Abstract
Loss of the 3p21.3-encoded semaphorins, SEMA3B and SEMA3F is implicated in lung cancer development. Although both antagonize VEGF binding/response to neuropilin (NRP) receptors, in lung cancer lines,SEMA3F is predominantly expressed and preferentially utilizes NRP2. In lung cancer patients, SEMA3F loss correlates with advanced disease and increased VEGF binding to tumor cells. In cell lines, VEGF enhances adhesion and migration in an integrin-dependent manner, and exogenous SEMA3F causes cells to round and lose extracellular contacts. Using retroviral infections, we established stable SEMA3F transfectants in two NSCLC cell lines, NCI-H157 and NCI-H460. When orthotopically injected into nude rats, both control lines caused lethal tumors in all recipients. In contrast, all animals receiving H157-SEMA3F cells, survived to 100 days, whereas all H157 controls succumbed. In H460 cells, which express NRP1 but not NRP2, SEMA3F did not prolong survival. This antitumor effect in H157 cells was associated with loss of activated αvβ3 integrin and adhesion to extracellular matrix components. In addition, H157-SEMA3F cells, and parental H157 cells exposed to SEMA3F-conditioned medium, showed loss of p42/p44 MAPK phosphorylation. Thus, in this in vivo lung cancer model, SEMA3F has potent antitumor effects, which may impinge on activated integrin and MAPK signaling.
Keywords: Semaphorin SEMA3F, lung cancer, integrin, MAPK, tumor suppressor gene
Introduction
Semaphorins are a large family of proteins [1] initially identified by their collapsing activity on growth cones [2,3]. Secreted semaphorins bind high-affinity neuropilin receptors (NRP1 and NRP2) [4,5], which associate with Plexins in signal transduction [6]. Neuropilins were independently identified as coreceptors for vascular endothelial growth factor (VEGF) [7], and the semaphorin/neuropilin system is an important regulator of cardiovascular development [8].
Two secreted semaphorins, SEMA3B and SEMA3F, are encoded in 3p21.3, a region that undergoes homozygous deletion and frequent loss of heterozygosity in lung cancer [9–11], suggesting that these genes have tumor-suppressor function. Accordingly, in an in vitro assay, transfection of SEMA3B into the lung cancer cell line, H1299, led to apoptosis and reduced colony formation [12]. Tumor-suppressor activity for SEMA3F was first shown by transfer of an 80-kb genomic fragment to mouse A9 fibrosarcoma cells [13] and was subsequently confirmed by direct gene transfection [14]. Similarly, overexpression of SEMA3F by adenovirus in HEK293 cells was shown to inhibit tumor formation and angiogenesis [15]. Moreover, during submission of our manuscript, Bielenberg et al. [16] reported that SEMA3F blocked metastases of melanoma cells injected subcutaneously into nude mice and that the tumors were poorly vascularized and encapsulated. In contrast, one report [14], using the small cell lung cancer line GLC45, found no effect on tumorigenicity by SEMA3F, although no other studies have tested the in vivo effects of SEMA3F in lung cancer cells.
We previously demonstrated that loss of SEMA3F staining was associated with advanced stage lung cancer and was an early event in premalignant lesions [17,18]. In vitro, we found that SEMA3F has a repulsing activity on breast cancer cells [19]. In addition, exogenous SEMA3F inhibited cell attachment and spreading, and these effects were counteracted by VEGF [20]. VEGF has been shown to promote the attachment and migration of tumor cells in an integrin-dependent manner, including the activation of αvβ3 integrin [21]. Interestingly, in endothelial cells, the secreted semaphorins, SEMA3A (collapsin) and SEMA3F, were reported to downregulate activated αvβ3 integrin [22], suggesting that similar results might occur in lung cancer.
We previously observed that plasmid-mediated SEMA3F expression led to complete loss of attached cells, and stable transfectants could not be obtained. In the present study, using a retroviral vector, we were able to establish stably transfected derivatives of the non small cell lung cancer cell lines, NCI-H157 and NCI-H460 (hereafter H157 and H460). For many tumor lines, subcutaneous injection can be inefficient, whereas orthotopic administration results in aggressive, metastatic tumors. Therefore, we directly instilled tumor cells through tracheal intubation into the lungs of nude rats. In this model system, the antitumor effects of SEMA3F on the NRP2-expressing H157 cells were dramatic, whereas H460 cells were not inhibited. Of note, H460 expresses NRP1, whereas NRP2 expression is barely detectable; among various semaphorin signaling components, this difference was the most striking.
In accordance with previously reported effects of VEGF and SEMA3A on αvβ3 integrin in melanoma and endothelial cells, respectively, SEMA3F antitumor activity was associated with loss of staining with the WOW-1 antibody, which binds only activated αvβ3 integrin. In addition, tumor suppression was associated with downregulation of p42/p44 MAPK phosphorylation.
Materials and Methods
Cell Culture
The lung cancer cell lines NCI-H157 (squamous carcinoma) and NCI-H460 (large cell carcinoma) were grown in RPMI 1640 plus 10% fetal calf serum (In Vitrogen, Cergy Pontoise, France). SEMA3F or vector-only expressing cells were also maintained in 600 µg/ml G418 (In Vitrogen). The packaging cell line PT67 (Clontech, Saint Quentin Fallavier, France) was similarly grown in DMEM.
Plasmid Constructions
The SEMA3F cDNA was amplified with the following primers (5′-GC-GAA-TTC-GCC-ACC-ATG-CTT-GTC-GCC-GGT-CTT-CT-3′) and (5′-GC-GTT-AAC-TCA-CAG-ATC-CTC-TTC-TGA-GAT-GAG-TTT-TTG-TTC-TGT-GTC-CGG-AGG-GTG-GTG-CCG-3′), which contains a Myc tag at the 3′ end. The fragment was cloned into the EcoRI and HpaI sites of the pLXSN retroviral vector (Clontech).
