Abstract
A RecA-independent chromosomal rearrangement in the upstream region of the streptolysin O (slo) gene of Streptococcus pyogenes which affects slo expression was identified. PCR analysis was used to demonstrate that this kind of rearrangement was found in several strains of different lineages. Chromosomal loci involved in the recombination were found to be 746 kb apart on the 1.85-Mb-long chromosome. The primary structure of the splicing region, the reproducibility of the rearrangement, and the fact that reconstructed recombinant molecules fused to erm and lacZ reporter genes affected their expression indicate that this event is not accidental but may play a role in the expression of the slo gene. In addition, the product of the recombining DNAs, including the splicing site, does not follow any example of a known recombination mechanism. The implications of this rearrangement for slo expression are discussed.
Gene order is a stable characteristic of all organisms analyzed, and the genetic map, the embodiment of this order, is expected to be shared by all members of a species. However, the discovery of transposable elements showed that the concept of fixed genomes slowly evolving by point mutations is an oversimplification. Indeed, it became clear that gene rearrangements, including not only those related to transpositions, are endemic in bacteria. DNA rearrangements in bacteria are classified into two groups: accidental or unprogrammed DNA rearrangements and programmed DNA rearrangements (3). Accidental rearrangements derive from a plethora of events at the chromosome, such as repair, transpositions, and insertion or excision of plasmids, phages, or other foreign DNA. Most of these rearrangements are detrimental to the cell. However, at the population level, where survival of the few means little to the survival of the whole, the overall picture attains a new quality and biological justification. By permanently generating cells with varied phenotypes, a mixed population of organisms is better equipped to survive drastic environmental changes even though the genetic cripples maybe poorly adapted to the current environment (5). On the other hand, programmed DNA rearrangements are part of a genetic program, and the outcome of such rearrangements is largely predictable; i.e., amplification or deletion of genes, assembly of genes from gene segments, and DNA rearrangements that alter gene expression.
The group A streptococcus (GAS) Streptococcus pyogenes is the causative agent of numerous human diseases, including pharyngitis, scarlet fever, erysipelas, impetigo, necrotizing fasciitis, and toxic shock-like syndrome. Additionally, GAS can also provoke the nonsuppurative sequelae of rheumatic fever and acute glomerulonephritis. From a genetic point of view, GAS represent a paradigmatic case that defies the standard definition of the species as it relates to bacteria. More than 100 serological groups based on the antiphagocytic M protein have been identified so far among S. pyogenes isolates. This remarkable polymorphism is paralleled at the genetic level; pulsed-field gel electrophoresis analysis of chromosomal DNA demonstrated a variety of restriction fragment patterns not only between different M types but also among strains belonging to the same M group (16, 28, 29). The most recent data obtained from GAS sequencing projects suggest that these differences are prevalently due to nucleotide sequence polymorphism, although certain differences in gene content have also been observed (6, 25; W. M. McShan et al., unpublished data).
The flexibility of genomes and examples of DNA rearrangements and their role in gene regulation have been documented in different prokaryotes (3, 5, 9, 11). In this study, we report on a novel type of rearrangement in the chromosome of S. pyogenes that, under laboratory growth conditions, influences the expression of slo, the gene responsible for synthesis of the cytolytic exotoxin streptolysin O.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are described in Table 1. S. pyogenes strains were grown in Todd-Hewitt medium (THY) containing 0.2% yeast extract, and when needed, horse serum was added to a final concentration of 5% (vol/vol). Unless otherwise stated, the following antibiotics were added to THY when needed: erythromycin (0.1 μg/ml), tetracycline (12.5 μg/ml), and kanamycin (40 μg/ml). Escherichia coli was grown in Luria-Bertani broth (LB). Both THY and LB were solidified when necessary with 1.5% agar. When required, the following antibiotics were added to LB: ampicillin (100 μg/ml), kanamycin (40 μg/ml), tetracycline (12.5 μg/ml), and erythromycin (500 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Escherichia coli K-12 | ||
| JM109 | F′/traD36 lacIq Δ(lacZ)M15 proA+B+/el4 (McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(r− m+)) relA1 supE44 recA1 | 33 |
| DH5α | F′/endA1 hsdR17 (r− m+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR (φ80dlac Δ[lacZ]M15) | 32 |
