma cell lines. To confirm the direct involvement of methylation in silencing of gene transcription, we treated glioma cell lines with the demethylating agent 5′-aza-2′-deoxycytidine and showed that PCDH-γ-A11 was reexpressed in the treated cells.
Materials and Methods
Tumor Samples and Isolation of Nucleic Acids
Tumor tissue was obtained from patients with brain tumors treated at the University of Bonn Medical Center (Bonn, Germany) and was classified according to the WHO grading system of brain tumors using standard histologic and immunohistologic methods [1]. The patient age ranged from 11 to 78 years. Our patient cohort included 29 females and 29 males (Table 1). The series included 34 astrocytomas (WHO grades II and III), 24 glioblastomas (WHO grade IV), and eight glioma cell lines (308D, A1207, LN229D, U178MG, U87MG, LN428, A172, and U373MG). Tissues were selected for extraction of DNA and RNA after careful examination of H&E staining of corresponding frozen sections to exclude contaminating necrotic debris or normal brain tissues, and to determine histologic characteristics of the tumor. DNA was extracted using standard proteinase K digestion and phenol/chloroform extraction. RNA was isolated with TRIZOL reagent (Invitrogen, La Jolla, CA) following the manufacturer's protocol. Contaminating residual genomic DNA was removed by digestion with RNase-free DNase (Roche, Mannheim, Germany). Biopsies of white matter and grey matter (cortex) were included as normal tissue controls for hybridization of differential methylation hybridization (DMH) microarrays and for methylation and expression analyses.
Table 1.
Methylation and Expression Data for PCDH-γ-A11 in Astrocytomas and Glioma Cell Lines.
| Tumor ID | S/A | Diagnosis | Methst | RelmRNA Expr | Tumor ID | S/A | Diagnosis | Methst | RelmRNA Expr |
| 1074D | M/44 | AII | m | nd | 3728 | M/74 | GBM | m | nd |
| 2307 | M/35 | AII | m | nd | 4006 | M/38 | GBM | m | 0.21 (± 0.05) |
| 2154 | M/30 | AII | m | 0.33 (± 0.07) | 4032 | M/35 | GBM | m | 0 |
| 2246 | F/11 | AII | m | 0.11 (± 0.01) | 4416 | M/61 | GBM | m | 1.1 (± 0.1) |
| 2316 | M/35 | AII | m | 0 | 4594 | F/52 | GBM | m | 9.7 (± 1.85) |
| 3022 R | F/29 | AII | m | 2.44 (± 0.34) | 4714 | F/39 | GBM | m | 0 |
| 3294 R | F/34 | AII | m | 0.8 (± 0.03) | 4732 | F/61 | GBM | m | 0.1 (± 0.01) |
| 3632 | M/55 | AII | m | 27.67 (± 4.51) | 4804 | M/57 | GBM | m | 0.21 (± 0.08) |
| 4948 | M/46 | AII | m | nd | 4936 | F/37 | GBM | m | nd |
| 5232 R | M/36 | AII | m | 0.88 (± 0.05) | 4944 | F/59 | GBM | m | 0.2 (± 0.04) |
