Abstract
Insulin-like growth factors (IGF) have mitogenic and antiapoptotic functions, and may be involved in tumor growth. The purpose of the study was to investigate the role of IGF components in seminoma compared to normal testis. Normal testicular tissues from autopsy cases and seminoma from surgery cases were obtained for microarray and real-time reverse transcription polymerase chain reaction (RT-PCR) analysis of IGF-1, IGF-2, IGF receptor type 1 (IGF-R1), IGF-R2, insulin receptor isoforms A (IR-A) and B (IR-B), and IGF-binding proteins (IGFBP) 1–6. IGF-2 was localized by immunohistochemistry. IGFBP-5 protein expression was evaluated by Western blot analysis. mRNA expression in microarray and real-time RT-PCR showed similar tendencies: IGF-1, IGF-R1, IGF-R2, IR-A, and IGFBP-2 were not different in both groups. IGF-2, IR-B, IGFBP-1, IGFBP-4, and IGFBP-6 mRNA were downregulated in seminoma. IGFBP-3 tended to be upregulated in pT1 seminoma, but downregulated in pT2 stages. IGFBP-5 and IGF-2 protein expression correlated with mRNA expression. In conclusion, downregulation of mainly inhibiting IGFBPs may allow a stimulated tumor growth. The downregulated IGF-2 does not seem to be involved in the growth regulation of seminoma. Constantly expressed genes (e.g., IGF-1, IGF-R1, IR-A, and IGFBP-2) may reflect an involvement in spermatogenesis, but may also play a major role in tumor growth as their expression is not downregulated despite the lack of spermatogenesis in tumor tissue.
Keywords: IGF, insulin receptor, germ cell tumor, seminoma, testis
Introduction
Insulin-like growth factors (IGF) exhibit strong mitogenic and antiapoptotic effects and therefore have an important impact on tumor growth. They do not only influence tumor growth, but also migration and adhesion of tumor cells. Many tumors are associated with elevated IGF plasma levels and exhibit an increased local expression of IGF or IGF receptor type 1 (IGF-R1) [1]. IGF-R1 is a tyrosine kinase receptor, which mediates growth-stimulating effects of IGF-1 and IGF-2, such as mitogenesis, cell transformation, cell differentiation, and antiapoptosis [2]. Contrary to IGF-R1, the type 2 receptor, IGF-R2, exclusively binds IGF-2 and serves for the internalization and degradation of IGF-2 [3]. Especially locally produced IGF-2 seems to have important proliferating effects on several cancer entities, such as ovarian, mammary, prostate, and thyroid cancers [4–8]. The mitogenic effects of IGF-2 do not only seem to be transmitted by IGF-R1, but also in an important degree by the isoform A of the insulin receptor (IR-A) [5,8]. This autocrine loop may be an additional stimulator for tumor growth.
IGF-binding proteins (IGFBP) can have both inhibiting and stimulating properties on IGF function. Different phosphorylation stages of the IGFBP influence their affinity for binding IGF, so that the IGF function may be raised or inhibited. The activity and availability of IGFBP may further be affected by different partly specific proteases. Some of the IGFBP have their own receptors (e.g., IGFBP-3 and IGFBP-5) that allow them to bind and directly affect the cell. The intrinsic action of IGFBP-1 may be mediated by α5 β1 integrin [9]. IGFBP-3 is the main intravascular carrier and storage for IGF-1. Receptor binding of IGF is anticipated by forming complexes with acid labile subunit (ALS). IGFBP-4 is a soluble extracellular binding protein that inhibits the IGF/IGF binding site interaction by sequestering IGF. IGFBP-6 differs from the other binding proteins insofar as it is binding IGF-2 with 100-fold higher affinity than IGF-1 [10].
Zhou and Bondy [11] localized the mRNA of the IGF system in normal testicular tissue. IGF-R1 and IGF-R2 mRNA were most abundant in the germinal epithelium, whereas IGF-1 and IGFBP-1 mRNA could not be detected with the applied in situ hybridization. In contrast, immunohistochemistry showed IGF-1 staining in Sertoli cells, primary spermatocytes, and some Leydig cells [12]. The authors also demonstrated positivity for IGF-R1 in secondary spermatocytes, early spermatids, Sertoli cells, and some Leydig cells. In germ cell tumors, IGF-2 mRNA could be found in two of three seminoma [13]. The strongest IGF-2 expression in this study was seen in one embryonal carcinoma. Drescher et al. [14] investigated the distribution of the IGF system in carcinoma in situ (CIS) of the testis using immunohistochemistry. They found the strongest staining for IGFBP-5 compared to the other IGFBP, and therefore postulated an important role for IGFBP-5 by assuming an proliferative advantage for the CIS cells.
Systematic studies on the IGF system in normal testicular tissue and seminoma have not been performed until now. To address this issue, the goal of this study was to quantitatively compare the expression of the IGF system in normal testicular tissues and seminoma with emphasis on the autocrine growth promoter, IGF-2, and the possibly stimulating IGFBP-5, and to derive their meaning for the tumor growth of seminoma.
Materials and Methods
Tissue Collection
Normal testicular tissue (n = 9) was collected from autopsy cases (age: 34–75 years). The selected patients died of myocardial infarction, heart failure, stroke, or ileus. Intact spermatogenesis was verified in histologic examination. Bodies were cooled at 4°C within 2 hours after death, and tissue samples were snap-frozen in liquid nitrogen with the beginning of autopsy. Seminoma tissue (n = 22) was collected from operation specimens of patients with pure classic seminoma (age: 21–62 years). All patients underwent surgery between 1998 and 2002 at the Department of Urology, University Hospital Mannheim (Mannheim, Germany). Patients' informed consent was taken prior to all investigations. Histologic diagnosis and evaluation of the tumor stage were performed by conventional light microscopy. Eight pT1 and 14 pT2 stages were identified. Whereas pT1 stages of seminoma are limited to the testis and epididymal tissues, pT2 stages are characterized by hemangioinvasion or lymphangioinvasion, or by infiltration of the tunica vaginalis. Tissue samples were taken after surgical removal, snap-frozen in liquid nitrogen within 30 minutes, and stored at -80°C until RNA extraction. Histologic characterization and purity of all samples were verified by frozen sections before RNA extraction.
Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA from 30 mg of normal testicular tissue (n = 9) and seminoma (n = 22) was extracted using the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) following the animal tissue protocol of the manufacturer's instructions. Tissue lysates were homogenized with the QIAshredder (Qiagen). RNA was dissolved in water and spectroscopically quantified at 260 nm with the Ultrospec 3100 Pro (Biochrom Ltd., Cambridge, UK). The purity of RNA was verified by optical density (OD) absorption ratio OD260 nm/OD280 nm between 1.80 and 2.06 (mean = 2.0). RNA quality was analyzed using the RNA 6000 Nano LabChip Kit (Agilent Technologies GmbH, Böblingen, Germany) and the Agilent 2100 bioanalyser (Agilent Technologies GmbH) for electrophoretic separation. The 28S/18S rRNA ratio of all samples was between 1.7 and 2.0 (mean = 1.84). RNA quality of samples gained by autopsy and surgery, respectively, did not differ, and only samples with good quality RNA were used for the following reverse transcription reaction.
Constant amounts of 1 µg of total RNA were reverse-transcribed with 200 U of M-MLV Reverse Transcriptase (Promega Corp., Madison, WI) and Random Primers (Promega Corp.) according to the manufacturer's instructions. All investigated samples were transcribed in the same reverse transcription reaction.
