Abstract
The spatial and genetic unit of bacterial population structure is the clone. Surprisingly, very little is known about the spread of a clone (spatial distance between clonally related bacteria) and the relationship between spatial distance and genetic distance, especially at very short scale (microhabitat scale), where cell division takes place. Agrobacterium spp. Biovar 1 was chosen because it is a soil bacterial taxon easy to isolate. A total of 865 microsamples 500 μm in diameter were sampled with spatial coordinates in 1 cm3 of undisturbed soil. The 55 isolates obtained yielded 42 ribotypes, covering three genomic species based on amplified ribosomal DNA restriction analysis (ARDRA) of the intergenic spacer 16S-23S, seven of which contained two to six isolates. These clonemates (identical ARDRA patterns) could be found in the same microsample or 1 cm apart. The genetic diversity did not change with distance, indicating the same habitat variability across the cube. The mixing of ribotypes, as assessed by the spatial position of clonemates, corresponded to an overlapping of clones. Although the population probably was in a recession stage in the cube (103 agrobacteria g−1), a high genetic diversity was maintained. In two independent microsamples (500 μm in diameter) at the invasion stage, the average genetic diversity was at the same level as in the cube. Quantification of the microdiversity landscape will help to estimate the probability of encounter between bacteria under realistic natural conditions and to set appropriate sampling strategies for population genetic analysis.
Assessing the genetic structure of bacterial populations has been the subject of growing interest. Bacteria are naturally clonal due to binary fission; however, due to the occurrence of horizontal gene transfer, the genetic structure of populations is likely to range from strictly clonal (organized in a clone-like manner) to panmictic (12, 16, 17, 27). Clonal multiplication and mating take place at a short spatial scale in the soil matrix and, when repeated isolates from a strain are sampled, the starting point in space and time and also the further development of bacterial spread and growth are unknown (3). Many population genetic studies among conspecific bacteria have used pooled collections of strains from widely separated locations and different sampling times. Although the clone is the unit of the genetic structure of bacterial population, the spread of a clone and its spatial organization are not known. The knowledge of the microspatial distribution of population seems important for several reasons. Mutational drift can be revealed at the microscale. Competition between genotypes for resources also takes place at a small scale and could lead to individualization of ecotypes (2). Furthermore, the occurrence of gene transfer partly depends on encounters between cells, which is determined by the patterns of spatial distribution of bacteria. Low population densities or isolation by distance may impose a clonal structure on bacterial populations by reducing the rate at which different genotypes encounter one another for mating (14). Spatial analysis of genetic divergence among local populations has always played a central role in population genetics and evolutionary biology (4). It seems to be less so for bacteria at the microhabitat level, and very few works have thus far targeted the small-scale organization level in soil (6, 14, 19, 33).
Works published on the genetic structure of bacterial populations have pointed out difficulties such as sampling strategies and points of data analysis (12, 16, 17, 30). Spatial and temporal patterns of clonal propagation of bacteria are not observable prior to sampling, as is the case with plants and many animals. Thus, the results obtained from any sampling design for bacteria involve unknown interactions of the pattern and scale of sampling with natural scales of bacterial distribution (14). Clonality was rarely tested on samples with clear coordinates because data on bacterial distribution are scarce, particularly at a small scale (11). Thus, samples are not strictly population samples in the sense of a local deme, the setting most propitious for recombination. Analyses of isolates from volumetrically small samples from a single microsite provide the strongest test for natural recombination (14). It was also early recognized that conclusions regarding population structure sometimes depended on factors such as how one analyzed the genetic data, if all isolates or one representative of each strain (undistinguishable isolates) should be taken into account, and how to compare the population structures of species from diverse ecological niches (12, 16, 17), not to mention the scale of study (27).
The objective of the present study was to evaluate the spatial spread of clones of a bacterial taxon in 1 cm3 of soil. We focused our study at the microscale level because it has been demonstrated that diversity was larger than expected in 1 g of soil (32) and that population diversity was still present in a 50-μm-diameter portion of soil, as shown for Nitrobacter sp. (10). The bacterial model chosen here was the soil bacterium Agrobacterium biovar 1, which is known to contain several genomic species (22). This choice was based on the existence of a specific medium that has been shown to allow easy isolation and counting (21) and on the fact that agrobacteria have large rrs-rrl intergenic spacer (IGS) 16S-23S allowing for accurate ribotyping (24). According to Tenover et al. (28), isolates indistinguishable by such a method belong to the same strain in a taxonomic sense and are assumed to be clonally related (they are referred to as clonemates here). In the present study, the diversity from all isolates recovered from microsamples 500 μm in diameter with spatial coordinates in the 1-cm3 cube was studied. More precisely, we were interested (i) in examining whether genetic distance between isolates, based on ARDRA patterns, correlates with the spatial distance between isolation sites in 1 cm3 of soil and therefore (ii) in evaluating the spatial size of a clone (isolates with same ARDRA pattern) in soil.
