Abstract
It has been proposed that human colorectal tumors can be classified into two groups: one in which methylation is rare, and another with methylation of several loci associated with a “CpG island methylated phenotype (CIMP),” characterized by preferential proximal location in the colon, but otherwise poorly defined. There is considerable overlap between this putative methylator phenotype and the well-known mutator phenotype associated with microsatellite instability (MSI). We have examined hypermethylation of the promoter region of five genes (DAPK, MGMT, hMLH1, p16INK4a, and p14ARF) in 106 primary colorectal cancers. A graph depicting the frequency of methylated loci in the series of tumors showed a continuous, monotonically decreasing distribution quite different from the previously claimed discontinuity. We observed a significant association between the presence of three or more methylated loci and the proximal location of the tumors. However, if we remove from analysis the tumors with hMLH1 methylation or those with MSI, the significance vanishes, suggesting that the association between multiple methylations and proximal location was indirect due to the correlation with MSI. Thus, our data do not support the independent existence of the so-called methylator phenotype and suggest that it rather may represent a statistical artifact caused by confounding of associations.
Keywords: CpG methylation, phenotype, colorectal, cancer, microsatellite instability
Introduction
Two different major pathogenetic mechanisms have been proposed for the development of colorectal cancers (CRCs) [1]. The first, so-called “classic pathway,” seems to be the most common and depends on multiple additive mutational events (germline and/or somatic) in tumor-suppressor genes and oncogenes, frequently involving chromosomal deletions in key genomic regions [2]. However, the “mutator pathway,” operationally recognizable by the presence of microsatellite instability (MSI), depends on early mutational loss of the mismatch repair system (germline and/or somatic), leading to accelerated accumulation of gene mutations in critical target genes and progression to malignancy. The distinction between these pathways seems to be more than academic because there is evidence that the tumors emerging from the mutator pathway have a specific “mutator phenotype” that includes preferential localization in the right colon, undifferentiated histology, lymphocyte infiltration, a better prognosis, and resistance to adjuvant therapy with 5-fluorouracil [3–5].
Recently, it has been discovered that in either pathogenetic pathways, loss of activity of key genes may occur through epigenetic, rather than genetic, means [6]. Indeed, although lack of expression of mismatch repair genes is generally found in sporadic tumors with MSI, the majority of such tumors does not show mutations in these DNA repair genes [7–9]. In fact, methylation of hMLH1 is the single most common recognizable form of MSI in sporadic colorectal tumors [10]. Recent work has shown that loss of tumor-suppressor and/or DNA repair gene function by promoter methylation can occur in many different genes in sporadic CRCs [11]. In 1999, Toyota et al. studied human CRC with a technique which they called “methylated CpG island amplification” and observed that tumors could be classified in two very distinct groups: one with simultaneous methylation of several loci, and another in which methylation of these loci was very rare. Moreover, they noted that a large proportion of proximal tumors belonged to the former group, and they proposed the existence of a “CpG island methylated phenotype (CIMP).” They also observed that CIMP+ tumors often also exhibited hMLH1 methylation and MSI, and they did remark that MSI was also correlated with proximal tumors but failed to point out that this chain of associations might lead to confounding of variables [12].
We have studied promoter methylation of the tumor-suppressor genes p16INK4a and p14ARF, the apoptosis-associated gene death-associated protein kinase (DAPK), and the DNA repair genes hMLH1 and O6-methylguanine-DNA-methyltransferase (MGMT), and also analyzed MSI in 106 human colorectal adenocarcinomas. We did not find any discontinuities in the distribution of the number of methylated genes in CRC. Moreover, after we removed tumors that had MSI from the statistical analysis, there was no longer a significant association between multiple methylated loci and proximal tumor location. Thus, our data do not support the existence of the so-called methylator phenotype and suggest that it rather represents a statistical artifact caused by confounding of associations.