The specific primers for quantitative real-time PCR were designed using publicly available sequences from the Nucleotide Sequence Database, NCBI [insulin receptor (IR): accession no. AH002851; IGFBP-1: accession no. M74587; IGFBP-3: accession no. NM_000598; IGFBP-5: accession no. NM_000599; and IGFBP-6: accession no. NM_002178] or used according to literature [15] (Table 1). A master mix was prepared as follows: 6.4 µL of water, 1.2 µL of MgCl2 (25 mM), 0.2 µL of forward primer (20 µM), 0.2 µL of reverse primer (20 µM), and 1.0 µL of LightCycler Fast Start DNA Master SYBR Green I (Roche Diagnostics, Mannheim, Germany). Nine microliters of the master mix was filled in glass capillaries and 1 µL of PCR template containing 25 ng of reverse-transcribed total RNA was added. To ensure an accurate quantification of the desired product, a high-temperature fluorescence measurement in a fourth segment of the PCR run was performed [16]. The following general real-time PCR protocol was employed: denaturation for 10 minutes at 95°C; 40 (IGF-2, IGFBP-2, IGFBP-3, and IGFBP-5), 45 (IGF-1, IGF-R1, IR, IR-A, IR-B, IGFBP-1, IGFBP-4, and IGFBP-6), or 80 (IGF-R2) cycles of a four-segment amplification and quantification program; a melting step by slow heating from 60°C to 99°C with a rate of 0.1°C/s and continuous fluorescence measurement; and a final cooling down to 40°C. The four-segment amplification and quantification program was carried out as follows: 15 seconds of denaturation at 95°C; 10 seconds of annealing at 58°C (IGFBP-1, IGFBP-2, IGFBP-3, IGFBP-5, and IGFBP-6), 60°C (IGF-R2, IR-A, and IR-B), 62°C (IGF-1, IGF-2, and IR), 63°C (IGF-R1), and 66°C (IGFBP-4), respectively; 15 seconds of elongation at 72°C; and 5 seconds of fluorescence acquisition at 80°C (IGFBP-1), 81°C (IGFBP-3), 84°C (IGF-R1), 85°C (IR, IR-A, IR-B, and IGFBP-5), 86°C (IGFBP-6), 87°C (IGF-R2), 88°C (IGF-1 and IGFBP-2), and 89°C (IGF-2 and IGFBP-4), respectively. Crossing point (CP) values were acquired by using the second derivative maximum method of the LightCycler Software 3.5 (Roche Diagnostics). CP is the number of PCR cycles when maximal acceleration of the fluorescence increase is reached. The earlier the fluorescence increases, the higher is the concentration of the measured mRNA. All CP of the 31 samples per investigated factor were detected in one run to eliminate interassay variance. Real-time PCR efficiencies were acquired by amplification of a standardized dilution series of the template cDNA and the given slopes in the LightCycler Software 3.5 (Roche Diagnostics). The corresponding efficiencies (E) were then calculated according to the equation: E = 10[-1/slope] [17]. The specificity of the products was documented with a high-resolution gel electrophoresis and analysis of the melting temperature, which is product-specific [15] (for specific melting temperatures, see Table 1). PCR products were purified with the Nucleo Spin Extraction Kit (Macherey-Nagel, Düren, Germany) and sent for commercial sequencing (GENterprise GmbH, Mainz, Germany). The results were compared to the known sequences in the NCBI database.
Table 1.
Primer Sequence, Product Length, Product-Specific Melting Temperature, Real-Time PCR Efficiency, Mean (n = 31) Coefficient of Variation in % (CV%), and Range of Crossing Points (CP) in Human Testis and Seminoma, Respectively, for the Investigated Factors.
| Factor | Primer Sequence | Length (bp) | Melting Temperature (°C) | PCR Efficiency | Mean CV% | CP Range |
| IGF-1 | For 5′-TCGCATCTCTTCTATCTGGCCCTGT-3′ | 240 | 90.4 | 1.73 | 6.97 | 28.98–33.91 |
| Rev 5′-GCAGTACATCTCCAGCCTCCTCAGA-3′ | ||||||
| IGF-2 | For 5′-GACCGCGGCTTCTACTTCAG-3′ | 203 | 91.8 | 1.89 | 11.3 | 20.31–31.05 |
| Rev 5′-AAGAACTTGCCCACGGGGTAT-3′ | ||||||
| IGF-R1 | For 5′-TTAAAATGGCCAGAACCTGAG-3′ | 314 | 87.2 | 2.15 | 1.45 | 31.06–34.78 |
| Rev 5′-ATTATAACCAAGCCTCCCAC-3′ | ||||||
| IGF-R2 | For 5′-TACAACTTCCGGTGGTACACCA-3′ | 144 | 88.3 | - | 6.90 | 55.71–64.68 |
| Rev 5′-CATGGCATACCAGTTTCCTCCA-3′ | ||||||
| IR | For 5′-GCTGAAGCTGCCCTCGAGGA-3′ | 290 + 254 | 87.6 | 1.88 | 4.58 | 22.23–27.53 |
| Rev 5′-CGGCCACCGTCACATTCCCA-3′ | ||||||
| IR-A | For 5′-GCTGAAGCTGCCCTCGAGGA-3′ | 210 | 87.0 | 1.87 | 3.94 | 22.61–27.32 |
| Rev 5′-CGAGATGGCCTGGGGACGAA-3′ | ||||||
| IR-B | For 5′-GCTGAAGCTGCCCTCGAGGA-3′ | 244 | 87.0 | 1.99 | 4.86 | 23.59–30.54 |
| Rev 5′-AGATGGCCTAGGGTCCTCGG-3′ | ||||||
| IGFBP-1 | For 5′-TCAAAAAATGGAAGGAGCCCT-3′ | 127 | 82.3 | 2.12 | 2.22 | 30.18–33.78 |
| Rev 5′-AATCCATTCTTGTTGCAGTTT-3′ | ||||||
| IGFBP-2 | For 5′-CACCGGCACATGGGCAA-3′ | 136 | 90.0 | 1.98 | 3.19 | 26.50–31.40 |
| Rev 5′-GAAGGCGCATGGTGGAGAT-3′ | ||||||
| IGFBP-3 | For 5′-CTACAAAGTTGACTACGAGTC-3′ | 139 | 84.3 | 1.73 | 5.74 | 24.47–30.05 |
| Rev 5′-ACTCAGCACATTGAGGAACTT-3′ | ||||||
| IGFBP-4 | For 5′-GCCCTGTGGGGTGTACAC-3′ | 342 | 92.7 | 1.55 | 7.18 | 31.60–36.87 |
| Rev 5′-TGCAGCTCACTCTGGCAG-3′ | ||||||
| IGFBP-5 | For 5′-AGCAAGTCAAGATCGAGAGAGA-3′ | 154 | 88.5 | 1.64 | 5.75 | 23.23–30.76 |
| Rev 5′-TTCTTTCTGCGGTCCTTCTTCA-3′ | ||||||
| IGFBP-6 | For 5′-AGAGGAGAATCCTAAGGAGAGT-3′ | 229 | 89.5 | 1.66 | 5.64 | 25.44–33.27 |
| Rev 5′-ATTGGGCACGTAGAGTGTTTGA-3′ | ||||||
Microarray
The gene expression profiles of 40 seminoma of different stages [pT1 (n = 22), pT2 (n = 14), and pT3 (n = 4)] were compared with normal testicular tissue (n = 3) using tissue homogenates and cDNA microarrays focusing on the expression of the IGF-associated genes. One of these normal testicular tissues and 17 of the seminoma specimens were part of the sample collective in Mannheim taken for real-time RT-PCR investigations. Tissue samples were taken from patients aged 25 to 58 years. All patients underwent surgery between 1995 and 2002 at the Department of Urology in Mannheim, Essen, or Münster. Patients' informed consent was taken prior to all investigations.