Production of SEMA3F Stable Transformants
Stable virus-producing cell lines were generated by transfecting PT67 packaging cells with the SEMA3F-pLXSN construct (or pLXSN vector alone) using Effectene Transfection Reagent (Qiagen, Courtaboeuf, France) as recommended by the manufacturer. Cells were selected in 400 µg/ml G418 for 10 days. Virus generated from overnight culture was used to infect H157 and H460 cells. Single G418-resistant colonies were isolated in 800 µg/ml G418 for 10 days and subsequently maintained at 600 µg/ml. DNA was prepared with the DNeasy Tissue kit (Qiagen) and screened by PCR using primers 59H8 (5′-TTC-AAC-TTC-CTG-CTC-AAC-3′) and 39G5 (5′-GAA-GAC-CAT-GCG-AAT-ATC-3′), as previously described [9]. Southern blot analysis was performed with the SEMA3F probe at 65°C, using a nylon membrane (HYBOND-N+; Amersham Pharmacia Biotech, Orsay, France) in Rapid-hyb buffer (Amersham). Washings were performed two times for 15 minutes in 0.2x SSC-SDS 0.1% at 65°C.
RNA Expression
Total RNA was extracted using the RNeasy Mini kit (Qiagen). RT-PCR was performed with Superscript II reverse transcriptase (In Vitrogen) using the procedure supplied by the manufacturer. We assessed levels of gene expression relative to GAPDH by quantitative real-time PCR and the ABI7000 (PE Biosystems, Courtaboeuf, France) quantitative PCR system with SYBR-Green chemistry. The PCR was carried out in 20-µl reaction volumes consisting of 1x PCR SYBR-Green buffer, 0.125 µM primers, 200 µM of each dNTP, 2 mM MgCl2, and 0.025 U/µl AmpliTaq Gold (PE Biosystems). The amplification parameters were: 50°C for 2 minutes, 95°C for 10 minutes, followed by 35 cycles at 95°C x 15 seconds, 60°C x 1 minute. Primer sequences are described in Table 1. The cycle at which a particular sample reaches an arbitrary fluorescent threshold (Ct) is proportional to the quantity of the input template.
Table 1.
Primers Sequences for Real-Time Quantitative RT-PCR.
| Forward | Reverse | ||
| GAPDH | 5′-TGC ACC ACC AAC TGC TTA GC-3′ | 5′-GGC ATG GAC TGT GGT CAT GAG-3′ | |
| SEMA3F | 5′-AGC AGA CCC AGG ACG TGA G-3′ | 5′-AAG ACC ATG CGA ATA TCA GCC-3′ | |
| PlexA1 | 5′-TGC AGT GCC AGA ATT CCT CG-3′ | 5′-TGC GCC TGG ATG TTC TGT G-3′ | |
| PlexA2 | 5′-GTG AGC CTT GCA GTG CAT CC-3′ | 5′-CCT CCA CCT CTG TCG GGT TC-3′ | |
| PlexA3 | 5′-GTT CCT TGG TGG CAT TGG TG-3′ | 5′-TTG GTG CCT GTC TCC TGG TC-3′ | |
| PlexB1 | 5′-GAC GTG AGA TAT GCG TCC GTG-3′ | 5′-CAC GGC TCC TCA AAT TGC TG-3′ | |
| PlexB2 | 5′-TTT CTG GGC ACC TCT GAT GG-3′ | 5′-GGT CGC GCT TGA CTC TCT TG-3′ | |
| CRMP1 | 5′-ATC GCC AAG GAC TGA CTG AG-3′ | 5′-GAG GAT GCT TCT CTG CAA CC-3′ | |
| CRMP2 | 5′-TTC TGT CAC CTT TCC CCC TA-3′ | 5′-AGA GAC AAG GGT GGT GAT GG-3′ | |
| CRMP3 | 5′-CAC CAG GCT TCT TGG TGA AC-3′ | 5′-GGT CCC TGA AGT CTC AGC AG-3′ | |
| CRMP4 | 5′-GGA AAG GGT GTT TGG GAA AT-3′ | 5′-AAT CCA GGC TCA GGT GTC AG-3′ | |
| VEGF | 5′-CAA GAC AAG AAA ATC CCT GTG G-3′ | 5′-CCT CGG CTT GTC ACA TCT G-3′ | |
| VEGFR1 | 5′-ATG CCA CCT CCA TGT TTG ATG-3′ | 5′-GAG GCC TTG GGT TTG CTG TC-3′ | |
| VEGFR2 | 5′-TTC TCT TGA TCT GCC CAG GC-3′ | 5′-AGG CTC CAG TGT CAT TTC CG-3′ | |
| NRP1 | 5′-ATC ACG TGC AGC TCA AGT GG-3′ | 5′-TCA TGC AGT GGG CAG AGT TC-3′ | |
| NRP2 | 5′-GGA TGG CAT TCC ACA TGT TG-3′ | 5′-ACC AGG TAG TAA CGC GCA GAG-3′ | |
| Int α5 | 5′-TGT CAC CAT CCT TAA TGG CTC A-3′ | 5′-GGC CAC TGC ATA GCC AAA GTA G-3′ | |
| Int αv | 5′-GAA CGC AGT CCC ATC TCA AAT C-3′ | 5′-GGC TCC TTT CAT TGA ATA GCC A-3′ | |
| Int α6 | 5′-CCT TTG ACA CCC CAT ATC TGG A-3′ | 5′-CGA GAC CGA TAA AAG CAG TTC A-3′ | |
| Int β1 | 5′-TGC CGG GTT TCA CTT TGC-3′ | 5′-GTG ACA TTG TCC ATC ATT TGG TAA A-3′ | |
| Int β3 | 5′-TGA CCC GCT TCA ATG AGG AA-3′ | 5′-CCT CCA GCC AAT CTT TTC ATC A-3′ | |
| Int β4 | 5′-GAG GAT GAC GAC TGC ACC TAC A-3′ | 5′-GGC AGT CCT TCT TCT TGT GCA C-3′ |
Western Blot Analysis
A total of 2.5 x 106 H157 and H460 stably transfected cells was washed twice with PBS, then lysed in 500 µl of lysis buffer [1% NP-40 (wt/vol), 1% DOC (wt/vol), 0.1% SDS, 0.15 M NaCl, 0.01 M NaH2PO4, 2 mM EDTA, 0.5 mM PMSF, 10 µg/ml leupeptin, 5 µg/ml pepstatin A, and 1.7 µg/ml aprotinin] and subsequently sonicated. Total protein was quantified using the BCA kit (PerBio Sciences, Bezons, France). Thirty micrograms of protein lysate was resolved on an 8% SDS-PAGE, followed by Western blot analysis. Primary antibodies and dilutions used were: monoclonal anti-c-MYC (1/2000; Sigma, St. Louis, MO), phospho p42/44 MAPK (1/1000; Cell Signaling, Montigny le Bretonneux, France), and p42/44 MAPK (1/4000; Promega, Madison, WI). Secondary antibodies were HRP-labeled (1/5000; PE Biosystems) and blots were revealed by ECL detection.