Materials and Methods
Sample Collection and Nucleic Acid Isolation
Primary tumor samples from 106 patients diagnosed with CRC were collected at the A. C. Camargo Cancer Hospital (São Paulo, Brazil). Informed consent was obtained from all patients, and this research was approved by the Research Ethics Committee of the A. C. Camargo Hospital and the Ludwig Institute for Cancer Research (São Paulo, Brazil). In 30 of the patients, we also obtained matching normal colon tissue. To avoid selection bias, the samples were collected on sequential surgical cases of CRC. Immediately after collection, the samples were snap-frozen in liquid nitrogen and kept as part of a tumor bank. For this study, H&E-stained sections from each tumor sample were histologically examined, and only those that were microdissected to contain more than 70% neoplastic cells were used for analysis. DNA was prepared from microdissected tissue by digestion with pronase in 1% SDS, followed by standard phenol-chloroform extraction and ethanol precipitation [13]. In all patients, we obtained medical information on the nature of the cancer, patient sex and age, tumor location, histologic features, and clinical evolution.
Bisulfite Treatment and Methylation-Specific PCR (MSP)
MSP is based on the chemical modification of genomic DNA with sodium bisulfite, which converts unmethylated cytosines (but not methylated cytosines) to uracil. Specific primers are then designed to distinguish between the sequence differences produced with methylated and unmethylated DNA in MSP [14]. We studied the methylation status of the following loci: DAPK, MGMT, hMLH1, p16INK4a, and p14ARF. Briefly, 1 µg of genomic DNA was denatured with NaOH (final concentration, 0.2 M), and 10 mM 6-hydroquinone (Sigma, St. Louis, MO). Sodium bisulfite, pH 5.0 (Sigma), was added to a final concentration of 3 M and the mixture was incubated at 50°C for 16 hours. The modified DNA was then purified using the Wizard DNA purification kit (Promega Corporation, Madison, WI), followed by precipitation with ethanol.
The primers and thermal cycle conditions for MSP of DAPK, MGMT, hMLH1, p16INK4a, and p14ARF were as detailed elsewhere [14–17]. The PCR mixture contained bisulfite-modified DNA, specific primers (final concentration, 0.6 µM each per reaction), 1 U of Taq polymerase (Phoneutria, Belo Horizonte, Brazil), and deoxynucleotide triphosphates (1.25 mM) in 1% Triton X-100, 500 mM KCl, 15 mM MgCl2, and 100 mM Tris-HCl (pH 8.4). Reactions were maintained at 950°C for 5 minutes before the addition of polymerase. Amplification was carried out using a PTC100 MJ Research, Inc. Thermal Cycler (Watertown, MA). About 10 µl of the amplified products was electrophoresed on 6% acrylamide gels and visualized by silver staining.
MSI
The Bethesda consensus panel, composed of two mononucleotide repeat microsatellites (BAT25 and BAT26) and three dinucleotide repeat microsatellites (D2S123, D5S346, and D17S250), was used to evaluate MSI [18]. The mononucleotide microsatellite BAT-26, which is part of the panel, has been reported to have close to 100% sensitivity and specificity as a marker of this phenomenon [19,20]. As an additional criterion for MSI, we also utilized a battery of nine tetranucleotide microsatellite loci and one trinucleotide microsatellite [21].
Statistical Analysis
The 2 x 2 cross-categorized frequency data were tested by Fisher's exact test using the online facility at http://faculty.vassar.edu/lowry/VassarStats.html. A probability value of < .05 was considered significant, and we applied the Bonferroni correction for multiple comparisons [22].