Testicular tissues used for microarray analyses were lysed in TRIzol (Life Technologies, Karlsruhe, Germany) and total cellular RNA was prepared using the RNeasy Mini Kit (Qiagen), according to the manufacturer's protocol. RNA quality was controlled spectrophotometrically and by gel electrophoresis. Preparation of cDNA targets starting from 5 to 10 µg of total RNA, fragmentation, hybridization to HG-U95Av2 arrays, washing, staining, and scanning were performed following the manufacturer's protocols. Expression values (signal and detection calls: A—absent; P—present; and M—marginal) were obtained by performing the absolute analysis using Affymetrix microarray Suite Version 5.0. This includes the background correction of the average of the lowest two percentiles of intensities on a four-by-four grid on the chip, the introduction of an “ideal mismatch” forced to be lower than the corresponding perfect match, and the usage of Tukey's biweight to elicit an expression value out of single probe intensity pairs. Global scaling was applied to allow comparison of gene signals across multiple microarrays. Annotation of the probe sets was taken from the annotation files provided on the Affymetrix homepage [18].
Immunohistochemistry
Immunohistochemistry was performed in nine normal testicular tissues and 22 seminoma using a mouse monoclonal antibody to human IGF-2 (clone W3D9; Serotec Ltd., Oxford, UK). Cross reactivity with human IGF-1 and insulin is < 0.01%. Tissue samples were fixed in Bouin's solution for 24 hours at room temperature (21°C) and embedded in paraffin wax after dehydration. Sections of 3 µm were incubated with the IGF-2 antibody at a 1:40 dilution at 4°C overnight. Immunoreactive sites were detected using incubation with a 1:1000 dilution of biotin SP-conjugated goat antimouse IgG/IgM (Jackson Immuno Research, West Grove, PA) for 30 minutes at room temperature (21°C) and subsequent incubation with streptavidin-conjugated alkaline phosphatase (Jackson Immuno Research) diluted to 1:250 at room temperature (21°C) for 30 minutes. Immunoreactive sites were developed using Fuchsine substrate chromogen (Dako, Glostrup, Denmark). Sections were counterstained with haematoxylin and mounted in Kaiser's glycerol gelatin (Merck, Darmstadt, Germany). Stained slides were examined to identify the cellular localization of IGF-2 immunoreactivity.
Western Blot
Samples of frozen tumor (n = 22) and normal testicular tissue (n = 9) were homogenized in lysis buffer (150 mM NaCl, 105 mM ethylenediaminetetraacetate, 3% glycerol, 1 mg/ml bovine serum albumin, 1% IGEPAL CA-630; Sigma-Aldrich Chemie, Steinheim, Germany). After determination of the protein concentration using a Bradford assay, a cocktail of proteinase inhibitors [1 µg/ml leupeptin (Amersham Pharmacia Biotech, Freiburg, Germany), 5 µg/ml aprotinin (Roth, Karlsruhe, Germany), 2 µg/ml pepstatin A (Amersham Pharmacia Biotech), and 0.5 mM pefabloc (Roche Diagnostics)] was added. From each sample, 20 µg of total protein was separated on 18% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. The membranes were blocked with 3% skim milk (Slimfast, Wiesbaden, Germany) and incubated overnight at room temperature (21°C) with a rabbit polyclonal anti-human IGFBP-5 antibody (Upstate, Lake Placid, NY) at a dilution of 1:1000. After thorough washing, an alkaline phosphatase-conjugated goat anti rabbit antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was applied at a 1:1000 dilution for 60 minutes at room temperature (21°C). Bands were visualized using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) (Roche Diagnostics) in AP buffer (100 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 50 mM MgCl2). The expression of IGFBP-5 was densitometrically analyzed with the AIDA 1D Software (AIDA; Raytest Isotopenmessgeräte GmbH, Straubenhardt, Germany).
Statistical Analysis
Evaluation of the real-time PCR results. Data were analyzed by the Relative Expression Software Tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR [19]. This software calculates an expression ratio in regard to the control group (normal testicular tissue). The expression ratio (R) is:
with E = efficiency. REST also indicates coefficients of variation (CV) in percent and standard deviations based on the CP of the target gene. The data are shown as a maximal cycle number of 40 minus the acquired CP ± SEM. The higher the 40 - CP values, the higher is the concentration of the target gene. As PCR amplification is an exponential process, a difference of two CP (ΔCP = 2) signifies approximately a regulation by a factor of E2 (with E=efficiency) and is indicated in the text according to the expression ratio calculated by REST.
Evaluation of microarray results. Exploratory data analysis, Wilcoxon paired nonparametric tests for difference, and the Mann-Whitney test for the nonparametric independent two-group comparisons were performed with the program SPSS 10 for Windows (SPSS, Inc., Chicago, IL) and Microsoft Excel 2000 (Microsoft Corp., Redmond, WA). Differences with P < .05 were regarded as statistically significant.
Evaluation of Western blot results. Data were tested with the Mann-Whitney rank sum test. Differences with P ≤ .001 were regarded as highly significant. The data are shown as OD, calculated as integral (Int) minus background (Bkg).
Results
Real-Time PCR
The investigated transcripts showed high real-time PCR efficiencies, low coefficients of variation (CV%), CP ranges from 20.31 to 36.87 and 65.87, respectively (Table 1), and a good linearity over the range of 0.2 to 50 ng of cDNA input.
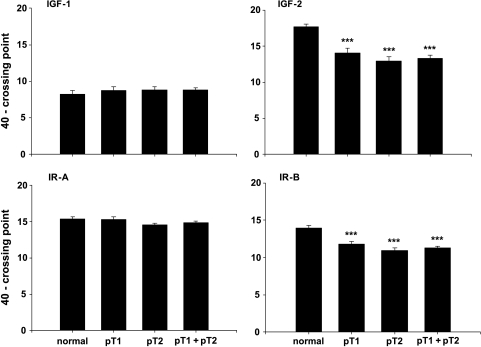
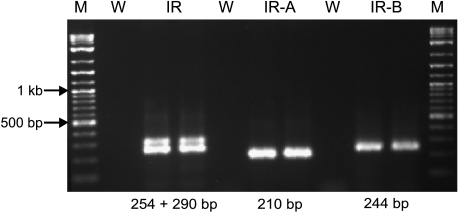
IGF-1 (Figure 1) and IGF-R1 (data not shown) mRNA showed no significantly different expressions in normal testis and seminoma. IGF-2 mRNA was significantly downregulated in seminoma by a factor of 16 (P < .001), with a maximal downregulation in pT2 stages by a factor of 21 (P < .001) (Figure 1). IGF-R2 expressions were not significantly different between groups and very low with CP between 55.71 and 65.68 (data not shown). Amplification with the IR primer pair showed the two isoforms of the insulin receptor with stronger expression of the smaller isoform, IR-A (Figure 2). Specific amplification of the two isoforms revealed a constant expression of IR-A in both normal testicular tissue and seminoma, and a downregulation of IR-B in seminoma (P < .001) with maximal downregulation by a factor of 8 (P < .001) in pT2 stages (Figure 1).
Figure 1.
Expression data (mRNA) for IGF-1, IGF-2, IR-A, and IR-B in normal testicular tissue and pT1, pT2, and pT1 + pT2 stages of seminoma. Data are shown as a maximal cycle number of 40 minus the acquired crossing point (40 - CP) ± SEM. Significance is indicated as follows: ***P < .001.
Figure 2.
Specific PCR products for IR (290 + 254 bp), amplifying both insulin receptor isoforms, IR-A (210 bp) and IR-B (244 bp). M = marker; W = water. Notice the pronounced expression of the smaller isoform in the IR product.