Immunofluorescence
For αvβ3 integrin immunofluorescence, cells were plated at sparse density on 0.5 µg/ml vitronectin (Chemicon, Souffolweyershein, France) and grown for 48 hours. Cells were fixed and stained with the WOW-1 Fab antibody (kindly provided by Prof. Sanford Shattil, La Jolla, CA), which selectively recognizes αvβ3 integrin because of its engineered RGD segment from the penton-based adenovirus protein as described [23]. α6 Antibody was from Chemicon (GoH3, 1/50 dilution). The secondary antibodies were antimouse (for α6) or antirat (for αvβ3) Alexa 488-conjugated antibody (1/250 dilution; Molecular Probes, Eugene, OR, USA).
Cell Sedimentation Manifold (CSM) Assays
The CSM (CSM, Inc., Phoenix, AZ) migration assay system is composed of 10 aligned channels for sedimentation of low cell numbers as discrete circles on Teflon-delineated wells of slides. Slides were coated with 1% bovine serum albumin for 1 hour, and 1.5 µl of cells (3 x 106 cells/ml) was applied per channel according to the manufacturer's protocol. Cells were initially left to sediment for 1 hour at 4°C followed by an additional period of 4 hours at 37°C. Cells were then maintained for 24 hours at 37°C followed by photography.
Cell Adhesion
Ninety-six well plates were pretreated with 0.5 µg/ml fibronectin, vitronectin, or laminin (Sigma-Aldrich, Saint Quentin Fallavier, France) for 1 hour at 37°C. Uncoated wells were used as controls. The wells were drained and nonspecific binding was blocked with PBS/3% BSA for 45 minutes at 37°C then emptied, and 1 x 103 cells in a volume of 100 µl were added and left to sediment x 1 hour at 37°C. Unattached cells were aspirated and the attached cells were fixed (10% formalin x 10 minutes at room temperature), stained (0.1% crystal violet x 10 minutes at room temperature), then rinsed three times with distilled water. Cells were left to dry overnight and counted under a microscope field. Statistical differences were calculated using the Student's t test.
Orthotopic Rat Tumor Model
Six- to 8-week-old female athymic nude rats, obtained from the National Cancer Institute (Washington, DC, USA), were maintained in pathogen-limited conditions at the Animal Resources Center, University of Colorado Health Sciences Center (Aurora, CO). One day prior to tumor cell installation, rats (six rats per transformant) were treated with 450 cGy of total body irradiation using a Co60 source. Control or SEMA3F-transformed H157 or H460 cells (1 x 107 cells in 100 µl of serum-free media) were instilled intratracheally into the left lung of anesthesized rats by administration through a 3-in. 22-gauge catheter (Popper & Sons, Inc., New Hyde Park, NY), according to a published procedure [24]. The procedure required less than 3 minutes and the animals recovered in less than 5 minutes without showing signs of stress or casualty. The entire experiment was repeated twice with essentially identical results. Survival was analyzed using the Kaplan-Meier survival model and log-rank test. Hazard ratios for treatment versus control were estimated with 95% confidence intervals. All statistical analyses were performed with SAS Software, Version 8.1 (SAS Institute, Cary, NC).
Results
Establishment of H157 and H460 Subclones Stably Expressing SEMA3F
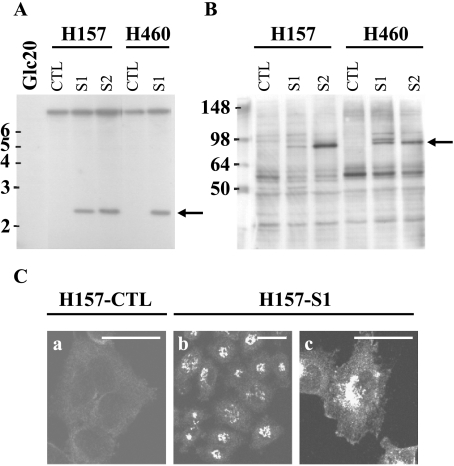
To study the effect of SEMA3F on in vivo lung tumorigenicity, we first established stable SEMA3F transfectants using the squamous carcinoma cell line, H157, and the large cell carcinoma cell line, H460. These were chosen because both express low levels of SEMA3F (Table 2). Following retroviral infection, Myc-tagged SEMA3F-expressing cells were selected in G418 and analyzed by Southern blot. As shown in Figure 1A, the intensity of the SEMA3F hybridization signal indicated that only one copy of SEMA3F had been integrated. Two independent SEMA3F-expressing subclones from each tumor cell line (hereafter H157-S1, H157-S2, H460-S1, and H460-S2) were chosen, as well as subclones containing only the pLXSN vector (i.e., H157 and H460-control or CTL). By real-time RT-PCR (Table 2), SEMA3F mRNA levels were approximately 8- to 16-fold higher in these various SEMA3F transfectants compared to either short-term normal bronchial epithelial (NHBE) cultures [18] or normal-appearing bronchial epithelial biopsies (unpublished observations). Compared to NHBE cells, the normal lung expresses higher levels of SEMA3F (i.e., four-fold to five-fold greater), which we suspect is due to the endothelial component based on expression results in HUVEC cells (unpublished observations). Thus, the retroviral infections resulted in overexpression of SEMA3F, albeit at levels that were tolerated by the recipient cells. Also, levels of exogenous SEMA3F mRNA were comparable in the H157 and H460 transfectants (Table 2).
Table 2.