MATERIALS AND METHODS
Soil sampling.
A clod of soil was sampled in an agricultural soil from La Côte Saint André (Isère, France) (silty loam, Alfisol) (9) planted for more than 10 years with corn. Water content and Agrobacterium biovar 1 abundance, as measured in the surrounding soil, were 9% (wt/wt) and 103 agrobacteria g−1 (CFU count), respectively. A cubic piece of soil (1-cm/side) was gently carved inside a cohesive clod and embedded in 2% agar (wt/vol) (Difco) to maintain its coherence. Thirty-two pieces (referred to here as subcubes [2.5 by 2.5 by 5 mm]) were then dissected as indicated in Fig. 1 and are spatially referenced in the cube. Each subcube was then further dissected into microsamples on a grid gauge (millimetric paper) under a binocular, and 24 to 40 microsamples (volumetric units [VUs]) fitting into the standard (0.5-mm side squares allowing us to spot 0.5-mm-diameter microsamples) were sampled arbitrarily (9). The spatial coordinates of the VUs were based on the subcube to which they belonged and not on their exact one. When possible, VUs that were juxtaposed in the undisturbed cube were marked. Maximum care was taken to avoid cross-contamination. Dissection of subcubes was carried out with a scalpel blade, which was sterilized after each use. An eventual risk of contamination occurred when we cut the initial cube from top to bottom to obtain parts 1 to 5, 2 to 6, 3 to 7, and 4 to 8. Subcubes a, b, c, and d never happened to be in contact during manipulations. Otherwise, each face of the blade was in contact with one subcube at a time. Then each of the 32 subcubes was dissected separately on a sterile gauge, and care was taken to ensure that each piece was dragged far enough from the initial spot to allow further dissection without moving previously dissected VUs, so as to avoid picking up bacteria eventually originating from another VU. The shapes of the VUs were very irregular, and points of possible contact with the gauge were very restricted. Very little loss of soil material to the dissecting surface was observed.
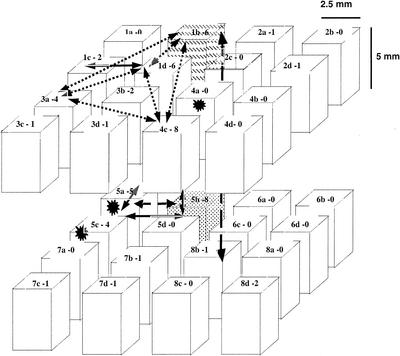
FIG. 1.
Spatial distribution of positive VUs in the 1-cm3 cube. The names of each subcube and the numbers of isolates obtained in the subcube are given as “name-number” at the top of each cube. The spatial spread of the clones (identical ribotype) is also indicated. Stars represent the positions of clonemates found in a single VU. The dashed subcubes contained clonemates. Otherwise, identical ribotypes are joined by arrows.
Each VU was transferred into an Eppendorf tube containing 500 μl of deionized sterile water and shaken for 1 h. The entire content of each Eppendorf tube was then poured onto 1ATe agar specific medium from Agrobacterium biovar 1 (21). Incubation was for 5 days at 28°C. All typical glistening black colonies with a metallic sheen were then suspended in 1 ml of water and streaked onto MG medium plates for purification and to check the identity as biovar 1 by testing the transformation of lactose to 3-keto-lactose (20). Isolates were transferred into 20% glycerol (vol/vol) and kept at −20°C.
An independent experiment was carried out with 40 VUs 500 μm in diameter taken in sieved soil (mesh, 4 mm) from the same cornfield sampled in June, whereas the cube was sampled in September. These samples were treated and analyzed as described above.
Diversity analysis.