| 7282 | M/29 | AII | m | 1.07 (± 0.39) | 4962 | F/70 | GBM | m | 0.07 (± 0.01) |
| 7342 | M/40 | AII | m | 0 | 5162 | F/78 | GBM | m | 8.16 (± 0.57) |
| 9306 | F/55 | AII | m | 0.18 (± 0.15) | 6236 | M/69 | GBM | m | 13.8 (± 1.14) |
| 9421 | F/37 | AII | m | 0.82 (± 0.2) | 6840 | M/54 | GBM | m | nd |
| 9640 | M/32 | AII | m | 0.2 (± 0.02) | 10120 R | M/40 | GBM | m | 98.47 (± 29.5) |
| 11092 | M/35 | AII | m | 0.52 (± 0.03) | 11418 | F/68 | GBM | m | 0.4 (± 0.01) |
| 11240 | M/25 | AII | m | 0.29 (± 0.05) | 11571 | F/70 | GBM | m | 16.74 (± 3.8) |
| 12020 R | F/21 | AII | m | 0.93 (± 0.26) | 11666 | M/42 | GBM | m | 0.05 (± 0.01) |
| 9004 R | F/32 | AII | nm | 26.7 (± 2.19) | 4530 | F/66 | GBM | nm | 0.08 (± 0.01) |
| 7564 | M/38 | AII | nm | 18.5 (± 2.49) | 4548 | F/60 | GBM | nm | 0 |
| 2392 R | F/41 | AIII | m | nd | 4400 | M/49 | GBM | nd | 0 |
| 2754 | F/50 | AIII | m | 1.03 (± 0.04) | 11637 | F/52 | GBM | nm | 3.1 (± 0.2) |
| 2814 | M/16 | AIII | m | 1.44 (± 0.01) | 308D | GBMcl | m | nd | |
| 7006 | M/42 | AIII | m | nd | A1207D | GBMcl | m | nd | |
| 7020 | M/32 | AIII | m | nd | LN229D | GBMcl | m | nd | |
| 9846 | F/50 | AIII | m | 0.05 (± 0.02) | U178MG | GBMcl | m | 0.13 (± 0.01) | |
| 10048 | M/49 | AIII | m | 0.18 (± 0.04) | U178MG+ | GBMcl | 10.4 (± 1.96) | ||
| 10760 | F/30 | AIII | m | 1.14 (± 0.12) | U87MG | GBMcl | m | 0.2 (± 0.01) | |
| 11060 | F/32 | AIII | m | 0.09 (± 0.01) | U87MG+ | GBMcl | 25.26 (± 7.16) | ||
| 11226 | F/32 | AIII | m | 0.18 (± 0.02) | LN428 | GBMcl | m | 0 | |
| 12442 | F/28 | AIII | m | nd | LN428 ++ | GBMcl | 4.13 (± 0.28) | ||
| 12490 | F/51 | AIII | m | 0.57 (± 0.15) | A172 | GBMcl | m | 0 | |
| 2446 | F/62 | AIII | nm | 14.6 (± 1.02) | A172 + | GBMcl | 2.6 (± 0.24) | ||
| 10414 | F/38 | AIII | nm | 5.62 (± 0.32) | U373MG | GBMcl | m | 0 | |
| 537D R | M/49 | GBM | m | nd | U373MG+ | GBMcl | 11.36 (± 2.71) | ||
| 1150D R | M/62 | GBM | m | nd | NB | WM,GM | 7.8 (± 1.49) | ||
M, male; F, female; R, recurrent; AII, diffuse astrocytoma of WHO grade II; AIII, anaplastic astrocytoma of WHO grade III; GBM, glioblastoma of WHO grade IV; GBMcl, glioma cell line; S/A, sex/age; m, methylated; nm, not methylated; (+) 0.5 µM 5-aza-2′-deoxycytidine; (++) 1 µM 5-aza-2′-deoxycytidine; nd, not determined; NB, pool of normal brain (white and grey matter) samples; WM, white matter; GM, grey matter (cortex); Methst, methylation status; RelmRNA Expr, relative transcription levels and standard error of PCDH-γ-A11 expression are indicated.