RNA Expression of Selected Genes in H157 and H460 Transfectants and Controls.
| H157 CTL | H157-S1 | H157-S2 | H460 CTL | H460-S1 | H460-S2 | |
| SEMA3F | 0.07 | 26.79 | 93.30 | 0.01 | 53.59 | 50.00 |
| PlexA1 | 0.30 | 0.17 | 0.18 | 0.12 | 0.09 | 0.20 |
| PlexA2 | 0.00 | 0.00 | 0.00 | 0.09 | 0.17 | 0.21 |
| PlexA3 | 4.12 | 3.35 | 2.54 | 1.79 | 1.67 | 1.92 |
| PlexB1 | 0.21 | 0.21 | 0.22 | 0.17 | 0.15 | 0.18 |
| PlexB2 | 2.92 | 2.92 | 5.08 | 1.79 | 2.21 | 3.85 |
| CRMP1 | 0.28 | 0.24 | 0.39 | 0.09 | 0.05 | 0.07 |
| CRMP2 | 5.44 | 5.44 | 7.69 | 7.18 | 3.35 | 5.83 |
| CRMP3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| CRMP4 | 0.01 | 0.12 | 0.00 | 1.18 | 0.17 | 0.21 |
| VEGF | 4.42 | 1.92 | 2.06 | 0.20 | 0.20 | 0.22 |
| VEGFR1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| VEGFR2 | 0.00 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 |
| NRP1 | 1.56 | 0.78 | 2.06 | 0.17 | 0.28 | 0.34 |
| NRP2 | 0.73 | 0.59 | 0.68 | 0.00 | 0.00 | 0.00 |
| Int α5 | 3.35 | 3.35 | 1.92 | 0.32 | 0.20 | 0.21 |
| Int αv | 9.47 | 9.47 | 9.47 | 2.37 | 0.90 | 0.90 |
| Int α6 | 3.59 | 1.46 | 3.35 | 0.42 | 0.45 | 0.39 |
| Int β1 | 1.79 | 4.12 | 2.92 | 1.27 | 1.67 | 0.96 |
| Int β3 | 3.13 | 8.84 | 6.70 | 0.00 | 0.01 | 0.01 |
| Int β4 | 0.12 | 0.21 | 0.28 | 0.01 | 0.00 | 0.00 |
RNA expression was estimated by real-time quantitative PCR and expressed as percentage of GAPDH expression. CTL is control clones (vector-only transfected cells).
Figure 1.
Establishment of H157 and H460 clones stably expressing SEMA3F. Southern blot (A) was performed with the SEMA3F cDNA probe and HpaI and EcoRI digested DNA from SEMA3F transformants and control (vector only) cells. DNA markers are indicated in kilobasepairs on the left. The arrow indicates the transfected SEMA3F DNA; the upper band is the endogenous SEMA3F. DNA from the Glc20 cell line was chosen as a negative control as these cells are homozygously deleted for SEMA3F [43]. SEMA3F protein was detected on the S1 and S2 subclones with an anti-Myc antibody by Western blot analysis (B) and immunofluorescence (C). The arrow indicates the SEMA3F band; protein markers are indicated in kilodaltons (B). (c) An enlargement of (b) Only background was detected by immunofluorescence in H157-CTL (a) Scale bar, 50 µm.
SEMA3F protein expression was verified by Western blot analysis (Figure 1B). A band of the expected size (∼80 kDa) was detected using the anti-Myc antibody specifically in transfected cells. In H460-S1, however, a doublet was observed, the nature of which is unknown. The intensity of the observed bands reflected the amount of SEMA3F mRNA. As expected for a secreted protein, strong immunofluorescence staining was concentrated in the Golgi region (H157-S1 in Figure 1C, parts b and c).
SEMA3F Inhibits Tumorigenicity of H157 Cells But Not H460
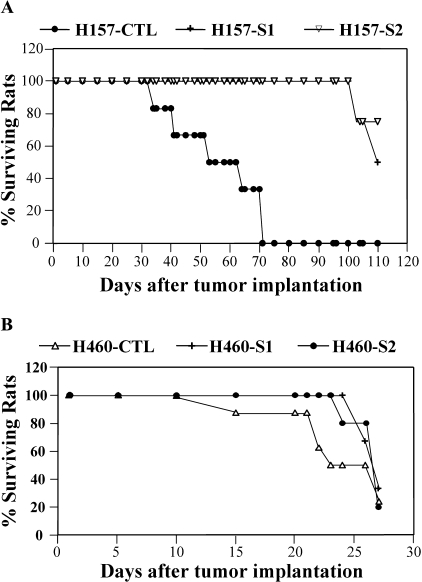
SEMA3F-expressing H157 and H460 cells, plus controls, were injected into the left lung of nude rats through tracheal administration. In two independent experiments, all animals receiving H157 control cells died with a median survival of 52 days, whereas no animal receiving SEMA3F-expressing cells (H157-S1 and H157-S2) had died by 100 days (P < .001; see Figure 2A). Lungs isolated from control animals showed massive primary tumors with multiple metastases in the right lung and mediastinal lymph nodes. In a few H157-S1- and H157-S2-injected rats followed to day 220, only small primary tumors were observed in the left lung. These were not analyzed for loss of SEMA3F. In contrast, exogenous expression of SEMA3F in H460 cells had no effect on the rat survival compared to controls (Figure 2B).
Figure 2.
SEMA3F inhibits tumorigenicity of H157 cells but not H460. Survival curves are presented for rats injected into the lung with SEMA3F transfectants or control H157 (A) and H460 (B) cells.
With these results, we looked for differences in components of the class 3 semaphorin signaling pathway (NRPs, Plexins, and CRMP) VEGF and VEGF receptors (VEGFR1 and VEGFR2) using quantitative RT-PCR (Table 2). Although some clonal variations were observed, these results demonstrated that H157- and H460-transfected cells expressed comparable levels of Plexin A1, Plexin A3, Plexin B1, and Plexin B2. Plexin A2 was not expressed in H157, whereas it was moderately expressed in H460. Although considerable complexity in Plexin-NRP interactions probably exists, the absence of Plexin A2 in H157 cells would be an unlikely explanation for the observed differential tumor suppression. CRMP1 expression was three-fold higher in H157 cells, although the pattern was reversed for CRMP4. VEGF levels were considerably higher (23-fold) in the SEMA3F-sensitive (H157) cells, whereas VEGFR1 and VEGFR2 were not, or only slightly, expressed in either cell line. In contrast to these results, the major observed difference involved NRP2, which was essentially absent in H460. In addition, NRP1, the lower-affinity receptor, was expressed at nine-fold higher levels in H157 cells. Thus, we suspect that SEMA3F receptor levels may explain the observed differential antitumor responsiveness.