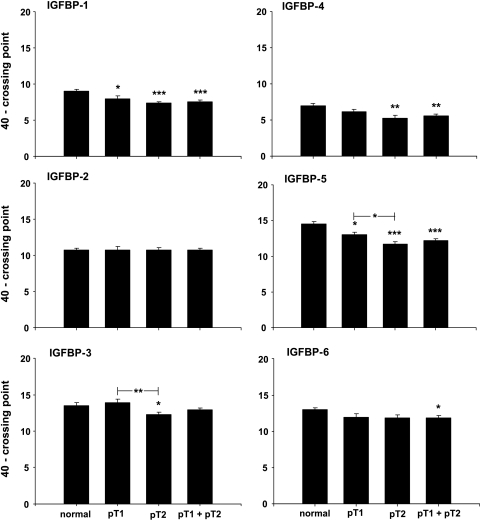
IGFBP-1 mRNA was downregulated by a factor of 3 (P < .001), with a maximal 3.5-fold (P < .001) downregulation in pT2 stages (Figure 3). There were no significant differences in IGFBP-2 expression between groups. IGFBP-3 expression showed a slight increase in pT1 stages but a significant downregulation by a factor of 2 (P < .05) in pT2 stages when compared to normal testis. Further, IGFBP-3 was significantly downregulated by a factor of 2.4 (P < .01) in pT2 stages, when compared to pT1 stages. IGFBP-4 was downregulated in seminoma by a factor of 1.8 (P < .01). Although the downregulation of IGFBP-4 was not significant between pT1 stages and the control group, it became significant (P < .01) when comparing pT2 stages with the control group (downregulation by a factor of 2.1). IGFBP-5 mRNA expression was downregulated by a factor of 3.1 (P < .001) in seminoma with a maximal downregulation by a factor of 4 (P < .001) in pT2 stages. IGFBP-5 mRNA was significantly downregulated by a factor of 1.9 (P < .05) in pT2 stages compared with pT1 stages. There is a significant downregulation by a factor of 1.7 (P < .05) for IGFBP-6 expression in seminoma, but there was no significant difference when comparing pT1 and pT2 stages separately with normal testicular tissue.
Figure 3.
Expression data (mRNA) for IGFBP-1, IGFBP-2, IGFBP-3, IGFBP-4, IGFBP-5, and IGFBP-6 in normal testicular tissue and pT1, pT2, and pT1 + pT2 stages of seminoma. Data are shown as a maximal cycle number of 40 minus the acquired crossing point (40 - CP) ± SEM. Significance is indicated as follows: *P < .05, **P < .01, and ***P < .001.
Microarray
Microarray analysis identified regulated genes in three normal testicular specimens and 40 seminoma of different stages [pT1 (n = 22), pT2 (n = 14), and pT3 (n = 4)]. The gene expression profiles of four samples of tumor stage pT1 were compared with those of three normal testicular tissue samples resulting in 12 single comparisons representative for the 43 samples investigated. The expression of 1490 probe sets was determined as differentially expressed in at least 80% of the comparisons with a P value of < .001 and > .999 for 635 increased changes and 855 decreased changes, respectively. Furthermore, we focused on the regulation of the expression of the following IGF-related genes: IGF-1, IGF-2, IGF-R1, IGF-R2, IGFBP-1, IGFBP-2, IGFBP-3, IGFBP-4, IGFBP-5, and IGFBP-6 (Table 2).
Table 2.
Expression Data (mRNA) for IGF-Related Genes Present on the Array in 43 Tissue Samples Regarding Different Tumor Stages.
| Gene | Probe Set ID | NCBI ID | Product | P-call (%) | NT | pT1 | pT2 | pT3 | ||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||||
| IGF-1 | 1501_at | X57025 | Complete cds | 91 | 801 | 518 | 775 | 424 | 572 | 584 | 335 | 281 |
| 1975_s_at | X03563 | Potential IGF-1 precursor | 88 | 906 | 371 | 1763 | 1303 | 1370 | 808 | 1755 | 1017 | |
| 38737_at | M14156 | Exon 4 | 77 | 304 | 191 | 241 | 169 | 194 | 143 | 104 | 26 | |
| IGF-R1 | 1335_at | X04434 | Complete cds | 2 | 271 | 91 | 125 | 129 | 99 | 73 | 412 | 424 |
| 34718_at | X04434 | Complete cds | 100 | 510 | 127 | 518 | 282 | 484 | 299 | 864 | 678 | |
| IGF-2 | 1591_s_at | J03242 | Complete cds | 0 | 96 | 19 | 76 | 71 | 60 | 63 | 92 | 89 |
| 2079_s_at | M13970 | Exon 1 of four | 26 | 244 | 4 | 339 | 142 | 315 | 92 | 559 | 133 | |
| 36782_s_at | J03242 | Complete cds | 0 | 29 | 32 | 13 | 9 | 20 | 16 | 59 | 88 | |
| IGF-R2 | 160027_s_a | Y00285 | Complete cds | 100 | 1295 | 98 | 1385 | 583 | 1250 | 397 | 893 | 380 |
| IGFBP-1 | 1232_s_at | M74587 | Complete cds | 42 | 90 | 24 | 93 | 42 | 80 | 38 | 95 | 20 |
| IGFBP-2 | 1741_s_at | S37730 | Precursor | 91 | 701 | 165 | 928 | 893 | 826 | 658 | 1570 | 1148 |
| 40422_at | X16302 | Complete cds | 100 | 1450 | 248 | 1791 | 1436 | 1794 | 1279 | 2713 | 1536 | |
| IGFBP-3 | 1586_at | M35878 | Complete cds | 79 | 426 | 96 | 934 | 663 | 581 | 365 | 513 | 520 |
| 37319_at | M35878 | Complete cds | 79 | 501 | 246 | 1376 | 832 | 950 | 570 | 980 | 1050 | |
| IGFBP-4 | 1737_s_at | M62403 | Complete cds | 88 | 1040 | 540 | 1424 | 736 | 915 | 432 | 1518 | 1370 |
| 39781_at | U20982 | Promoter and complete cds | 100 | 3741 | 1876 | 4529 | 2072 | 2939 | 1163 | 3552 | 3412 | |
| IGFBP-5 | 1396_at | L27560 | 3′ UTR | 81 | 2282 | 2229 | 839 | 685 | 576 | 510 | 945 | 1204 |
| 1601_s_at | L27559 | Partial exon 4 | 0 | 19 | 5 | 23 | 15 | 26 | 25 | 61 | 40 | |
| 1677_at | M65062 | Complete cds | 7 | 151 | 64 | 149 | 76 | 159 | 81 | 245 | 144 | |
| 1678_g_at | M65062 | Complete cds | 7 | 101 | 130 | 33 | 30 | 32 | 26 | 66 | 29 | |
| 38650_at | L27560 | 3′ UTR | 88 | 3003 | 3248 | 987 | 714 | 689 | 631 | 1237 | 1779 | |
| 41420_at | AF055033 | Complete cds | 9 | 112 | 149 | 45 | 34 | 47 | 28 | 70 | 63 | |
| IGFBP-6 | 1736_at | M62402 | Complete cds | 65 | 566 | 459 | 525 | 634 | 379 | 274 | 315 | 248 |
IGF-1 and IGF-R1 were constantly expressed in seminoma and normal testicular tissue. Two of three probe sets for IGF-2 were under the threshold level of the microarray. The third probe set covered exon 1 of four and showed a slight increase in expression from normal testis to seminoma (P = .066) with a significant difference between normal testicular tissue and tumor stage pT1 (P = .046), as well as between pT1 and pT3 (P = .009). The IGF-R2 gene was constantly expressed in all 43 samples.