DMH Analysis
The DMH procedure was performed as described [17]. CpG-rich DNA fragments were isolated from the human CGI (CpG island) library and screened for the presence of BstUI (NEB, Beverly, MA) and HpaII (NEB) restriction sites. A total of 7680 suitable fragments was PCR-amplified using plasmid primer and spotted onto UltraGAPS microarrays (Corning, Acton, MA). For amplicon generation, 2 µg of genomic DNA of five astrocytoma (WHO grade II) samples and a pool of four normal brain samples (white matter) were digested with MseI (NEB) according to the manufacturer's protocol. After ligation of linker H12/H24, fragments were digested with methylation-sensitive restriction enzymes BstUI and HpaII and amplified for 20 cycles with H24 as primer. PCR fragments were labeled by direct incorporation of Cy3-dCTP (normal brain) and Cy5-dCTP (tumor) using fluorescent dyes (Amersham, Buckinghamshire, UK) and the Klenow fragment (Invitrogen). Labeled amplicons were purified with Microcon YM-30 columns (Millipore, Bedford, MA) and equal amounts of Cy3 and Cy5 label and 20 µg of human CotI DNA were combined for hybridization on DMH microarrays. Hybridization and analysis procedure were carried out as described [18]. Data from single-copy sequences were normalized and loci with a Cy5/Cy3 ratio greater than 2 were scored as hypermethylated.
Bisulfite Treatment and Bisulfite Sequencing
DNA was treated with sodium bisulfite as described [19]. Primers used for PCR amplification were PCDH-γ-A11 fw-5′-ATTTGGTTATTTGGTGATTAAGGTG-3′ and PCDH-γ-A11 rev-5′-AAAATTTCAAAATTAACCAAAAACT-3′. The 312-bp PCR product (acc. no. NM 018914; 2010–2321 bp) contains 26 putative CpG methylation sites and was cloned with the pGEM-T Vector System kit (Promega, Madison, WI). Individual bacterial clones were amplified by PCR using vector-specific primers. PCR products were purified with the high pure PCR purification kit (Roche), sequenced using the BigDye Prism DNA sequencing kit (Applied Biosystems, Foster City, CA), and analyzed on an automated DNA sequencer 377 (Applied Biosystems).
Combined Bisulfite Restriction Analysis (COBRA)
PCR products generated with PCDH-γ-A11 primers from bisulfite-treated tumor and control DNA were digested with the methylation-sensitive restriction enzyme BstUI and subsequently run on 2.5% agarose gels adjacent to an untreated aliquot of the respective PCR product. BstUI restriction sites (CGCG) are conserved when the respective DNA samples are methylated; unmethylated DNA samples are changed by bisulfite treatment into TGTG and are not recognized by the enzyme. The 312-bp amplicon contains three BstUI restriction sites, yielding fragments of 151, 104, 35, and 22 bp if all sites are methylated.
Cell Culture and Treatment with 5-Aza-2′-deoxycytidine
Glioma cell lines U178MG, U87MG, LN428, A172, and U373MG were cultured in DMEM medium (Biochrom, Berlin, Germany) supplemented with 10% fetal calf serum (Gibco, Paisley, UK) at 37°C and 5% CO2. To induce demethylation, cells were treated with 0.5 and 1 µM 5-aza-2′-deoxycytidine (Sigma, St. Louis, MO) for 3 days with changes of medium every other day and, finally, total RNA was extracted for PCDH-γ-A11 expression analysis.
cRT-PCR
RNA competitor molecules with internal deletions for PCDH-γ-A11 and the housekeeping gene PBGD (porpho-obilinogen desaminase) were generated by in vitro mutagenesis and in vitro transcription as described [20]. To achieve a quantitative assessment, predetermined amounts of the specific standard RNA covering an equimolar range of the corresponding mRNA transcript were added to 250 ng of sample RNA prior to reverse transcription using the SuperScript Preamplification System (Invitrogen). One microliter of cDNA was amplified with primers PCDH-γ-A11 fw 5′-caaagattcaggccagaacg-3′ (exon 1) and PCDH-γ-A11 rev 5′-ccaagatcatggcttgcagc-3′ (exon 3) spanning intronic regions, yielding products of 227 bp for the endogenous transcript and 212 bp for the competitor transcript. For PBGD, the following primers were used: PBGD fw 5′-CCAGGACATCTTGGATCTGG-3′ and PBGD rev 5′-TAAGCTGCCGTGCAACATCC-3′, endogenous product size 389 bp, competitor 367 bp. Every PCR was carried out in triplicates. One primer for each gene was labeled with a fluorescent dye. PCR products were separated on 4.5% denaturating acrylamide gels on an ABI 377 DNA sequencer and analyzed using the Genescan software (Applied Biosystems). The expression levels of target RNA were calculated using the following algorithm: (PCDH-γ-A11endogenous / PCDH-γ-A11competitor) / (PBGDendogenous / PBGDcompetitor).