Changes in Cell Morphology and Downregulation of Activated αvβ3 Integrin by SEMA3F
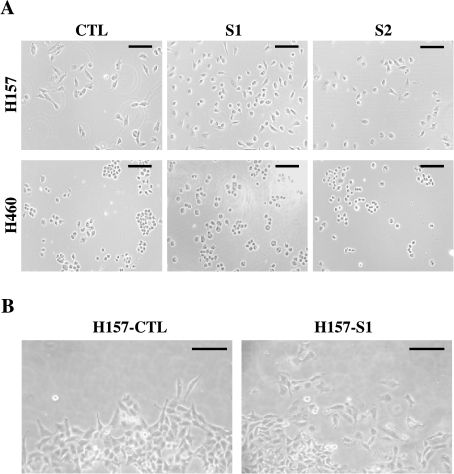
Compared to controls, H157 cells transfected with SEMA3F appeared more rounded when plated on culture dishes (Figure 3A). This involved loss of protrusions and large lamellipods. A few hours after plating, this effect was lessened and no difference was observed with confluent cells. We used a cell sedimentation device (CSM, Inc.) to examine cell migration. Concentrated cells were deposited onto a circular area of a coated microscope slide and examined after 24 hours. At the periphery, H157-S1 cells were isolated whereas control cells retained close intercellular contacts (Figure 3B).
Figure 3.
Changes in cell morphology of SEMA3F-transfected H157 cells. Cell morphology was observed for H157 control and SEMA3F-transfected cells (A) several hours after plating. Cells were also observed at time 24 hours, at the periphery of an island of concentrated cells obtained with the CSM device (B). Scale bar, 100 µm.
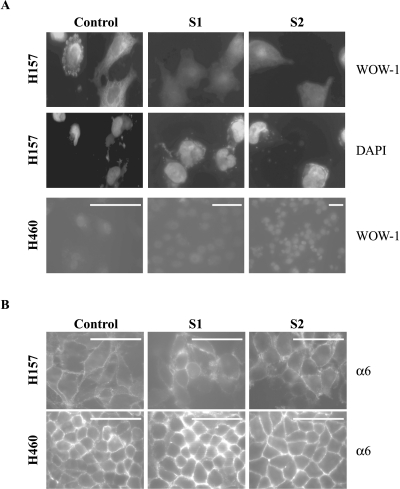
Recently, SEMA3A and SEMA3F were shown to antagonize VEGF-induced integrin activation in endothelial cells [22]. Because integrins contribute to signaling from various tyrosine kinase receptors and contribute as well to tumorigenesis, metastasis, and angiogenesis (for review, see Ref. [25]), we looked for effects of integrin activation in SEMA3F-transfected cells. For this purpose, we used the recombinant antibody WOW-1, which specifically recognizes the active form of αvβ3 integrin [23]. H157 and H460 cells were plated on vitronectin (the major substrate for αvβ3 integrin) and αvβ3 integrin activation was followed by immunofluorescence. Although H460 cells did not express αvβ3 integrin at the RNA level (Table 2), activated αvβ3 integrin membrane staining was detected in H157 control cells (Figure 4A), whereas this staining was abolished in the SEMA3F transfectants. This effect was specific, as neither the expression nor localization of total α6 integrin subunit was affected (Figure 4B). Analysis of various integrins by RT-PCR (Table 2) demonstrated that most family members were expressed at much lower levels in H460 cells. Only β1 integrin was comparably expressed at the mRNA level. Unfortunately, we did not have antibodies directed against the activated form of β1 integrin.
Figure 4.
Downregulation of activated αvβ3 integrin by SEMA3F. Immunocytochemistry was performed on H157 and H460 control and SEMA3F-transfected cells with WOW-1, an anti-activated αvβ3 integrin antibody (A), or an anti-α6 integrin antibody (B). H157 cells were stained with DAPI (A). No change in staining was noticed with the anti-α6 integrin, whereas loss of αvβ3 staining was observed for H157-S1 and H157-S2. Scale bar, 50 µm.
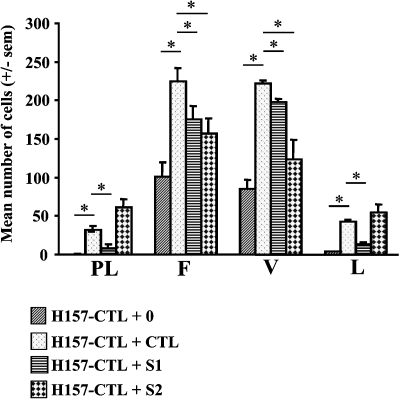
To pursue this observation, we performed adhesion tests on plastic, fibronectin, vitronectin, and laminin substrates. Adhesion on each substrate is integrin-dependent, although different α-β subunits (e.g., αvβ3 and α5β1) are involved. More control H157 cells attached to fibronectin and vitronectin than to plastic or laminin in serum-free medium demonstrating substrate-specific adhesion (Figure 5). With conditioned medium (containing serum) from H157 control cells, adhesion to all substrates was enhanced. In contrast, when the conditioned media from the control cells were replaced by medium from H157-S1 or H157-S2 cells, adhesion to fibronectin and vitronectin was significantly reduced (Figure 5) (for H157 cells treated by H157-S1 and -S2 conditioned media, P values for adhesion on fibronectin are .04385 and .03877, respectively and, for adhesion on vitronectin, P values are .0031 and .01802 respectively). Thus, we suggest that SEMA3F downregulates the active form of αvβ3 integrin in H157 cells. In contrast, conditioned media from the SEMA3F-expressing H460 clones had no effect on the adherence of H460 cells to the various substrates (data not shown).
Figure 5.
SEMA3F reduced the adhesion of H157 control cells to fibronectin and vitronectin. H157 control (CTL) cells were grown on different substrates: plastic (PL), fibronectin (F), vitronectin (V), and laminin (L) with a serum-free medium (+0) or control (+CTL), S1 (+S1), or S2 (+S2) conditioned media. The number of attached cells on each substrate is expressed as the mean number of cells. Assays were done in triplicate for two independent experiments. *P value < .05.