Ribotyping was carried out with 1 μl of bacterial suspension in 20% glycerol (vol/vol) by PCR amplification with the primers F38 5′-CCGGGTTTCCCCATTCGG-3′ and F72 5′-TGCGGCTGGATCCCCTCCTT-3′ (24) targeting the rrs-rrl IGS, followed by restriction with the six enzymes HaeIII, HpaII, NdeII, RsaI, TaqI, and CfoI (Gibco). Genetic distances were calculated according to the method of Nei and Li (23) by using the DistAFLP software (http://pbil.univ-lyon1.fr/ADE-4/microb) and then analyzed by principal coordinates analysis (8) with ADE-4 software (29). ARDRA allows the delineation of genomic species of Agrobacterium biovar 1 based on the correlation between genetic distance given by ARDRA and the genomic distance used to delineate genomic species (21, 22). Representative strains of eight genomic species clustering with Agrobacterium biovar 1 (22, 25) were included in the present study: S56 (genomic species 1, G1), CIP28-75 (genomic species 2, G2), CIP111-78 (genomic species 3, G3), B6 (genomic species 4, G4), NCPPB925 (genomic species 6, G6), NCPPB1641 (genomic species 7, G7), C58 (genomic species 8, G8), and O363 (genomic species 9, G9).
Spatial coordinates.
The subcubes were 2.5 by 2.5 by 5 mm (Fig. 1). Spatial distances between VUs obtained from different subcubes were assumed to be the euclidian distances between subcube centers as follows: d = a1/3 + b1/3 + c1/3, with a, b, and c, being the distances between two subcube centers on the three-dimensional axis. Within subcubes, spatial distances are given as follows: a distance of 1.5 mm corresponded to isolates from separated VUs, a distance of 0.5 mm corresponded to isolates coming from juxtaposed VUs, and a distance of 0.25 mm corresponded to isolates coming from the same VU.
RESULTS
Spatial organization of isolates in the cube.
A total of 865 VUs representing 5% of the VUs contained in the initial 1 cm3 have been explored. A total of 55 agrobacterium-like isolates have been obtained, representing ca. 5% of the culturable agrobacteria expected in the cube. Of 865 VUs, 42 yielded agrobacterial isolates, showing that only 5% of the soil microunits tested were colonized by culturable agrobacteria. The number of isolates obtained per VU varied from 0 to 6, but most positive VUs yielded only one agrobacterial isolate (Table 1). The positive VUs came from 18 subcubes out of the 32 subcubes (2.5 by 2.5 by 5 mm) forming the cube (Fig. 1). Spatial distances between isolates in the cube ranged from 0 mm (when they came from the same VU) to 10.3 mm, and both short and long distances were represented (data not shown).
TABLE 1.
Distribution of isolates among the 32 subcubes and the 856 VUs
| No. of isolates | No. of
|
|
|---|---|---|
| Subcubes (n = 32) | VUs (n = 856) | |
| 0 | 14 | 823 |
| 1 | 8 | 35 |
| 2 | 3 | 4 |
| 3 | 0 | 2 |
| 4 | 2 | 0 |
| 5 | 1 | 0 |
| 6 | 2 | 1 |
| 7 | 0 | 0 |
| 8 | 2 | 0 |
In an independent experiment carried out on 40 VUs from sieved soil, 8 contained agrobacteria. A total of 13 agrobacterium isolates were obtained from the spreading of one VU (VU A), 31 were obtained from another VU (VU B), and 34 were obtained from another VU (VU C). The five other VUs had one or two agrobacterium isolates. VUs B and C only were studied further for diversity.
Ribotype diversity.