Results
Aberrant Methylation within the First Exon of PCDH-γ-A11
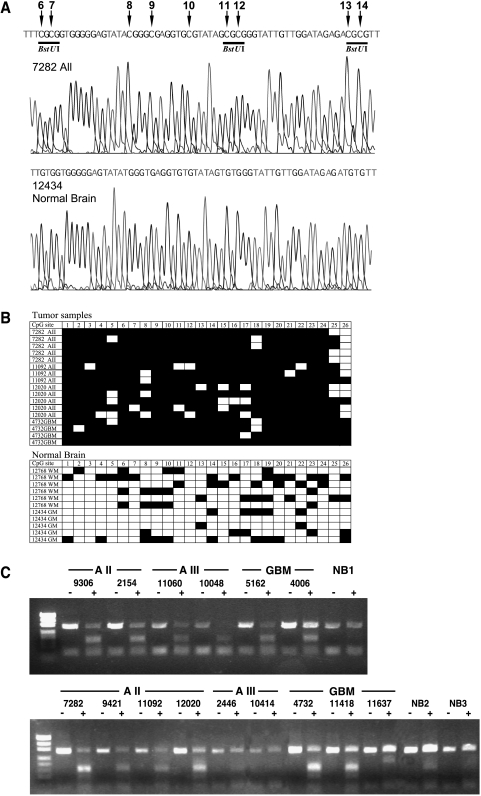
Using methylation microarray analysis, we have identified a CG-rich fragment within the first exon of PCDH-γ-A11 (acc. no. NM 018914; 1839–2162 bp), which was found to be hypermethylated in astrocytomas of WHO grade II but not in normal brain tissues. A total of 57 astrocytomas and eight cell lines were included in the methylation analysis. To investigate the extent of de novo methylation within the first exon of the PCDH-γ-A11 gene and to validate the microarray data, bisulfite sequencing of three astrocytomas of WHO grade II (7282, 11092, and 12020), one GBM sample (WHO grade IV; 4732), and two normal brain samples (white matter 12768 and grey matter 12434) was conducted. Figure 1 A shows representative sequencing profiles of CpG sites 6 to 14 within the region analyzed in astrocytoma sample 7282 and cortical brain sample 12434 (acc. no. NM 018914; 2010–2321 bp). The tumor exhibits complete methylation of all the studied CpG sites, whereas the normal brain showed a complete conversion of these CpG sites, indicating a complete lack of methylation in this region. Comparison of the methylation pattern of the 26 CpG sites within the 312-bp bisulfite PCR fragment clearly showed significant accumulation of de novo methylation in the selected tumor samples. However, moderate methylation in a few alleles was also apparent in normal brain tissues (Figure 1 B). The overall percent methylation was 89% in the tumor samples and 23% in the normal samples. The three BstUI restriction sites flanked by the primer sequences were found to be frequently hypermethylated in the tumor samples in comparison to normal brain and thus was an ideal region to employ the COBRA technology to scan the methylation status of PCDH-γ-A11 methylation in an expanded set of tumor samples. De novo methylation of these BstUI sites identified by restriction of the 312-bp bisulfite PCR product was found in 18 of 20 (90%) astrocytomas of WHO grade II, in 12 of 14 (86%) anaplastic astrocytomas of WHO grade III, in 20 of 23 (87%) glioblastomas of WHO grade IV, and in 8 of 8 (100%) glioma cell lines. Much less hypermethylation was observed in the 10 normal brain samples (two grey matter and eight white matter samples) at these restriction sites. In Figure 1 C, representative data of the methylation analysis by restriction digest are illustrated for 15 tumor samples and three white matter controls (NB 1–3). Tumor samples displayed high levels of restriction fragments (indicating the presence of methylated alleles), whereas little to no restriction fragments were found in normal brain tissues as well as in astrocytomas AIII 2446, 10414, and GBM 11637 (indicating the presence of little methylated alleles). These samples were therefore scored as unmethylated. The undigested PCDH-γ-A11 band in the BstUI-treated samples is likely contributed by residual normal brain tissue (nontumoral cells in the sample, e.g., endothelial cells), or may reflect the heterogenous nature of PCDH-γ-A11 methylation among the tumor cells. However, the two assays employed in this study both point to the unique frequent hypermethylation found in the first exon of PCDH-γ-A11 in grades II and III astrocytomas and not in normal brain tissue.