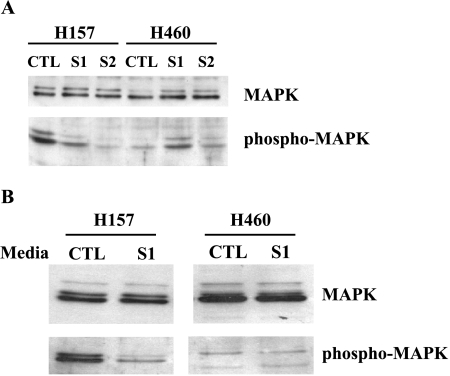
Downregulation of Phospho-MAPK and Cell Proliferation by SEMA3F
In neurons, movement of growth cones away from a SEMA3A source involves changes in MAPK phosphorylation [26,27]. Therefore, we examined phosphorylation of p42/p44 MAPK in the SEMA3F transfectants. As shown in Figure 6A, H157-S1 and S2 cells were found to have reduced levels of phospho-p42/p44 MAPK compared to controls. To exclude the possibility that this was due to clonal variation, we treated H157 parental cells with cultured supernatants from either control or H157-S1 cells. As shown in Figure 6B, phospho p42/p44 MAPK was downregulated by conditioned media from H157-S1 but not control cells. In contrast, the endogenous low level of phospho-p42/p44 MAPK in H460 cells was either unchanged or even increased in the SEMA3F-expressing clones (Figure 6A). The observed increase in H460-S1 MAPK phosphorylation was likely due to clonal variation because, when added to the parental cells, SEMA3F-containing media had no effect on p42/44 MAPK phosphorylation (Figure 6B).
Figure 6.
Downregulation of MAPK phosphorylation by SEMA3F. Western blot analysis of protein extracts of H157 and H460 control and SEMA3F-transfected cells with anti-p42/44 MAPK and anti-phosphorylated p42/44 MAPK antibodies (A). Parental cells were treated either with control or S1-conditioned media for 6 hours (B), and Western blot analysis was performed as in (A).
Discussion
The role of secreted class 3 semaphorins (which includes SEMA3F [1]) as tumor-suppressor proteins has emerged during the last few years. SEMA3F was identified by positional cloning of genes from a homozygous 3p21.3 deletion identified in lung cancer cell lines. Although SEMA3F has activities consistent with tumor-suppressor function in vitro [12], there has been no report demonstrating in vivo activity in lung cancer. The results described above, using an orthotopic model of lung cancer with H157 cells, clearly establish that SEMA3F has potent antitumor function and provide clues as to the involved mechanisms. To our knowledge, this is the first in vivo evidence of such activity in lung cancer.
These results are in agreement with two previous reports demonstrating that SEMA3F inhibited subcutaneous tumors formed by either mouse A9 fibrosarcoma cells, HEY ovarian carcinoma, or HEK293 cells in nude mice [14,15]. However, whereas these latter studies demonstrated an antitumor activity of SEMA3F, the importance of SEMA3F loss has not been defined in either rodent sarcomas, human ovarian cancer, or HEK293 cells (which were derived from normal embryonic kidney). Moreover, it has been our experience that lung cancer cell lines are considerably more tumorigenic when administered orthotopically compared to subcutaneous injections. Nevertheless, taken together, these results are consistent and strongly support the role of SEMA3F as a tumor-suppressor gene.
In contrast to H157, SEMA3F-transfected H460 cells readily formed tumors. Indeed, in other model systems, SEMA3F tumor inhibition was specific; no activity was found in the small cell lung carcinoma cell line, GLC45 [14]. Moreover, SEMA3F had reduced in vitro antitumor activity compared to SEMA3B in H1299 lung cancer cells [12]. One simple explanation for these results could be the absence of NRP2—the highest affinity SEMA3F receptor. However, NRP2 was expressed in GLC45 [14], although the levels were not provided. In the present study, we looked for differences that might explain the SEMA3F sensitivity of H157 cells compared to H460. We found that variations in levels of VEGF, VEGF receptors, and downstream NRP signaling components (i.e., Plexin and CRMP family members) appeared to be insufficient to explain the observed biologic effects. In contrast, the major difference was NRP2, which is essentially absent in H460. However, formal proof will require exogenous NRP2 expression in this cell line. Moreover, we cannot exclude the difference in aggressiveness of these two cell lines.
Visually, SEMA3F-expressing H157 cells showed substantial changes not observed in H460. H157 cells lost lamellipods and were rounded, effects that appeared identical to those in the breast cancer cell lines, MCF7 and C100 [20], as well as in endothelial cells treated with exogenous SEMA3F [15]. This “collapsing activity” has also been demonstrated for SEMA3A in COS7 cells [28], as well as for COS7, 3T3, and SKBR3 cells treated with SEMA4D [29]. Moreover, with the CSM device, we observed that SEMA3F-transfected H157 cells showed reduced intercellular contacts at the periphery of the islet. We and others have previously shown that secreted semaphorins negatively affect adhesion and migration of tumor cell lines, and that exogenous VEGF can block these effects [19,20,30–32]. Strikingly, it was recently reported that SEMA3F inhibits VEGF-induced αvβ3 and α5β1 integrin activation in endothelial cells [22]. These findings are in agreement with the report of Byzova et al. [21] demonstrating that VEGF enhances tumor cell adhesion and migration in an integrin-dependent manner with αvβ3 integrin being one of the important components.
In SEMA3F-transfected H157 cells, we found loss of activated αvβ3 integrin staining. These results were substantiated with the demonstration of reduced adhesion to fibronectin and vitronectin. During submission of our manuscript, similar results were reported using another system: inhibition of metastasis and integrin function with SEMA3F-overexpressing melanoma cells in nude mice [16]. Integrins mediate cellular adhesion and migration on extracellular matrix (ECM) components [25,33–35]. They also regulate cell cycle entry, and withdrawal, apoptosis, and metastasis. For instance, strategies that inhibit the αv subunit result in loss of melanoma tumorigenicity, activation of p53, and blocking of Bcl-2 expression [36–39]. Furthermore, αvβ3 integrin binds MMP-2 and facilitates matrix degradation and invasion [40]. αvβ3 Integrin plays a very important role in cancer through angiogenesis. It regulates vascular cell survival in vivo, and αvβ3 antagonists disrupt angiogenesis and induce tumor regression (see review in Ref. [25]). Thus, the observed loss of activated αvβ3 integrin staining (in H157 cells) in response to SEMA3F is likely to be an important mechanism of its antitumor activity.
Because semaphorins affect MAPK activity [15,26,27,41], we examined the phosphorylation of p42/p44 MAPK in transfected cells. Signaling through MAPK represents a critical focal point in the response of multiple receptors involved in tumorigenesis [42]. Importantly, the H157 SEMA3F transfectants had reduced levels of phopho-p42/p44 MAPK and the same was true when H157 parental cells were exposed to conditioned medium from the SEMA3F transfectants, excluding clonal variation as an explanation. In neurons, the opposite effect was reported for SEMA3A [26], and repulsion of neuronal progenitors, as well as protein synthesis regulation, involve the activation of p42/p44 MAPK [27,41]. However, in HUVEC cells, SEMA3F has been shown to inhibit VEGF as well as bFGF-induced p42/p44 MAPK phosphorylation [15]. Thus, semaphorin effects on MAPK phosphorylation show cell type specificity, probably reflecting the complexity of growth factor signaling and cell extracellular matrix interactions.