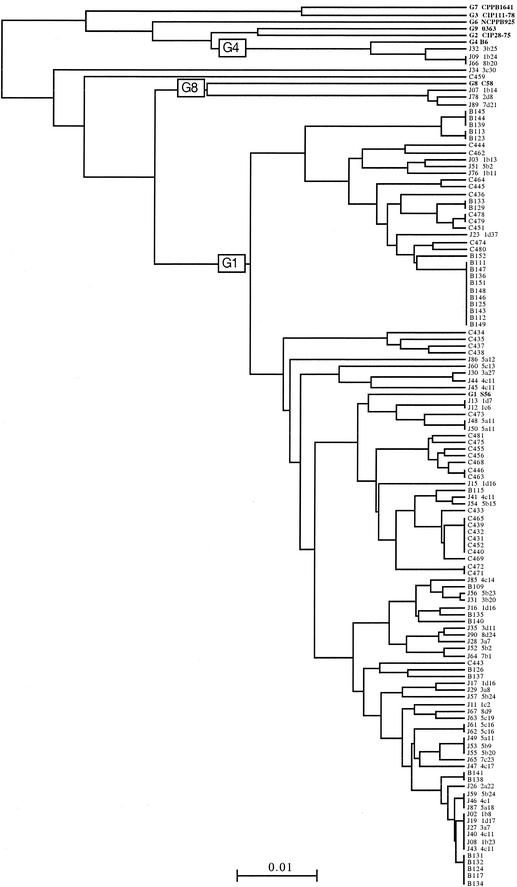
All agrobacterium-like isolates were found to be bona fide biovar 1 agrobacteria according to the 3-ketolactose test and according to the phylogeny based on ARDRA analysis. Among the 55 agrobacterium isolates obtained in the 1-cm3 cube, 42 ribotypes could be distinguished. VU B yielded 12 ribotypes among 31 isolates, and VU C yielded 25 ribotypes among 34 isolates. A dendrogram of ribotype relatedness between isolates and reference strains was constructed (Fig. 2) that showed diversity at the species and infraspecific level. Three genomic species were identified: genomic species G1, G4, and G8. In subcube 1b (Fig. 1), one isolate belonged to genomic species G8 (1b14), another to genomic species G4 (1b24), and four others to genomic species G1 (1b8, 1b11, 1b13, and 1b23). This latter genomic species was the most common encountered in the cube and in the two other individual VUs studied, VUs B and C. Clonemates (i.e., isolates with identical IGSs) were observed within the 1-cm3 cube of soil, as well as within VUs B and C (Fig. 2). In the 1-cm3 cube the most common clone was found at a frequency of 11%; it was found at frequencies of 32% in VU B and 18% in VU C. Most strains were represented by 1 isolate in the cube, whereas VUs B and C yielded one major clone with 10 and 6 isolates, respectively, and other clones with smaller numbers of isolates. The overall estimated frequencies of clonemates (i.e., the total number of isolates with clonemates/total number of isolates, as defined by Istock et al. [14]) were 36% in the cube, 65% in VU B, and 35% in VU C.
FIG. 2.
Dendrogram of genetic distances between ribotypes. Isolates with the letter “B” or “C” belong to VU B or VU C, respectively. Isolates from the cube are indicated by the letter “J.” G1 to G9 indicate representative strains of genomic species as determined by Popoff et al. (25) in biovar 1.
Spatial organization of ribotype diversity.
We first studied the spatial spread of clones. Seven ribotypes comprising several clonemates (from 2 to 6) were found within different VUs in the cube. The spatial spread of the clonemates delineates the space occupied by a clone in the cube (Fig. 1). Clonemates could be found as coisolates in a same VU (J40-4c11 and J43-4c11, J48-5a11 and J50-5a11), as well as up to 10.3 mm apart (1b24 and 8b20). The spatial distances for each of the seven clones were 0.25, 0.5, 2.5, 1.5 to 2.5, 0.25 to 7.9, 2.5 to 10.3, and 7.5 mm.
We then tried to find a spatial organization of any genetic structure within the most abundant genomic species, G1. A principal coordinate analysis using the genetic distances between all G1 ribotypes pointed to one genetic structure representing 9% of the total genetic variance (data not shown). This structure was mainly due to two groups of isolates that consisted of three and five isolates, respectively. Isolates from each group were very closely related (ranging from 0.010 to 0.016 and 0.000 to 0.032 nucleotide substitutions per site [nss−1], respectively). The average genetic distance between these groups was among the largest found in this population, ranging from 0.073 to 0.108 nss−1. One was represented by isolates J76, J51, and J03 from subcubes 1b and 5b; the other was represented by isolates J12, J13, J15, J48, and J50 from subcubes 1c, 1d, and 5a. Thus, as with the clones, these two groups of closely related isolates were found to be spread among different subcubes. Isolates from the different groups were not found in the same subcubes. Nevertheless, the spatial distances between isolates from different groups were not different from the spatial distances between isolates of the same group (Fig. 1).