Results
Frequency of Methylation in Primary Colorectal Tumors and Corresponding Nonmalignant Tissues
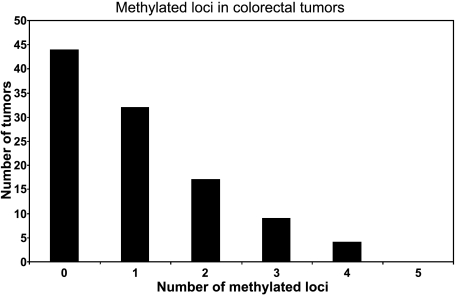
We used MSP to determine the frequency of methylation of DAPK, MGMT, hMLH1, p16INK4a, and p14ARF in 106 microdissected primary CRCs. These loci were chosen because they are among the most frequently methylated in CRC [23]. No aberrant methylation of any of these loci was detected in 30 samples of nonmalignant colon tissues. However, a total of 109 methylation events was detected in 106 tumors. In addition, the unmethylated form of all genes was detected in 100% of samples in both tumors and nonmalignant tissues. This was not unexpected because, inevitably, all tumor specimens contain a small proportion of normal cells. Moreover, some loci may be heterozygous for methylation and thus possess a nonmethylated allele. As shown in Figure 1, the most frequently methylated locus was MGMT (32/109; 29.4%), followed by DAPK (21/109; 19.3%), p16INK4a (20/109; 18.3%), hMLH1 (19/109; 17.4%), and p14ARF (17/109; 15.6%). We identified at least one methylated promoter region in 58.5% (62/106) of the tumors (Figure 1). Overall, 41.5% (44/106) of the tumors had no methylated genes, 30.2% (32/106) had only one methylated gene, 16.0% (17/106) had two methylated genes, 8.5% (9/106) had three methylated genes, and 4% (4/106) had four methylated genes (Figure 2). We checked all loci for pairwise association using Fisher's exact test. The only significant association found, following application of the Bonferroni correction for multiple comparisons, was between the methylation of p16INK4a and p14ARF. However, this finding was not pursued further because numerous other studies have previously tested the possibility of this association and ruled it out [17,24,25].
Figure 1.
Hypermethylation of the promoter region of five genes (DAPK, MGMT, hMLH1, p16INK4a, and p14ARF) in 106 microdissected primary CRCs. Each column is a different tumor. Back squares indicate methylated loci. The top row shows (marked with an X) tumors with MSI (MSI+).
Figure 2.
Bar graph of the proportion of methylated loci in 106 CRCs.
MSI
We scored tumors for MSI (MSI+) following the Bethesda guidelines [18] (i.e., if there were alterations in two or more of the mononucleotide repeat microsatellites Bat25 and Bat26, or the dinucleotide repeat microsatellites D2S123, D5S346, and D17S250). As expected from previous reports [19,20], deletions in the BAT26 were seen in all tumors with instability. Moreover, we also looked for extra alleles in a battery of one trinucleotide and nine tetranucleotide microsatellite loci [21]. There was complete concordance between the two criteria. Fourteen of 106 tumors (13.2%) displayed MSI. This value is compatible with that found in other studies [26,27]. As expected, the presence of MSI was very highly associated with the location of the tumor proximal to the splenic flexure of the colon (Table 1). However, it showed no significant correlation with recurrence within 3 years of diagnosis (Table 1).
Table 1.
Values of P (Two-Tailed) from Several Pairwise Comparisons Using Fisher's Exact Test.
| hMLH1 Methylation | MSI | Location | Recurrence within 3 Years | |||||||||
| + | - | + | - | Proximal | Distal | + | - | |||||
| MSI | + | 8 | 7 | |||||||||
| - | 11 | 80 | ||||||||||
| P = .0008* | ||||||||||||
| Location | Proximal | 9 | 13 | Proximal | 9 | 13 | ||||||
| Distal | 8 | 62 | Distal | 5 | 65 | |||||||
| P = .004* | P = .0005* | |||||||||||
| Recurrence within 3 years | + | 1 | 12 | + | 2 | 11 | + | 3 | 10 | |||
| - | 14 | 60 | - | 12 | 62 | - | 19 | 54 | ||||
| P = .295 | P = .652 | P = .563 | ||||||||||
| ≥2 Methylated loci | + | 14 | 16 | + | 7 | 23 | + | 10 | 15 | +4 | 21 | |
| - | 5 | 71 | - | 8 | 68 | - | 12 | 55 | - | 9 | 66 | |
| P = .000006* | P = .086 | P = .029 | P = .415 | |||||||||
| ≥3 Methylated loci | + | 8 | 5 | + | 4 | 9 | + | 6 | 4 | + | 0 | 7 |
| - | 11 | 82 | - | 11 | 82 | - | 16 | 66 | - | 13 | 80 | |
| P = .0002* | P = .083 | P = .011* | P = .365 | |||||||||
Proximal = proximal to the splenic flexure; Distal = distal to the splenic flexure.
Statistically significant after Bonferroni correction for multiple comparisons.