In summary, we have described the tumor-suppressor activity of SEMA3F in H157 lung cancer cells in an in vivo model. We suggest that this activity is mediated by reductions in activated integrins and MAPK signaling in this cell line. In the presence of normal levels of SEMA3F, the propensity of developing lung cancer cells to interact with the stroma might be diminished. This is in agreement with our previous studies demonstrating that SEMA3F loss occurred early during human lung tumorigenicity [18] and was also associated with more aggressive lung tumors [17]. Recent published data [14–16], together with the results presented here, strongly support the role of SEMA3F as a tumor-suppressor gene in diverse cell types, and raise the possibility that pharmacologic approaches to reexpress SEMA3F have therapeutic potentials.
Acknowledgements
We greatly acknowledge S. Shattil for his integrin antibody.
Footnotes
This work was supported by Ligue Contre le Cancer and ARC (J.R., S.K., and P.N.) and by the University of Colorado Lung Cancer SPORE, CA58187 (S.K., V.P., D.C., R.G., and H.D.). We are very grateful for the support of S.K. by the UICC and the Société Française du Cancer (SFC).
Patrick Nasarre, Daniel Chan, and Vincent Potiron contributed equally as second authors.
References
- 1.Semaphorin Nomenclature Committee, author. Unified nomenclature for the semaphorins/collapsins. Cell. 1999;97:551–552. doi: 10.1016/s0092-8674(00)80766-7. [DOI] [PubMed] [Google Scholar]
- 2.Luo Y, Raible D, Raper A. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- 3.Kolodkin A, Matthes D, Goodman C. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. doi: 10.1016/0092-8674(93)90625-z. [DOI] [PubMed] [Google Scholar]
- 4.Kolodkin A, Levengood D, Rowe E, Tai Y, Giger R, Ginty D. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Chédotal A, He Z, Goodman CS, Tessier-Lavigne M. Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron. 1997;19:547–559. doi: 10.1016/s0896-6273(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 6.Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- 7.Soker S, Takashima S, Miao H, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 8.Klagsbrun M, Takashima S, Mamluk R. The role of neuropilin in vascular and tumor biology. Adv Exp Med Biol. 2002;515:33–48. doi: 10.1007/978-1-4615-0119-0_3. [DOI] [PubMed] [Google Scholar]
- 9.Roche J, Boldog F, Robinson M, Robinson L, Varella-Garcia M, Swanton B, Waggoner B, Fishel R, Franklin W, Gemmill R, et al. Distinct 3p21.3 deletions in lung cancer, analysis of deleted genes and identification of a new human semaphorin. Oncogene. 1996;12:1289–1297. [PubMed] [Google Scholar]
- 10.Sekido Y, Bader S, Latif F, Chen JY, Duh FM, Wei MH, Albanes JP, Lee CC, Lerman MI, Minna JD. Human semaphorins A (V) and (IV) reside in the 3p21.3 small cell lung cancer deletion region and demonstrate distinct expression patterns. Proc Natl Acad Sci USA. 1996;93:4120–4125. doi: 10.1073/pnas.93.9.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang R, Hensel C, Garcia D, Carlson H, Kok K, Daly M, Kerbacher K, Van Den Berg A, Veldhuis P, Buys C, et al. Isolation of the human semaphorin III/F gene (SEMA3F) at chromosome 3p21, a region deleted in lung cancer. Genomics. 1996;32:39–48. doi: 10.1006/geno.1996.0074. [DOI] [PubMed] [Google Scholar]
- 12.Tomizawa Y, Sekido Y, Kondo M, Gao B, Yokota J, Roche J, Drabkin H, Lerman MI, Gazdar AF, Minna JD. Inhibition of lung cancer cell growth and induction of apoptosis after reexpression of 3p21.3 candidate tumor suppressor gene SEMA3B. Proc Natl Acad Sci USA. 2001;98:13954–13959. doi: 10.1073/pnas.231490898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todd M, Xiang R, Garcia D, Kerbacher K, Moore S, Hensel C, Liu P, Siciliano M, Kok K, van den Berg A, et al. An 80 kb P1 clone from chromosome 3p21.3 suppresses tumor growth in vivo. Oncogene. 1996;13:2387–2396. [PubMed] [Google Scholar]
- 14.Xiang R, Davalos AR, Hensel CH, Zhou XJ, Tse C, Naylor SL. Semaphorin 3F gene from human 3p21.3 suppresses tumor formation in nude mice. Cancer Res. 2002;62:2637–2643. [PubMed] [Google Scholar]
- 15.Kessler O, Shraga-Heled N, Lange T, Gutmann-Raviv N, Sabo E, Baruch L, Machluf M, Neufeld G. Semaphorin-3F is an inhibitor of tumor angiogenesis. Cancer Res. 2004;64:1008–1015. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 16.Bielenberg D, Hida Y, Shimizu A, Kaipainen A, Kreuter M, Kim C, Klagsbrun M. Semaphorin SEMA3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J Clin Invest. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brambilla E, Constantin B, Drabkin H, Roche J. Semaphorin SEMA3F localization in malignant human lung and cell lines: a suggested role in cell adhesion and cell migration. Am J Pathol. 2000;156:939–950. doi: 10.1016/S0002-9440(10)64962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lantuéjoul S, Constantin B, Drabkin H, Brambilla C, Roche J, Brambilla E. Expression of VEGF, semaphorin SEMA3F, and their common receptors neuropilins NP1 and NP2 in preinvasive bronchial lesions, lung tumours, and cell lines. J Pathol. 2003;200:336–347. doi: 10.1002/path.1367. [DOI] [PubMed] [Google Scholar]
- 19.Nasarre P, Kusy S, Constantin B, Castellami V, Drabkin H, Bagnard D, Roche J. Semaphorin SEMA3F has a repulsing activity on breast cancer cells and inhibits E-cadherin-mediated cell adhesion. Neoplasia. 2005;7(2):180–189. doi: 10.1593/neo.04481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasarre P, Constantin B, Rouhaud L, Harnois T, Raymond G, Drabkin HA, Bourmeyster N, Roche J. Semaphorin SEMA3F and VEGF have opposing effects on cell attachment and spreading. Neoplasia. 2003;5:83–92. doi: 10.1016/s1476-5586(03)80020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byzova TV, Goldman CK, Pampori N, Thomas KA, Bett A, Shattil SJ, Plow EF. A mechanism for modulation of cellular responses to VEGF: activation of the integrins. Mol Cell. 2000;6:851–860. [PubMed] [Google Scholar]