Finally, the spatial organization of the overall genetic diversity of the most abundant species, G1, was studied at the cube level by plotting the genetic distances and the spatial distances for each isolate versus all of the others (all belonging to genomic species G1). None of the plots showed a significant relationship between genetic and spatial distances (data not shown). Moreover, the genetic diversity of genomic species G1 inside the cubes was found to be very similar to those observed in two independently sampled VUs, B and C. The mean genetic distances were 0.037 ± 0.031 nss−1 inside the cube, 0.032 ± 0.020 nss−1 in VU B, and 0.045 ± 0.028 nss−1 in VU C. Thus, there was also no evident relationship between spatial distances and genetic distances within a species at the VU level, the cube level, and probably at the larger scale. It should be noted that the diversity within VUs could not be reliably determined in the cube itself because the numbers of isolates from the same VU were very low.
DISCUSSION
Assessing the spatial and genetic structure of populations and gene dissemination in soil is still challenging. Inferences on the genetic structure of isolate populations from collections have often been impeded by the fact that isolates had no clear spatial reference. In studies of different bacterial types in bulk soil, i.e., Nitrobacter-like bacteria (10), Bacillus subtilis (14), Bacillus thuringiensis, Bacillus cereus (33), putative clonemates were isolated from relatively distant locations, from centimeters to kilometers. The spread of clones was nevertheless not specifically studied. Since the clone is a conspicuously important genetic unit of the bacterial population structure, knowledge of its spatial distribution should lead to the definition of the spatial unit of population structure. Our results provide evidence of the spread of clones and data about the spatial organization of genetic diversity on a model soil taxon, Agrobacterium spp., by intensively sampling 1 cm3 of undisturbed soil and exhaustively exploring each sampled VU.
Spatial description validity.
The experiment reported here is pioneering in its scale and in the large number of microsamples exhaustively explored (n = 865), which permits a sound description of an undisturbed soil cube. The volume ratio of the size of a bacterium (1 μm3) to the 1 cm3 explored is 1012. Thus, the explored volume of soil cannot be considered negligible and looks reasonable in order to obtain a good description of the bacterial microlandscape. Arguments can be raised about the reproducibility in the case of this type of study in soil. Seven clones were detected, five being obviously spread in different VUs, covering ca. 1 cm. There were thus seven different repeats of the same type of spatial distribution. Owing to the care taken during sampling and to the low population level, this was probably not due to incidental cross-contamination. It is clear, however, that the exploration of another cube would not give the exact same spatial distribution nor even the same ribotypes. However, new evidence concerning the spread of clonemates and the mixing of genotypes cannot be discarded.
Fragmented habitat and ribotype diversity.
Our results provide data about bacterial microlandscape showing the potential isolation between bacteria from the same species. Concretely, the distance between cells can be estimated for 103 bacteria randomly distributed in the soil cube to be ca. 1 mm. This distance is likely to be underestimated since a patchy pattern of isolate distribution has been demonstrated with various soil bacteria (10, 11) and might also occur with agrobacteria.
On the other hand, the present study points to an important genetic diversity even at the smallest scale. This study is in agreement with the diversity observed at the small scale (500 50-μm microsamples) with nitrifiers (10), as well as with the fact that at least 30 different 2,4-dichlorophenoxyacetic acid-degrading populations occurred in the original 1 g of a cultivated soil (5). Moreover, in the present study, the high genetic diversity did not change with distance (within 1 cm), and there were instances in which it did not change with sample sizes ranging from 500-μm VUs to a 1-cm3 cube. In a work on Burkholderia cepacia and Burkholderia picketti carried out in soil and sediments, the bacterial genetic diversity varied according to habitat variability (18, 19). In addition, the role of a structured environment in bacterial population structure was experimentally demonstrated in Pseudomonas populations (26): the total genetic diversity with respect to colony morphology was clearly greater in the structured (nonshaken culture) than in the nonstructured habitat (shaken culture). More generally, as shown for other organisms (31), it is highly probable that habitat fragmentation caused by lowering competition helps to maintain diversity. Based on the conclusions of these earlier studies, the results presented here indicate that there is the same habitat variability across the cube and down to 500 μm. Since several strains could coexist in the same VU, this could indicate that VUs contained smaller ecologically relevant domains (i.e., niches). This would in turn suggest that hypothetical telluric parameters relevant to the occurrence of such a high genetic diversity would intervene at a very short scale. Soil texture and soil microstructure, for example, span the same microspatial scale. Seemingly, pores with diameters of <6 μm have been recognized as being protective microhabitats (13).
High diversity at a small scale and size of a clone.