Figure 1.
Methylation analysis of a CpG island within the first exon of the PCDH-γ-A11 gene. (A) Bisulfite sequencing profile of a WHO grade II astrocytoma (7282) and normal brain (grey matter 12434). Methylated CpG sites 6 to 14 are indicated by arrows. Underlined are three BstUI restriction sites that are methylated and therefore conserved during bisulfite treatment in this tumor sample and are used for the restriction-based methylation assay. (B) Bisulfite sequencing analysis of CpG sites 1 to 26 of individual clones of the 312-bp PCDH-γ-A11 PCR product of four tumor samples (AII 7282, AII 11092, AII 12020, and GBM 4732) and two normal brain samples (white matter 12768, grey matter 12434). (▪) Methylated CpG site; (□) unmethylated CpG site. (C) Restriction-based methylation assay of the first exon of PCDH-γ-A11 in astrocytomas. The 312-bp bisulfite PCR products without (-) and after treatment (+) with BstUI restriction enzyme are shown. Restricted fragments of the PCR product are only apparent in tumor samples and not in the normal brain tissues (NB1, white matter 12284; NB2, white matter 12692; NB3, white matter 12768). Tumor samples AIII 2446, AIII 10414, and GBM 11637 also show no methylation in this assay.
PCDH-γ-A11 Expression Analysis in Glioma Tissues and 5-Aza-2′-Deoxycytidine-Treated Glioma Cell Lines
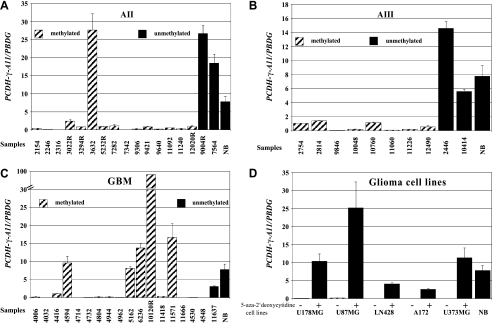
A cRT-PCR approach with specific RNA competitor molecules for PCDH-γ-A11 and the housekeeping gene PBGD was used to determine the expression level of PCDH-γ-A11 in 17 astrocytomas of WHO grade II, 10 anaplastic astrocytomas of WHO grade III, 19 glioblastomas of WHO grade IV, five glioma cell lines, and five normal brain samples (two white matter and three grey matter specimens) of which RNA was available. Relative PCDH-γ-A11 mRNA levels have been determined by correlation to PBGD transcript levels (Table 1). Fourteen of 17 (82%) astrocytomas of WHO grade II, 8 of 10 (80%) anaplastic astrocytomas of WHO grade III, and 14 of 19 (74%) glioblastomas of WHO grade IV exhibited no or low expression of PCDH-γ-A11 compared to the mean of all five normal brain samples (Table 1 and Figure 2). Expression profiles with standard error are shown in Figure 2, A–C. Tumor samples with PCDH-γ-A11 methylation are indicated by dashed columns, whereas tumor and control (NB) tissues with no methylation are represented by black columns. It is interesting to note that few GBM samples, especially the recurrent case 10120, showed dramatically elevated transcription of PCDH-γ-A11 even in the presence of methylated PCDH-γ-A11 alleles. In all analyzed glioma cell lines (U178MG, U87MG, LN428, A172, and U373MG), no or low PCDH-γ-A11 expression was observed. After treatment of the cells with 0.5 or 1 µM 5-aza-2′-deoxycytidine, a significant increase of the PCDH-γ-A11 transcript was detected (Figure 2 D).