Association between Methylation and Clinical Features
We searched for associations between the number of loci found to be methylated and some clinical characteristics of the tumors (i.e., we tested if a “methylator phenotype” could be recognized). Because we had not found any discontinuities in the distribution of the number of methylated loci per tumor (Figure 1) as previously claimed by Toyota et al. [12,28], we lacked a clear criterion for defining a “high-methylation group.” Thus, we did the analysis using as “high-methylation group” the category of three or more methylated loci (criterion 1) and repeated it with the category of two or more methylated loci (criterion 2). We used Fisher's exact test to assess, for each of the categories of ≥2 and ≥3 methylated loci, an association with location of the tumor (distal versus proximal colon), recurrence within 3 years of diagnosis, and MSI. We found no significant association of the level of methylation with recurrence rate or with MSI (Table 1). However, tumor location was significantly associated with three or more methylated loci (P = .011) and also with two or more methylated loci (P = .029), although the former was no longer significant after applying the Bonferroni correction for multiple comparisons (Table 1). This led us to use Fisher's exact test to ascertain whether there was any association between location and methylation at each of the five loci in isolation. There was no association of proximal location with DAPK, MGMT, p14ARF, or p16INK4a, but there was a highly significant association with the methylation of hMHL1 that persisted after application of the Bonferroni correction for multiple contrasts (Table 1). As expected, methylation of hMLH1 was also highly associated with MSI and with the categories of ≥2 and ≥3 methylated loci (Table 1).
Discussion
CRC is a common malignancy that is expected to afflict approximately 106,000 people and to cause 57,000 deaths in the United States in 2004 [29]. There is an urgent need for markers that can be used in the establishment of a prognosis and that can guide in choosing the most appropriate treatment. In this sense, the discovery of the “mutator pathway” of CRC, operationally signaled by the presence of MSI (MSI+), was a major development. MSI+ CRC has characteristic biologic properties that include preferential proximal location, undifferentiated histology, and a relatively better prognosis [3,4]. Recent data suggest that fluorouracil-based adjuvant chemotherapy is of no benefit to patients with MSI+ CRC [5]. If these findings are confirmed, there will be a strong case for testing all CRCs for MSI [30].
Hypermethylation of the promoter region of specific genes can be profitably used as a molecular marker of cancer cells in the detection of micrometastases, diagnosis of recurrences, and even as a screening tool for discovering primary tumors [6]. An advantage of DNA methylation is that it constitutes a positive and stable marker that cannot be masked by the presence of normal tissues and thus offers extraordinary sensitivity in cancer detection through the use of MSP. Moreover, methylation might provide a new therapeutic target in CRC [6]. It is less clear whether multiple methylations have prognostic value. Toyota et al. [12,28] proposed the existence of a “CIMP” in human CRC, which included simultaneous methylation of several genes, preferential proximal location in the colon, and also an association with MSI. Although several other authors have supported this concept [31–33], the exact nature of such methylator phenotype is still poorly defined [27]. In particular, there is considerable overlap between the well-known phenotype associated with MSI and the proposed methylator phenotype. Because there exists a correlation between multiple methylations and MSI, the possibility of statistical confounding must be considered.
We have examined here the hypermethylation of the promoter region of five genes (DAPK, MGMT, hMLH1, p16INK4a, and p14ARF) in 106 microdissected primary CRCs. A histogram depicting the frequency of methylated loci in the series of tumors showed a continuous, monotonically decreasing distribution (Figure 2) that was quite different from the discontinuity that had been previously described by Toyota et al. [12]. Our results agree well with other authors who examined large numbers of tumors and who also did not find any discontinuities [27,33]. There was a significant association between the presence of three or more methylated loci and the tumor location proximal to the splenic flexure (Table 1). However, if we removed from analysis the tumors that display hMLH1 methylation or those with MSI, the significance of association of “high methylation” with location vanishes (P = .32 and P = .26, respectively). Likewise, the data of Yamashita et al. [27] also show that the significant association observed between “high methylation” and right-side location becomes nonsignificant (after correction for multiple testing) on removal of the MSI+ tumors. Thus, it appears that the association of “high methylation” with proximal location is not direct, but indirect, due to correlation with MSI. However, the association of MSI with location is known to occur very strongly in hereditary nonpolyposis colorectal cancer (HNPCC), in which 70% of tumors are right-sided [3] and in which methylation is uncommon [10].