There were no significant differences in the expression of IGFBP-1 and IGFBP-2 between the different groups. IGFBP-2 mRNA expression correlated significantly (r = 0.64 and r = 0.564, P ≤ .001) with IGF-R1 mRNA. The expression of IGFBP-3 was measured on two probe sets and showed a significant increase (P ≤ .05) from normal testicular tissue to seminoma tissues with tumor stage pT1 in one probe set and decreasing tendencies during further tumor progression. Similar tendencies were identified for the expression of IGFBP-4 with a slight increase in pT1 stages and a decrease (P ≤ .05) from pT1 to pT2 stages. The expression of both genes (IGFBP-3 and IGFBP-4) correlated significantly in all 43 samples. Considering the different probe samples giving inconsistent data (Table 2), the expression of IGFBP-5 showed a decreasing tendency in seminoma when compared to normal testicular tissue. The hybridization intensity in three of five probe sets with positive IGFBP-5 was significantly decreased in seminoma stages pT1 and pT2 when compared to normal testicular tissue. IGFBP-6 was constantly expressed in seminoma and normal testicular tissue.
DLK1 (delta Drosophila homolog-like 1 gene) and the insulin-like hormone INSL3 are expressed by Leydig cells [20,21], whereas follicle-stimulating hormone receptor (FSH-R) and androgen receptor (AR) are expressed by Sertoli cells [22,23] and therefore act as Leydig cell and Sertoli cell markers, respectively. IGF-R1 and IGF-2 correlated positively with FSH-R, whereas IGFBP-3 showed a negative correlation with FSH-R and AR (Table 3). IGFBP-5 correlated positively with both FSH-R/AR and DLK3/INSL3.
Table 3.
Pearson's Correlation Coefficient for Expression of IGF-Related Genes and Sertoli/Leydig Cell Markers in 43 Samples Measured by Microarrays.
| FSH-R | AR | AR | DLK1 | INSL3 | |
| IGF-1 | |||||
| IGF-R1 | 0.422** | ||||
| IGF-2 | 0.354* | ||||
| IGF-R2 | |||||
| IGFBP-1 | |||||
| IGFBP-2 | |||||
| IGFBP-3 | -0.328* | -0.361* | |||
| -0.318* | -0.367* | ||||
| IGFBP-4 | |||||
| IGFBP-5 | 0.510*** | 0.357* | 0.416** | 0.516*** | 0.703*** |
| 0.405** | 0.437** | 0.351* | 0.453** | 0.675*** | |
| 0.415** | 0.454** | 0.514*** | 0.672*** | ||
| 0.502*** | 0.695*** | ||||
| IGFBP-6 | |||||
Different correlation coefficients indicate results of multiple probe sets.
P ≤ .05.
P ≤ .01.
P ≤ .001.
Table 4 summarizes the results from the microarrays in comparison with real-time RT-PCR.
Table 4.
Comparison of Microarray and Real-Time PCR Data: mRNA Expression in Seminoma Compared to Normal Testis Regarding Data from the Literature.
| Gene | Microarray | RT-PCR | Literature | |
| NT (n = 3), pT1 (n = 22), pT2 (n = 14), pT3 (n = 4) | NT (n = 9), pT1 (n = 8), pT2 (n = 14) | Testis [11] | CIS [14] | |
| IGF-1 | Constant | Constant | mRNA not detected | IHC positive |
| IGF-R1 | Constant | Constant | mRNA in germinal epithelium | |
| IGF-2 | 2x no expression, 1x increase in pT1 and pT3 | Decreased | mRNA in vessels, peritubular tissue, and connective tissue | IHC negative |
| IGF-R2 | Constant | Constant | mRNA in germinal epithelium | |
| IGFBP-1 | Constant | Decreased | Not detected | IHC weak |
| IGFBP-2 | Constant | Constant | In Leydig and Sertoli cells | IHC positive |
| IGFBP-3 | Increased in pT1, slightly decreased in pT2 and pT3 | Slightly increased in pT1, decreased in pT2 | mRNA in vessels | IHC weakly positive |
| IGFBP-4 | Slightly increased in pT1, decreased in pT2 | Decreased | mRNA in vessels, Leydig cells, and connective tissue | IHC weak |
| IGFBP-5 | Decreased in pT1 and pT2 | Decreased | mRNA in connective tissue and little in Leydig cells | IHC positive |
| IGFBP-6 | Constant | Decreased | mRNA peritubular and interstitial | IHC weak |
IHC = immunohistochemistry.
Immunohistochemistry
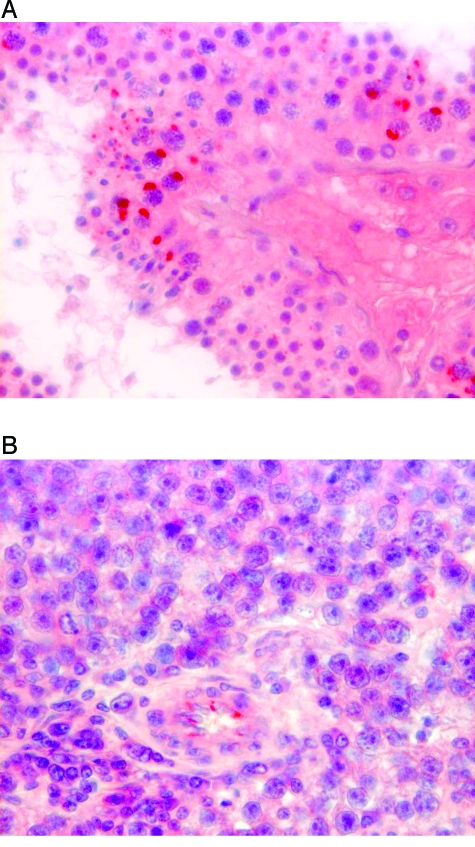
Normal testicular tissue showed strong cytoplasmatic IGF-2 staining in primary and secondary spermatocytes and rarely in some Sertoli cells in terms of paranuclear dots (Figure 4A) accumulating in a cuneiform fashion. Spermatogonia and spermatids were negative. All tubules were positive with approximately 30% to 50% of the spermatocytes per tubule expressing IGF-2. Typical staining was also seen in tumor adjacent tissues of seminoma cases, if there was still any intact spermatogenesis. Most of the tumor cases showed “burned out” tubules with nonmaturating germ cells or Sertoli cells only in the adjacent tumor-free tissue. In two cases, parts of the epididymis could be assessed as well: the rete testis and the efferent ductuli showed a strong luminal staining of the epithelia in a vesicular manner, and the epididymal duct showed a distinct staining of basal cells of its epithelial cell layer.
Figure 4.
Representative immunohistochemically-stained tissue sections of normal testicular tissue and seminoma. (A) Strong IGF-2 staining in terms of paranuclear dots in primary and secondary spermatocytes and some Sertoli cells; no staining of spermatogonia and seminoma. (B) Weak IGF-2 staining of tumor cells and a distinct staining of endothelial cells of some smaller capillaries; original magnification, x 250.
Tumor cells in all investigated seminoma were weakly positive with a rather diffuse cytoplasmatic staining for IGF-2 (Figure 4B). The staining appeared less intensive than in normal testicular tissue. Some endothelial cells of smaller capillaries showed a clear positive staining for IGF-2, also in terms of a paranuclear vesicle.
Western Blot
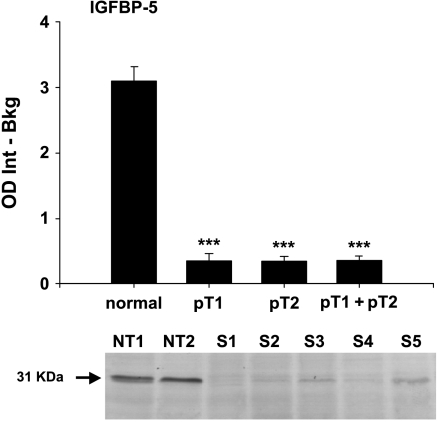
Western blot analysis of IGFBP-5 showed a weak protein expression in seminoma and a strong protein expression in normal testicular tissue (P < .001). Both pT1 and pT2 stages are significantly downregulated (both P < .001) when compared separately with normal testicular tissue. IGFBP-5 protein expression in pT1 stages compared to pT2 stages was not significantly different but showed a slight decrease in pT2 stages (median pT1 = 0.26, median pT2 = 0.24). Figure 5 shows a representative blot with the IGFBP-5-specific band at 31 kDa.