Figure 2.
Relative transcript levels of PCDH-γ-A11 analyzed by cRT-PCR in (A) astrocytomas of WHO grade II, (B) anaplastic astrocytomas of WHO grade III, (C) glioblastomas of WHO grade IV, and (D) glioma cell lines with (+) and without (-) 5′-aza-2-deoxycytidine treatment. NB, normal brain (mean of two white matter and three grey matter samples).
Correlation of Methylation Status with Transcriptional Activity of PCDH-γ-A11
To determine if CpG island methylation affects the expression of PCDH-γ-A11, we correlated the methylation status of 27 astrocytomas (WHO grades II and III) and 18 glioblastomas (WHO grade IV) with their transcript levels of PCDH-γ-A11. Of the 23 WHO grades II and III cases exhibiting hypermethylation at the first exon of PCDH-γ-A11, 20 (87%) showed low expression, 2 (9%) showed no expression, and 1 tumor sample (AII 3632, 4%) had high PCDH-γ-A11 expression levels compared to normal brain. Conversely, of the four cases where the first exon of PCDH-γ-A11 was unmethylated, all (100%) showed expression levels comparable to the control samples where PCDH-γ-A11 was also unmethylated (Table 1 and Figure 2). Statistical analysis of these data revealed a significant inverse relationship between PCDH-γ-A11 methylation and expression (Fisher's exact test; P = .00028) in astrocytomas of WHO grades II and III. However, such relationship was not observed in the glioblastoma samples analyzed in this study. From 15 glioblastoma cases with PCDH-γ-A11 hypermethylation, eight (53%) revealed low expression, two (13%) revealed no expression, and five (33%) exhibited significant PCDH-γ-A11 transcription when compared to normal brain samples. Three glioblastoma samples with unmethylated alleles showed no or low expression levels.
Discussion
Structural changes in the genome of gliomas have been studied over the last years and a series of candidate genes has been identified [2]. A critical evaluation of the current data, however, shows that most of these alterations are restricted to advanced stages of these neoplasms (e.g., glioblastoma multiforme, WHO grade IV). Recently, there is increasing evidence that altered methylation patterns of tumor-associated genes are already present in the early stages of astrocytic tumors such as astrocytomas of WHO grade II and oligoastrocytomas of WHO grade II [13,21]. In an attempt to identify new candidate genes that might be involved in the early development of astrocytic gliomas, we performed a microarray-based methylation analysis of WHO grade II astrocytomas. These experiments yielded a DNA sequence within the first exon of PCDH-γ-A11, which was methylated in astrocytomas of WHO grade II but not in normal brain samples. In this study, we investigated the methylation and expression status of PCDH-γ-A11, a member of the protocadherin-γ family, in an expanded series of astrocytomas of different malignancy grades, in glioma cell lines, and in normal brain samples. We detected extensive hypermethylation in tumor tissues in comparison to normal brain controls. Although this is the first time that PCDH-γ-A11 was found to be hypermethylated in tumor samples, previous studies identified CpG islands in each variable region of the protocadherin gene cluster, suggesting epigenetic modifications to be involved in the regulation of these genes [5]. Candidate gene approaches have provided evidence for a role of epigenetic modifications in the process of transcriptional silencing of the cadherin family members in neoplastic cells [22].