Unexpectedly, we found that at a small scale in soil, agrobacterial clones were spread and did not form tight microcolonies. The spatial spread of clones resulting from processes, such as active and passive transport (including the influence of water), that have not yet been specifically investigated can be hypothesized. Large distances between identical cells and high diversity at the same locations can be reconciled as results of the same process. There are population density increases followed by cell death, in other words, cycles of local invasion and recess, that spread the cells far from their initial parent cell (7, 23) and which, as a consequence, modify local diversity by shuffling clones. Our results suggest that a clone can spread at a distance of at least 1 cm, which is obviously an underestimation since we only explored a volume of 1 cm3. The distance between cells of a given clone could be the result of clonal spreads or relicts of former colonization. The latter hypothesis is the most probable in the present case since the sampling occurred in September, a season that corresponds to a phase of recess for agrobacteria (15). Moreover, the fact that the abundance was low and that only a few identical cells were found in a VU adds weight to this hypothesis.
Cells obviously did not survive (or did not remain culturable) in the form of a dense clone (microcolony) in the cube. Interestingly, the observed diversity in the cube was nevertheless high, as if most clones had one surviving cell left. This would agree with the theory that states that, in a clonal model, it is the clone that persists genetically (30): the size of each clone is affected and not the diversity. In the two independent VUs studied, VUs B and C, on the contrary, the presence of clonemates for several ribotypes and the high agrobacterium abundance would indicate that the population was in a period of local invasion. This spatial configuration is probably most propitious for mating. A sampling strategy based on exhaustive exploration of a microsample allowed us to compare diversity between samples, thus avoiding the problem of dominant strains as with classical suspension dilution methods, which hides the presence of single and/or rare genotypes.
Diversity organization and sampling strategy.
Distances covered by bacteria can be quite large, although it is difficult to imagine that transport through strict telluric was could concern the scale of continents. Genetic divergence may increase with geographic distance as observed by Cho and Tiedje (1), both because environmental variations (and associated selective effects) become more heterogeneous with large distances and because the low migration rates do not constrain divergence by random drift. However, the probability of finding two isolates of the same strain is low because clones are quite rare and because the genetic diversity is great. As a consequence, sampling strategies could obscure any conclusion on clonality.
The structure of the population at a given site at a single time point is the result of the integration of all migration, clonal reproduction, and mating that occurred during all of the time preceding the sampling (34). Since soil is a highly structured environment, the importance of connectivity in the matrix to allow encounters between bacteria points out the importance of soil physics knowledge in understanding soil microbiological functioning. We believe that studying the spatial genetic structure of a bacterial type at different scales, from the microhabitat to the continental scale, and then obtaining diversity data from intensive sampling at the short scale in soil will help evaluate the processes generating and maintaining diversity and thus to understand gene dissemination in the natural environment.
Acknowledgments
This work was supported by a grant from the MRNT-ACI Ecologie Quantitative.
REFERENCES
- 1.Cho, J. C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 3.Dijkshoorn, L., B. M. Ursing, and J. B. Ursing. 2000. Strain, clone, and species: comments on three basic concepts of bacteriology. J. Med. Microbiol. 49:397-401. [DOI] [PubMed] [Google Scholar]
- 4.Diniz-Filho, J. A. F., and M. Pires de Campos Telles. 2000. Spatial pattern and genetic diversity estimates are linked in stochastic models of population differentiation. Genet. Mol. Biol. 23:541-544. [Google Scholar]
- 5.Dunbar, J., S. White, and L. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felske, A., and A. D. L. Akkermans. 1998. Spatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soils. Microb. Ecol. 36:31-36. [DOI] [PubMed] [Google Scholar]
- 7.Franck, S. A., and P. Amarasekare. 1998. Increasing resource specialization among competitors shifts control of diversity from local to spatial processes. Ecol. Lett. 1:3-12. [Google Scholar]
- 8.Gower, J. C. 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325-338. [Google Scholar]
- 9.Grundmann, G. L., A. Dechesne, F. Bartoli, J. L. Chassé, J. P. Flandrois, and R. Kizungu. 2001. Spatial modeling of nitrifier micro habitats in soil. Soil Sci. Soc. Am. J. 65:1709-1716. [Google Scholar]
- 10.Grundmann, G. L., and P. Normand. 2000. Microscale diversity of the genus Nitrobacter in soil on the basis of analysis of genes encoding rRNA. Appl. Environ. Microbiol. 66:4543-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hattori, T. 1988. Soil aggregates as microhabitats of microorganisms. Institute of Agricultural Research, Tohoku University, Tohoku, Japan.