Figure 5.
Protein expression of IGFBP-5 in normal testicular tissue and pT1, pT2, and pT1 + pT2 stages of seminoma. Data are shown as the integral of optical density minus background (OD Int - Bkg). Significance is indicated as follows: ***P < .001. Underneath, a representative blot with specific bands at 31 kDa for normal testicular tissue (NT1 and NT2) and seminoma (S1–S5).
Discussion
The differential expression of IGF, their binding proteins, and receptors in normal versus seminoma tissue has been investigated. Protein and mRNA of IGFBP-5 showed the same expression pattern in normal testicular tissue and seminoma. Regarding the immunohistochemical expression profile of IGF-2, the strong IGF-2 expression in normal testicular tissue and the lower expression in tumor tissue, as seen in real-time PCR, seem to be comprehensible. For these two factors, mRNA expression is in accordance with protein expression.
IGF-R2 mRNA is only very weakly expressed in testicular tissue and seminoma. CP of 55 to 65 show that this receptor is rarely produced in this tissue. However, Zhou and Bondy [11] could verify IGF-R2 mRNA by in situ hybridization in normal testicular tissue. High CP can also be caused by inhibition of the PCR reaction. But melting curve and gel analysis showed no primer dimers, and sequencing resulted in a clear specific product. However, we cannot exclude tissue-specific inhibition of the reaction by unknown factors.
Studies in roe deer and goats suggest a specific role for IGF in the paracrine control of spermatogenesis [24,25]. The pronounced expression of IGF-2 in terms of paranuclear vesicles in primary and secondary spermatocytes, as well as in some Sertoli cells, implicates a similar role in human testis. This concept is supported by the correlation of IGF-2 and FSH-R expression, with the latter being a Sertoli cell marker. Parts of the epididymis could be analyzed in two cases. The strong, luminal, and vesicular staining in the rete testis and the efferent ductuli suggests a reabsorption of IGF-2 during the epididymal passage. Possibly IGF-2 is also involved in maintaining the function of the epididymal epithelium.
IGF-2 mRNA was previously demonstrated by Northern blot analysis in a small subset of germ cell tumors. In two of three seminoma, IGF-2 was present. The most abundant IGF-2 mRNA expression, however, was seen in one embryonal carcinoma case [13]. There are some references that propose a crucial role for IGF-2 in tumor growth [5,6,8]. In our study, mRNA expression results are somewhat contradictory. In the microarray data, two probe sets covering the complete coding sequence did not detect any IGF-2 expression, whereas one probe set covering exon 1 of four showed a slight but significant increase in pT1 and pT3 stages. In contrast, in real-time PCR, IGF-2 mRNA is 16-fold downregulated in seminoma compared to normal testicular tissue. Immunohistochemistry is not a quantitative method, but showed a weaker staining of tumor cells compared to normal testis. Concerning IGF-2, the microarray seems to be less sensitive than real-time PCR, and we regard the real-time PCR and immunohistochemical results as more reliable. Therefore, it seems to be unlikely that the tumor-stimulating effects of IGF-2 play a major role in the growth regulation of seminoma. These findings correspond to an immunohistochemical study of Drescher et al. [14], who found no IGF-2 staining in CIS—the precursor cells of testicular germ cell tumors.
IGF-2 is bound by IGFBP-6 with a 100-fold higher affinity than IGF-1 [10]. IGFBP-6 has primarily inhibiting effects on tumor growth [26–28]. In seminoma, IGFBP-6 is constantly expressed in microarray data and slightly downregulated in RT-PCR data. This downregulation, however, is not as pronounced as the IGF-2 decreases. Thus, the ratio of IGFBP-6 to IGF-2 increases and therefore intensifies the inhibition of tumor growth.
However, IGF-2 is the most strongly expressed mRNA, when compared to the other IGF components, even in the tumor tissue despite downregulation. RT-PCR cannot discriminate which cells produce the measured target gene expression. High expression levels may be caused by the tumor cells themselves, or by any interstitial or vascular tissue. The particular staining of endothelial cells of tumor capillaries in seminoma supports the finding of Ritter et al. [29] that IGF-2 is involved in tumor angiogenesis and strengthens the role of IGF-2 in tumor growth.
IGF-2 may act as an autocrine stimulator of tumor growth through the activation of IR-A [5,8]. The constant expression of IR-A and the pronounced downregulation of IR-B increase the relative abundance for the IGF-2 binding isoform A of the insulin receptor and therefore the mitogenic potential.
IGFBP generally have, with some exceptions, a rather inhibiting effect on IGF action in tumor tissues [1]. Thus, a downregulation of IGFBP results in less inhibited IGF action, increased binding of IGF to their receptors, and therefore increased antiapoptosis, mitogenesis, and tumor growth. In the case of a comparison between functional testicular tissue and proliferating tumor tissue, a downregulation in the tumor tissue may just reflect the lack of spermatogenesis. This may, in particular, apply to IGFBP-5, whose expression strongly correlates with Sertoli and Leydig cell markers. IGFBP-5 can have IGF-inhibiting and IGF-stimulating effects [30–32]. Its downregulation in seminoma may mean both a stimulated tumor growth by less inhibition, or an inhibited tumor growth by less stimulated mitogenesis. In an immunohistochemical study of CIS in the testis [14], IGFBP-5 was identified as the most strongly expressed IGFBP. The authors presume a prominent role of IGFBP-5 in the onset of testicular germ cell tumor. Regarding the results of our study, we can confirm the finding of IGFBP-5 to be the most strongly expressed IGFBP. Considering the significant difference in mRNA expression between pT1 and pT2 stages, IGFBP-5 may indeed also be involved in the progression of tumor growth in seminoma—but rather because of its downregulation, not because of its strong expression. Further studies (e.g., cell cultures) are needed to elucidate the specific function of IGFBP-5 in seminoma.
In transgenic mice, overexpression of IGFBP-1 resulted primarily in an inhibition of IGF action [33]. In prostate cancer cell lines (LNCaP cells), IGFBP-1 reduces cell growth and induces apoptosis by an IGF-dependent mechanism [34]. The mainly IGF-inhibiting role of IGFBP-1 implies that the downregulation of IGFBP-1 in seminoma may be involved in increased IGF action and tumor growth.
IGFBP-3 is the main intravascular carrier and storage for IGF-1. Receptor binding of IGF is anticipated by forming complexes with ALS. The successful treatment of breast cancer with Tamoxifen, an antiestrogen, induces elevated IGFBP-3 level and decreased IGFBP-3 proteolysis [35]. IGFBP-4 is a soluble, extracellular binding protein that inhibits the IGF/IGF binding site interaction by sequestering IGF. In malignant M12 epithelial cells of the prostate, overexpression of IGFBP-4 caused delayed tumor cell growth and increased apoptosis [36]. The correlation of both IGFBP expressions suggests a common regulation. The downregulation of both IGFBP-3 and IGFBP-4 in seminoma may contribute to a less inhibited tumor growth. The significant downregulation of IGFBP-3 in pT2 stages compared to pT1 stages may indicate this participation in tumor progression. The negative correlation of IGFBP-3 with the Sertoli cell markers argues for an IGFBP-3 regulation that is independent of functional spermatogenesis. The slight increase of IGFBP-3 expression in pT1 stages compared to normal testicular tissue may represent an attempt of counterregulation to inhibit a stimulated tumor growth.