A significant correlation (Fisher's exact test, P = .00028) between hypermethylation of PCDH-γ-A11 and decreased expression was identified in astrocytomas of WHO grade II and III. This association was not evident in glioblastomas of WHO grade IV, a subset of gliomas known to be histologically and genetically heterogenous. Although 66% of the glioblastoma samples with methylated alleles showed reduced or lack of expression, in line with epigenetic silencing of gene transcripts, 33% showed high transcript levels of PCDH-γ-A11 in the presence of methylation. This indicates that in these cases, the methylation of the analyzed CpG island is not sufficient to silence PCDH-γ-A11 transcription and that other 5′ regions of the PCDH-γ-A11 gene might be involved in its regulation. Alternatively, the tumor might contain two populations of cells, some of which are methylated and do not express high levels of PCDH-γ-A11 transcript whereas others contain high levels of PCDH-γ-A11 transcript. It also cannot be excluded that the inhibitory effect of CpG methylation by chromatin condensation in this area is overcome by a downstream event (e.g., binding of transcriptional activators or lack of histone modification). Of all the three cases that showed no PCDH-γ-A11 transcription in the absence of methylation in the analyzed region, it is possible that the accessibility of PCDH-γ-A11 regulatory region or availability of specific transcription factors is somehow impeded, leading to its nonexpression. Even though low levels of methylation were found in normal brain samples, the extent of methylation is distinctly lower than that found in tumor tissues. A similar observation in other candidate genes was also reported by Rood et al. [23] and Hong et al. [24].
Members of the protocadherin-γ cluster are generally assumed to play a role in synaptic connections between neurons. The expression of PCDH-γ-A11 detected in normal brain tissues and some of the analyzed tumor samples indicates that these genes may also be involved in cell-cell interactions in the glial cell compartment. Epigenetic regulation of another member of the protocadherin gene family, cadherin-related neuronal receptors (CNR), has been reported by Tasic et al. [25] in human cell lines HEC-1-B and NT2/D1. The expression and methylation analysis of the PCDH-γ-A11 gene presented in this study provides evidence that transcription is indeed blocked by methylation as its expression can be reactivated by 5-aza-2′-deoxycytidine treatment. However, it remains unclear if this is also the mode for determining the specific set of procadherin-γ genes expressed in a given cell as postulated by Yagi [26].
Based on the possible function of the PCDH-γ-A11 gene product, it is tempting to assume that transcriptional inactivation of this gene may affect the adhesion of glial cells to neighboring cells, thus promoting invasive growth of gliomas. Protocadherin genes cluster on chromosome 5q, where nearly 60 family members are known to locate [5,6]. The investigation of the putative role of other protocadherin-γ family members in glial neoplasms will constitute an important aspect of future studies.
We conclude that hypermethylation of the first exon of PCDH-γ-A11 is an early event in astrocytoma development and results in a substantial decrease in gene expression. Because PCDH-γ-A11 functions as a cell-cell contact molecule, it may be involved in the infiltration properties of these neoplasms.
Acknowledgements
We thank Denise Ehrentraut for excellent technical assistance and Ulrich Klatt for photographic work.
Footnotes
This study was supported by grants from the Deutsche Forschungsgemeinschaft (SFB400C2 to T.P.; SFB400C7 to A.W.), the Lise-Meitner-Habilitation Program (A.W.), National Cancer Institute grants (CA-69065 and CA-86701), and a start-up fund from the Ohio State University Comprehensive Cancer Center (T.H.-M.H and P.S.Y). T. H.-M. Huang is a consultant of Epigenomics, Inc.