In conclusion, our data do not support the existence of a methylator phenotype and suggest that it may represent a statistical artifact caused by confounding with the phenotype of tumors displaying MSI.
Acknowledgements
We are grateful to Glaura Franco (Department of Statistics, Universidade Federal de Minas Gerais) for her invaluable advice and to Rosiane Faleiro for help with the statistical analysis. Neuza Antunes Rodrigues, Kátia Barroso Gonçalves, and Mirian Rodrigues Costa provided expert technical support.
Abbreviations
- CRC
colorectal cancer
- CIMP
CpG island methylated phenotype
- MSI
microsatellite instability
- MSP
methylation-specific PCR
Footnotes
This work was supported by the Conselho Nacional de Pesquisas (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo a Pesquisa no Estado de São Paulo (FAPESP).
References
- 1.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 2.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 3.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 4.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 5.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 7.Bubb VJ, Curtis LJ, Cunningham C, Dunlop MG, Carothers AD, Morris RG, White S, Bird CC, Wyllie AH. Microsatellite instability and the role of hMSH2 in sporadic colorectal cancer. Oncogene. 1996;12:2641–2649. [PubMed] [Google Scholar]
- 8.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 9.Senba S, Konishi F, Okamoto T, Kashiwagi H, Kanazawa K, Miyaki M, Konishi M, Tsukamoto T. Clinicopathologic and genetic features of nonfamilial colorectal carcinomas with DNA replication errors. Cancer. 1998;82:279–285. [PubMed] [Google Scholar]
- 10.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 11.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 12.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahrendt SA, Chow JT, Xu LH, Yang SC, Eisenberger CF, Esteller M, Herman JG, Wu L, Decker PA, Jen J, et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst. 1999;91:332–339. doi: 10.1093/jnci/91.4.332. [DOI] [PubMed] [Google Scholar]
- 14.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzenellenbogen RA, Baylin SB, Herman JG. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood. 1999;93:4347–4353. [PubMed] [Google Scholar]
- 16.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 17.Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- 18.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for Cancer Detection and Familial Predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 19.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 20.Hoang JM, Cottu PH, Thuille B, Salmon RJ, Thomas G, Hamelin R. BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines. Cancer Res. 1997;57:300–303. [PubMed] [Google Scholar]
- 21.Pena SDJ. Single-tube single-colour multiplex PCR amplification of ten polymorphic microsatellites (ALF10): a new powerful tool for DNA profiling. Pure Appl Chem. 1999;71:1683–1690. [Google Scholar]
- 22.Sokal RR, Rohlf FJ. Biometry. 3rd ed. New York: W. H. Freeman and Co.; 1995. p. 887. [Google Scholar]
- 23.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 24.Burri N, Shaw P, Bouzourene H, Sordat I, Sordat B, Gillet M, Schorderet D, Bosman FT, Chaubert P. Methylation silencing and mutations of the p14ARF and p16INK4a genes in colon cancer. Lab Invest. 2001;81:217–229. doi: 10.1038/labinvest.3780230. [DOI] [PubMed] [Google Scholar]
- 25.Dominguez G, Silva J, Garcia JM, Silva JM, Rodriguez R, Munoz C, Chacon I, Sanchez R, Carballido J, Colas A, et al. Prevalence of aberrant methylation of p14ARF over p16INK4a in some human primary tumors. Mutat Res. 2003;530:9–17. doi: 10.1016/s0027-5107(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 26.Ward RL, Cheong K, Ku SL, Meagher A, O'Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–3736. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell. 2003;4:121–131. doi: 10.1016/s1535-6108(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 28.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ American Cancer Society, author. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 30.de la Chapelle A. Microsatellite instability. N Engl J Med. 2003;349:209–210. doi: 10.1056/NEJMp038099. [DOI] [PubMed] [Google Scholar]
- 31.van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51:797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehall VL, Wynter CV, Walsh MD, Simms LA, Purdie D, Pandeya N, Young J, Meltzer SJ, Leggett BA, Jass JR. Morphological and molecular heterogeneity within nonmicrosatellite instability high colorectal cancer. Cancer Res. 2002;62:6011–6014. [PubMed] [Google Scholar]
- 33.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]