There are several evidences that IGF-R1 overexpression is associated with tumor cell proliferation, adhesion, and metastasis [1]. But as it is constantly expressed in normal testicular tissue and seminoma, it does not seem to play a major role for the growth of seminoma. However, germ cells are a very active and proliferating tissue, and it is known that the IGF system is involved in spermatogenesis [11,12]. Furthermore, IGF-R1 expression correlates with FSH-R expression, a Sertoli cell marker, which is a hint that IGFR1 takes part in spermatogenesis. In the rather homogenously growing classic seminoma, Sertoli cells no longer exist. A missing downregulation in tumor tissue despite lack of spermatogenesis could therefore indicate a major role in tumor growth. Similar to IGF-R1, IGF-1 and IR-A do not show any significant differences in mRNA expression between normal tissue and seminoma.
There seem to be inhibiting and stimulating influences on IGF function for IGFBP-2: in non small cell lung cancer cell lines, IGFBP-2 inhibited the binding of IGF-1 and IGF-2 to their receptor IGF-R1 [37], and may thereby inhibit tumor growth. In contrast, glioblastoma cell lines overexpressing IGFBP-2 showed an increase in invasion-related genes and an increased invasion rate in cell culture experiments. It therefore seems to contribute to glioma progression [38]. Similar to this finding, IGFBP-2 is overexpressed in prostate carcinoma compared to normal prostate tissue [39], indicating an involvement in tumor growth. Due to its smaller molecular size, IGFBP-2 can bring IGF-1 and IGF-2 into extracellular avascular spaces where IGFBP-3 would be sterically too big. There are also evidences that IGFBP-2 may be found in the nucleus and thereby influence IGF—independently of apoptosis and cell survival [40]. There are no differences in IGFBP-2 expression between normal testicular tissue and seminoma, although IGFBP-2 mRNA is expressed in Leydig cells and Sertoli cells [11] that are not retrievable in tumor tissue. The highly significant correlation between IGF-R1 and IGFBP-2 expression suggests a major role in seminoma for these factors.
In conclusion, we assume an important paracrine or autocrine role of IGF-2 for human spermatogenesis. Genes correlating with Sertoli and Leydig cell markers, such as IGF-2 or IGFBP-5, seem to be involved in spermatogenesis and may therefore be downregulated in seminoma. Concerning the tumor growth of seminoma, the decrease of mainly IGF-inhibiting IGFBP may contribute to stimulated tumor growth. Constantly expressed genes, such as IGF-1, IGF-R1, IR-A, and IGFBP-2, may reflect, on one hand, an involvement in spermatogenesis and, on the other hand, a major role in tumor growth as they do not decrease in seminoma despite the missing spermatogenesis.
Acknowledgements
We are grateful to S. Kliesch (Department of Urology of the University, Münster), M. Bergmann (Institute of Veterinary Anatomy, Histology, and Embryology of the University, Giessen), and C. von Ostau (Department of Urology of the University Duisburg-Essen) for providing the tissue specimens. Furthermore, we thank L. Klein-Hitpass for his excellent performance of the microarrays. We further thank Alexandra Kappeler for excellent technical assistance.
Abbreviations
- CP
crossing point
- DLK1
delta Drosophila homologue-like 1 gene
- IHC
immunohistochemistry
- INSL3
insulin-like hormone 3
- IR
insulin receptor
- IR-A
insulin receptor, isoform A
- IR-B
insulin receptor, isoform B
- NT
normal testicular tissue
Footnotes
The microarray studies were supported by the German National Genome Research Cancer Network (NGFN/BMBF).
References
- 1.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 2.Grimberg A, Cohen P. Role of insulin-like growth factors and their binding proteins in growth control and carcinogenesis. J Cell Physiol. 2000;183:1–9. doi: 10.1002/(SICI)1097-4652(200004)183:1<1::AID-JCP1>3.0.CO;2-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludwig T, Le Borgne R, Hoflack B. Roles for mannose-6-phosphate receptors in lysosomal enzyme sorting, IGF-II binding and clathrin-coat assembly. Trends Cell Biol. 1995;5:202–206. doi: 10.1016/s0962-8924(00)89000-5. [DOI] [PubMed] [Google Scholar]
- 4.Kalli KR, Falowo OI, Bale LK, Zschunke MA, Roche PC, Conover CA. Functional insulin receptors on human epithelial ovarian carcinoma cells: implications for IGF-II mitogenic signaling. Endocrinology. 2002;143:3259–3267. doi: 10.1210/en.2001-211408. [DOI] [PubMed] [Google Scholar]
- 5.Sciacca L, Costantino A, Pandini G, Mineo R, Frasca F, Scalia P, Sbraccia P, Goldfine ID, Vigneri R, Belfiore A. Insulin receptor activation by IGF-II in breast cancers: evidence for a new autocrine/paracrine mechanism. Oncogene. 1999;18:2471–2479. doi: 10.1038/sj.onc.1202600. [DOI] [PubMed] [Google Scholar]
- 6.Guo N, Ye JJ, Liang SJ, Mineo R, Li SL, Giannini S, Plymate SR, Sikes RA, Fujita-Yamaguchi Y. The role of insulin-like growth factor-II in cancer growth and progression evidenced by the use of ribozymes and prostate cancer progression models. Growth Horm IGF Res. 2003;13:44–53. doi: 10.1016/s1096-6374(02)00121-1. [DOI] [PubMed] [Google Scholar]
- 7.Schaffer BS, Lin MF, Byrd JC, Park JH, Macdonald RG. Opposing roles for the insulin-like growth factor (IGF)-II and mannose 6-phosphate (Man-6-P) binding activities of the IGF-II/Man-6-P receptor in the growth of prostate cancer cells. Endocrinology. 2003;144:955–966. doi: 10.1210/en.2002-220751. [DOI] [PubMed] [Google Scholar]
- 8.Vella V, Pandini G, Sciacca L, Mineo R, Vigneri R, Pezzino V, Belfiore A. A novel autocrine loop involving IGF-2 and the insulin receptor isoform-A stimulates growth of thyroid cancer. J Clin Endocrinol Metab. 2002;87:245–254. doi: 10.1210/jcem.87.1.8142. [DOI] [PubMed] [Google Scholar]
- 9.Kelley KM, Oh Y, Gargosky SE, Gucev Z, Matsumoto T, Hwa V, Ng L, Simpson DM, Rosenfeld RG. Insulin-like growth factor-binding proteins (IGFBPs) and their regulatory dynamics. Int J Biochem Cell Biol. 1996;28:619–637. doi: 10.1016/1357-2725(96)00005-2. [DOI] [PubMed] [Google Scholar]
- 10.Baxter RC. Insulin-like growth factor (IGF)-binding proteins: interactions with IGFs and intrinsic bioactivities. Am J Physiol Endocrinol Metab. 2000;278:E967–E976. doi: 10.1152/ajpendo.2000.278.6.E967. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Bondy C. Anatomy of the insulin-like growth factor system in the human testis. Fertil Steril. 1993;60:897–904. doi: 10.1016/s0015-0282(16)56294-3. [DOI] [PubMed] [Google Scholar]
- 12.Vannelli BG, Barni T, Orlando C, Natali A, Serio M, Balboni GC. Insulin-like growth factor-I (IGF-I) and IGF-I receptor in human testis: an immunohistochemical study. Fertil Steril. 1988;49:666–669. doi: 10.1016/s0015-0282(16)59837-9. [DOI] [PubMed] [Google Scholar]