- 22.Serini G, Valdembri D, Zanivan S, Morterra G, Burkhardt C, Caccavari F, Zammataro L, Primo L, Tamagnone L, Logan M, et al. Class 3 semaphorins control vascular morphogenesis by inhibiting integrin function. Nature. 2003;424:391–397. doi: 10.1038/nature01784. [DOI] [PubMed] [Google Scholar]
- 23.Kiosses WB, Shattil SJ, Pampori N, Schwartz MA. Rac recruits high-affinity integrin alphavbeta3 to lamellipodia in endothelial cell migration. Nat Cell Biol. 2001;3:316–320. doi: 10.1038/35060120. [DOI] [PubMed] [Google Scholar]
- 24.Chan DC, Earle KA, Zhao TL, Helfrich B, Zeng C, Baron A, Whitehead CM, Piazza G, Pamukcu R, Thompson WJ, et al. Exisulind in combination with docetaxel inhibits growth and metastasis of human lung cancer and prolongs survival in athymic nude rats with orthotopic lung tumors. Clin Cancer Res. 2002;8:904–912. [PubMed] [Google Scholar]
- 25.Varner JA, Cheresh DA. Integrins and cancer. Curr Opin Cell Biol. 1996;8:724–730. doi: 10.1016/s0955-0674(96)80115-3. [DOI] [PubMed] [Google Scholar]
- 26.Guirland C, Suzuki S, Kojima M, Lu B, Zheng JQ. Lipid rafts mediate chemotropic guidance of nerve growth cones. Neuron. 2004;42:51–62. doi: 10.1016/s0896-6273(04)00157-6. [DOI] [PubMed] [Google Scholar]
- 27.Bagnard D, Sainturet N, Meyronet D, Perraut M, Miehe M, Roussel G, Aunis D, Belin MF, Thomasset N. Differential MAP kinases activation during semaphorin 3A-induced repulsion or apoptosis of neural progenitor cells. Mol Cell Neurosci. 2004;25:722–731. doi: 10.1016/j.mcn.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi T, Fournier A, Nakamura F, Wang LH, Murakami Y, Kalb RG, Fujisawa H, Strittmatter SM. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell. 1999;99:59–69. doi: 10.1016/s0092-8674(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 29.Barberis D, Artigiani S, Casazza A, Corso S, Giordano S, Love CA, Jones EY, Comoglio PM, Tamagnone L. Plexin signaling hampers integrin-based adhesion, leading to Rho-kinase independent cell rounding, and inhibiting lamellipodia extension and cell motility. FASEB J. 2004;18:592–594. doi: 10.1096/fj.03-0957fje. [DOI] [PubMed] [Google Scholar]
- 30.Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M. Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. J Cell Biol. 1999;146:233–242. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catalano A, Caprari P, Rodilossi S, Betta P, Castellucci M, Casazza A, Tamagnone L, Procopio A. Cross-talk between vascular endothelial growth factor and semaphorin-3A pathway in the regulation of normal and malignant mesothelial cell proliferation. FASEB J. 2003;18:358–360. doi: 10.1096/fj.03-0513fje. [DOI] [PubMed] [Google Scholar]
- 32.Bachelder R, Lipscomb E, Lin X, Wendt M, Chadborn N, Eickholt B, Mercurio A. Competing autocrine pathways involving alternative neuropilin-1 ligands regulate chemotaxis of carcinoma cells. Cancer Res. 2003;63:5230–5233. [PubMed] [Google Scholar]
- 33.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 34.Ben-Ze'ev A. Cytoskeletal and adhesion proteins as tumor suppressors. Curr Opin Cell Biol. 1997;9:99–108. doi: 10.1016/s0955-0674(97)80158-5. [DOI] [PubMed] [Google Scholar]
- 35.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 36.Felding-Habermann B, Mueller BM, Romerdahl CA, Cheresh DA. Involvement of integrin alpha V gene expression in human melanoma tumorigenicity. J Clin Invest. 1992;89:2018–2022. doi: 10.1172/JCI115811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders LC, Felding-Habermann B, Mueller BM, Cheresh DA. Role of alpha V integrins and vitronectin in human melanoma cell growth. Cold Spring Harb Symp Quant Biol. 1992;57:233–240. doi: 10.1101/sqb.1992.057.01.028. [DOI] [PubMed] [Google Scholar]
- 38.Mitjans F, Sander D, Adan J, Sutter A, Martinez JM, Jaggle CS, Moyano JM, Kreysch HG, Piulats J, Goodman SL. An anti-alpha v-integrin antibody that blocks integrin function inhibits the development of a human melanoma in nude mice. J Cell Sci. 1995;108:2825–2838. doi: 10.1242/jcs.108.8.2825. [DOI] [PubMed] [Google Scholar]
- 39.Stromblad S, Becker JC, Yebra M, Brooks PC, Cheresh DA. Suppression of p53 activity and p21WAF1/CIP1 expression by vascular cell integrin alphavbeta3 during angiogenesis. J Clin Invest. 1996;98:426–433. doi: 10.1172/JCI118808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks PC, Stromblad S, Sanders LC, von Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP, Cheresh DA. Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin alphavbeta3. Cell. 1996;85:683–693. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- 41.Campbell DS, Holt CE. Apoptotic pathway and MAPKs differentially regulate chemotropic responses of retinal growth cones. Neuron. 2003;37:939–952. doi: 10.1016/s0896-6273(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 42.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kok K, van den Berg A, Veldhuis P, van der Veen A, Franke M, Schoenmakers E, Hulsbeek M, van der Hout A, de Leij L, van de Ven W, Buys C. A homozygous deletion in a small cell lung cancer cell line involving a 3p21 region with a marked instability in yeast artificial chromosomes. Cancer Res. 1994;54:4183–4187. [PubMed] [Google Scholar]