References
- 1.Kleihues P, Cavenee WK. Tumours of the Nervous System. Lyon, France: Oxford University Press; 2000. [Google Scholar]
- 2.von Deimling A, Fimmers R, Schmidt M, Bender B, Fassbender F, Nagel J, Jahnke R, Kaskel P, Duerr EM, Koopmann J, et al. Comprehensive allelotype and genetic analysis of 466 human nervous system tumors. J Neuropathol Exp Neurol. 2000;59:544–558. doi: 10.1093/jnen/59.6.544. [DOI] [PubMed] [Google Scholar]
- 3.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, Colman DR, Huang JK, Wang Y, Shapiro L, et al. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J Neurosci. 2003;23:5096–5104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–562. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 5.Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Su H, Bradley A. Molecular mechanisms governing PCDH-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002;16:1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyooka S, Toyooka KO, Harada K, Miyajima K, Makarla P, Sathyanarayana UG, Yin J, Sato F, Shivapurkar N, Meltzer SJ, et al. Aberrant methylation of the CDH13 (H-cadherin) promoter region in colorectal cancers and adenomas. Cancer Res. 2002;62:3382–3386. [PubMed] [Google Scholar]
- 8.Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis. 2002;23:1139–1148. doi: 10.1093/carcin/23.7.1139. [DOI] [PubMed] [Google Scholar]
- 9.Mareel M, Bracke M, Van Roy F. Cancer metastasis: negative regulation by an invasion-suppressor complex. Cancer Detect Prev. 1995;19:451–464. [PubMed] [Google Scholar]
- 10.Braungart E, Schumacher C, Hartmann E, Nekarda H, Becker KF, Hofler H, Atkinson MJ. Functional loss of E-cadherin and cadherin-11 alleles on chromosome 16q22 in colonic cancer. J Pathol. 1999;187:530–534. doi: 10.1002/(SICI)1096-9896(199904)187:5<530::AID-PATH293>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 11.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 12.Behrens J. The role of cell adhesion molecules in cancer invasion and metastasis. Breast Cancer Res Treat. 1993;24:175–184. doi: 10.1007/BF01833258. [DOI] [PubMed] [Google Scholar]
- 13.Costello JF, Plass C, Cavenee WK. Aberrant methylation of genes in low-grade astrocytomas. Brain Tumor Pathol. 2000;17:49–56. doi: 10.1007/BF02482735. [DOI] [PubMed] [Google Scholar]
- 14.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura M, Watanabe T, Klangby U, Asker C, Wiman K, Yonekawa Y, Kleihues P, Ohgaki H. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001;11:159–168. doi: 10.1111/j.1750-3639.2001.tb00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlmann K, Rohde K, Zeller C, Szymas J, Vogel S, Marczinek K, Thiel G, Nurnberg P, Laird PW. Distinct methylation profiles of glioma subtypes. Int J Cancer. 2003;106:52–59. doi: 10.1002/ijc.11175. [DOI] [PubMed] [Google Scholar]
- 17.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 18.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 19.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waha A, Watzka M, Koch A, Pietsch T, Przkora R, Peters N, Wiestler OD, von Deimling A. A rapid and sensitive protocol for competitive reverse transcriptase (cRT) PCR analysis of cellular genes. Brain Pathol. 1998;8:13–18. doi: 10.1111/j.1750-3639.1998.tb00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Gomez P, Bello MJ, Alonso ME, Arjona D, Lomas J, de Campos JM, Isla A, Rey JA. CpG island methylation status and mutation analysis of the RB1 gene essential promoter region and protein-binding pocket domain in nervous system tumours. Br J Cancer. 2003;88:109–114. doi: 10.1038/sj.bjc.6600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- 24.Hong C, Bollen AW, Costello JF. The contribution of genetic and epigenetic mechanisms to gene silencing in oligodendrogliomas. Cancer Res. 2003;63:7600–7605. [PubMed] [Google Scholar]
- 25.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 26.Yagi T. Diversity of the cadherin-related neuronal receptor/protocadherin family and possible DNA rearrangement in the brain. Genes Cells. 2003;8:1–8. doi: 10.1046/j.1365-2443.2003.00614.x. [DOI] [PubMed] [Google Scholar]