- 12.Haubold, B., M. Travisano, P. B. Rainey, and R. R. Hudson. 1998. Detecting linkage disequilibrium in bacterial populations. Genetics 150:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heijnen, C. E., and J. A. Van Veen. 1991. A determination of protective microhabitats for bacteria introduced into soil. FEMS Microbiol. Ecol. 85:73-80. [Google Scholar]
- 14.Istock, C. A., K. E. Duncan, N. Ferguson, and X. Zhou. 1992. Sexuality in a natural population of bacteria: Bacillus subtilis challenges the clonal paradigm. Mol. Ecol. 1:95-103. [DOI] [PubMed] [Google Scholar]
- 15.Krimi, Z., A. Petit, C. Mougel, Y. Dessaux, and X. Nesme. 2002. Seasonal fluctuations and long-term persistence of pathogenic populations of Agrobacterium spp. in soils. Appl. Environ. Microbiol. 68:3358-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenski, R. E. 1993. Assessing the genetic structure of microbial populations. Proc. Natl. Acad. Sci. USA 90:4334-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maynard-Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McArthur, J. V., D. A. Kovacic, and M. H. Smith. 1988. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc. Natl. Acad. Sci. USA 85:9621-9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McArthur, J. V., L. G. Leff, and M. H. Smith. 1992. Genetic diversity of bacteria along a stream continuum. J. N. Am. Benthol. Soc. 11:269-277. [Google Scholar]
- 20.Moore, L. W., C. I. Kado, and H. Bouzar. 1988. Agrobacterium, p. 16-36. In N. W. Shaad (ed.), Laboratory guide for identification of plant pathogenic bacteria, 2nd ed. APS Press, St. Paul, Minn.
- 21.Mougel, C., B. Cournoyer, and X. Nesme. 2001. Novel tellurite-amended media and specific chromosomal and Ti plasmid probes for direct analysis of soil populations of Agrobacterium biovars 1 and 2. Appl. Environ. Microbiol. 67:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mougel, C., J. Thioulouse, G. Perriere, and X. Nesme. 2002. A mathematical method for determining genome divergence and species delineation using AFLP. Int. J. Syst. E vol. Microbiol. 52:573-586. [DOI] [PubMed] [Google Scholar]
- 23.Nei, M., and W.-H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponsonnet, C., and X. Nesme. 1994. Identification of Agrobacterium strains by PCR-RFLP analysis of pTi and chromosomal regions. Arch. Microbiol. 161:300-309. [DOI] [PubMed] [Google Scholar]
- 25.Popoff, M. Y., K. Kersters, M. Kiredjian, I. Miras, and C. Coynault. 1984. Position taxonomique de souches de Agrobacterium d'origine hospitalière. Ann. Microbiol. 135A:427-442. [PubMed] [Google Scholar]
- 26.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 27.Souza, V., L. Eguiarte, G. Avila, R. Cappello, C. Gallardo, J. Montoya, and D. Pinero. 1994. Genetic structure of Rhizobium etli biovar phaseoli associated with wild and cultivated bean plants (Phaseolus vulgaris and Phaseolus coccineus) in Morelos, Mexico. Appl. Environ. Microbiol. 60:1260-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thioulouse, J., D. Chessel, S. Dolédec, and J. M. Oliver. 1997. ADE-4: a multivariate analysis and graphical display software. Statistics Comput. 7:75-83. [Google Scholar]
- 30.Tibayrenc, M., F. Kjellberg, J. Arnaud, B. Oury, S. F. Breniere, M. L. Darde, and F. J. Ayala. 1991. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc. Natl. Acad. Sci. USA 88:5129-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tilmann, D. 1994. Competition and biodiversity in spatially structured habitats. Ecology 75:2-16. [Google Scholar]
- 32.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vilas-Boas, G., V. Sanchis, D. Lereclus, M. V. Lemos, and D. Bourguet. 2002. Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 68:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wise, M. G., L. J. Shimkets, and J. V. McArthur. 1995. Genetic structure of a lotic population of Burkolderia (Pseudomonas) cepacia. Appl. Environ. Microbiol. 61:1791-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]