- 13.Engstrom W, Hopkins B, Schofield P. Expression of growth regulatory genes in primary human testicular neoplasms. Int J Androl. 1987;10:79–84. doi: 10.1111/j.1365-2605.1987.tb00168.x. [DOI] [PubMed] [Google Scholar]
- 14.Drescher B, Lauke H, Hartmann M, Davidoff MS, Zumkeller W. Immunohistochemical pattern of insulin-like growth factor (IGF) I, IGF II and IGF binding proteins 1 to 6 in carcinoma in situ of the testis. Mol Pathol. 1997;50:298–303. doi: 10.1136/mp.50.6.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffl M, Mircheva Georgieva T, Penchev Georgiev I, Ontsouka E, Hageleit M, Blum JW. Real-time RT-PCR quantification of insulin-like growth factor IGF-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2 and 3 in the bovine species. Domest Anim Endocrinol. 2002;22:91–102. doi: 10.1016/s0739-7240(01)00128-x. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl M. Development and validation of an externally standardised quantitative insulin-like growth factor-1 RT-PCR using LightCycler SYBR Green I technology. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-Time PCR, Methods and Applications. Berlin: Springer-Verlag; 2001. pp. 281–292. [Google Scholar]
- 17.Rasmussen R. Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-Time PCR, Methods and Applications. Berlin: Springer-Verlag; 2001. pp. 21–34. [Google Scholar]
- 18.Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl MW, Horgan GW, Dempfle L. Relative Expression Software Tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen CH, Erb K, Westergaard LG, Kliem A, Teisner B. Fetal antigen 1, an EGF multidomain protein in the sex hormone-producing cells of the gonads and the microenvironment of germ cells. Mol Hum Reprod. 1999;5:908–913. doi: 10.1093/molehr/5.10.908. [DOI] [PubMed] [Google Scholar]
- 21.Ivell R, Bathgate RA. Reproductive biology of the relaxin-like factor (RLF/INSL3) Biol Reprod. 2002;67:699–705. doi: 10.1095/biolreprod.102.005199. [DOI] [PubMed] [Google Scholar]
- 22.Griswold MD, Heckert L, Linder C. The molecular biology of the FSH receptor. J Steroid Biochem Mol Biol. 1995;53:215–218. doi: 10.1016/0960-0760(95)00049-6. [DOI] [PubMed] [Google Scholar]
- 23.Bremner WJ, Millar MR, Sharpe RM, Saunders PT. Immunohistochemical localization of androgen receptors in the rat testis: evidence for stage-dependent expression and regulation by androgens. Endocrinology. 1994;135:1227–1234. doi: 10.1210/endo.135.3.8070367. [DOI] [PubMed] [Google Scholar]
- 24.Flores JM, Sanchez MA, Gonzalez M, Pizarro M. Caprine testicular hypoplasia associated with sexual reversion decreases the expression of insulin-like growth factor II (IGF-II) mRNA in testes. Anim Reprod Sci. 1998;52:279–288. doi: 10.1016/s0378-4320(98)00104-3. [DOI] [PubMed] [Google Scholar]
- 25.Wagener A, Blottner S, Goritz F, Streich WJ, Fickel J. Differential changes in expression of α and FGF, IGF-1 and -2, and TGF-alpha during seasonal growth and involution of roe deer testis. Growth Factors. 2003;21:95–102. doi: 10.1080/08977190310001621023. [DOI] [PubMed] [Google Scholar]
- 26.Seurin D, Lassarre C, Bienvenu G, Babajko S. Insulin-like growth factor binding protein-6 inhibits neuroblastoma cell proliferation and tumour development. Eur J Cancer. 2002;38:2058–2065. doi: 10.1016/s0959-8049(02)00240-x. [DOI] [PubMed] [Google Scholar]
- 27.Drivdahl RH, Sprenger C, Trimm K, Plymate SR. Inhibition of growth and increased expression of insulin-like growth factor-binding protein-3 (IGFBP-3) and -6 in prostate cancer cells stably transfected with antisense IGFBP-4 complementary deoxyribonucleic acid. Endocrinology. 2001;142:1990–1998. doi: 10.1210/endo.142.5.8158. [DOI] [PubMed] [Google Scholar]
- 28.Gallicchio MA, Kaun C, Wojta J, Binder B, Bach LA. Urokinase type plasminogen activator receptor is involved in insulin-like growth factor-induced migration of rhabdomyosarcoma cells in vitro. J Cell Physiol. 2003;197:131–138. doi: 10.1002/jcp.10352. [DOI] [PubMed] [Google Scholar]
- 29.Ritter MR, Dorrell MI, Edmonds J, Friedlander SF, Friedlander M. Insulin-like growth factor 2 and potential regulators of hemangioma growth and involution identified by large-scale expression analysis. Proc Natl Acad Sci USA. 2002;99:7455–7460. doi: 10.1073/pnas.102185799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andress DL, Birnbaum RS. Human osteoblast-derived insulin-like growth factor (IGF) binding protein-5 stimulates osteoblast mitogenesis and potentiates IGF action. J Biol Chem. 1992;267:22467–22472. [PubMed] [Google Scholar]
- 31.Marshman E, Green KA, Flint DJ, White A, Streuli CH, Westwood M. Insulin-like growth factor binding protein 5 and apoptosis in mammary epithelial cells. J Cell Sci. 2003;116:675–682. doi: 10.1242/jcs.00263. [DOI] [PubMed] [Google Scholar]
- 32.Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003;278:29676–29685. doi: 10.1074/jbc.M301965200. [DOI] [PubMed] [Google Scholar]
- 33.Murphy LJ. Overexpression of insulin-like growth factor binding protein-1 in transgenic mice. Pediatr Nephrol. 2000;14:567–571. doi: 10.1007/s004670000347. [DOI] [PubMed] [Google Scholar]
- 34.Ngo TH, Barnard RJ, Leung PS, Cohen P, Aronson WJ. Insulin-like growth factor I (IGF-I) and IGF binding protein-1 modulate prostate cancer cell growth and apoptosis: possible mediators for the effects of diet and exercise on cancer cell survival. Endocrinology. 2003;144:2319–2324. doi: 10.1210/en.2003-221028. [DOI] [PubMed] [Google Scholar]
- 35.Gronbaek H, Tanos V, Meirow D, Peretz T, Raz I, Flyvbjerg A. Effects of Tamoxifen on insulin-like growth factors, IGF binding proteins and IGFBP-3 proteolysis in breast cancer patients. Anticancer Res. 2003;23:2815–2820. [PubMed] [Google Scholar]
- 36.Damon SE, Maddison L, Ware JL, Plymate SR. Overexpression of an inhibitory insulin-like growth factor binding protein (IGFBP), IGFBP-4, delays onset of prostate tumor formation. Endocrinology. 1998;139:3456–3464. doi: 10.1210/endo.139.8.6150. [DOI] [PubMed] [Google Scholar]
- 37.Reeve JG, Morgan J, Schwander J, Bleehen NM. Role for membrane and secreted insulin-like growth factor-binding protein-2 in the regulation of insulin-like growth factor action in lung tumors. Cancer Res. 1993;53:4680–4685. [PubMed] [Google Scholar]
- 38.Wang H, Wang H, Shen W, Huang H, Hu L, Ramdas L, Zhou YH, Liao WS, Fuller GN, Zhang W. Insulin-like growth factor binding protein 2 enhances glioblastoma invasion by activating invasion-enhancing genes. Cancer Res. 2003;63:4315–4321. [PubMed] [Google Scholar]
- 39.Richardsen E, Ukkonen T, Bjornsen T, Mortensen E, Egevad L, Busch C. Overexpression of IGBFB2 is a marker for malignant transformation in prostate epithelium. Virchows Arch. 2000;442:329–335. doi: 10.1007/s00428-003-0786-2. [DOI] [PubMed] [Google Scholar]
- 40.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]