| Streptococcus pyogenes | ||
| NZ131 (M49) | Clinical isolate | Laboratory collection |
| 364 (M3) | Clinical isolate | Laboratory collection |
| 455 (M5) | Clinical isolate | Laboratory collection |
| SF370 (M1) | Clinical isolate | Laboratory collection |
| NY5 (M10) | Clinical isolate | 26 |
| OK50 | NZ131 with 102-bp-long deletion in Ex region | This work |
| OK94 | Obtained by homologous recombination between plasmid pOK66 and chromosomal (NZ131) slo upstream region | This work |
| OK95 | Obtained by homologous recombination between plasmid pOK67 and chromosomal (NZ131) Ex sequences | This work |
| OK96 | Obtained by homologous recombination between pOK67 and chromosomal (NZ131) slo upstream region | |
| OK86, OK87, OK88, OK89, OK90, OK91, OK103 | Strains obtained by integration of pCAMP17-based constructs OK73, pOK74, pOK77, pOK78, pOK83, pOK84, and pOK102, respectively, into chromosome of NZ131 | This work |
| Plasmids | ||
| pUC18 & pUC19 | Recombinant E. coli vectors | 33 |
| pT7Blue | Vector designed for cloning and sequencing of PCR products | Novagen, catalog no. 69820-1 |
| pGEM-T Easy | Vector designed for cloning and sequencing of PCR products | Promega Corp., catalog no. 1360 |
| p7erm | Insertional vector in streptococci; origin of replication from pUC18 | 14 |
| p7tet | erm gene in p7Erm replaced with tet | 14 |
| pCAMP17 | Recombinant plasmid for construction of lacZ reporter fusions; it integrates into serine tRNA gene of S. pyogenes chromosome | 8 |
| pOK18 | Recombinant plasmid carrying proximal part of slo gene and its upstream region on a HindIII fragment cloned into the HindIII site of vector pALTER-1 | This work |
| pOK23 | Recombinant plasmid harboring reconstructed fusion of Ex and slo DNAs, contains mutated translation initiation codon of slo gene | This work |
| POK23-1 | Same as pOK23 with recreated slo gene ATG initiation codon | This work |
| pOK66 | Recombinant plasmid carrying approximately 650 bp of the slo gene upstream region fused to the erm reporter gene | This work |
| pOK67 | Recombinant plasmid carrying 539 bp of Ex DNA region fused to the erm reporter gene via 190 bp | This work |
| pOK73, pOK77, pOK78, pOK83, pOK84, pOK102 | Recombinant plasmids carrying slo gene upstream region of different lengths fused to lacZ reporter gene of vector pCAMP17 | This work |
| pOK74 | Product of P98 × P99 PCR on pOK23 as a substrate, cut with PstI and BamHI, and cloned into pCAMP17 | This work |
Transformation of bacterial strains.
The plasmids and linear DNA were introduced into S. pyogenes strains by electrotransformation (13, 23). Transformation of E. coli was performed by the standard CaCl2 method as described before (15).
PCR analysis.
The oligonucleotide primers used are described below. Template DNA was purified by CsCl-ethidium bromide gradient centrifugation. In most cases, 40 cycles of amplification were carried out in a Pelkin-Elmer DNA thermocycler with strand denaturation (1 min at 94°C), annealing (1 min at 59°C), and elongation (1 min at 72°C). The total volume of the PCR mixture was usually 50 μl, and it consisted of Taq polymerase (1 U), Taq polymerase buffer (1×), Mg2+ (2 mM), deoxynucleoside triphosphates (200 μM each), and primers (1 μM each). Following amplification, a 10-μl aliquot was analyzed by agarose gel electrophoresis. When needed, the PCR products were purified with a PCR purification kit (Qiagen Inc.)
DNA manipulations and sequencing.
Plasmid isolation, restriction, and ligation, Southern transfer and hybridization, and other recombinant DNA techniques were performed by standard procedures (20). Plasmids from E. coli were also isolated with a plasmid isolation kit (Qiagen Inc.). For cloning PCR fragments, specialized cloning vectors with T-nucleotide overhangs, pT7 Blue (Novagen) and pGEM-T Easy (Promega), were used. All sequencing reactions were performed by the dideoxy termination method (21).
Construction of deletion covering the recombination active site in the Ex region.
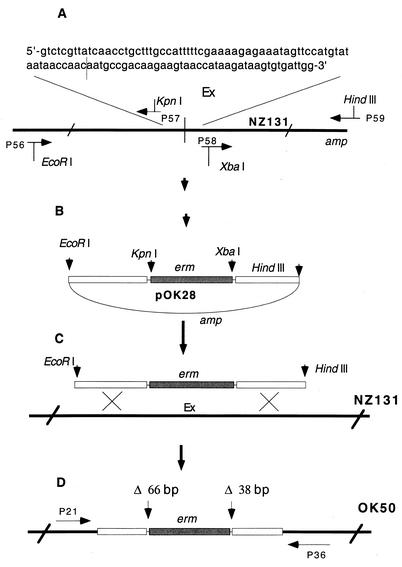
The deletion, covering 102 bp around the junction point of Ex DNA (temporary designation for “element X,” later found to map within Spy1528 [see Fig. 3]), was constructed as illustrated in Fig. 1. Two DNA fragments of 970 bp and 870 bp at distances of 65 bp and 37 bp from the junction point, respectively, were produced by PCR on NZ131 chromosomal DNA as the substrate. The primers used were designed to create suitable restriction sites (EcoRI, KpnI, XbaI, and HindIII) at the ends of the amplified fragments (Fig. 1A). After being cut with appropriate restriction enzymes and purified, the fragments were cloned in two consecutive steps into the multiple cloning site of plasmid pOK26, a derivative of pUC18 with an erythromycin resistance gene located within the multiple cloning site (not presented). The construct obtained, pOK28, was treated with EcoRI and HindIII enzymes, and the insert was separated from the vector by gel electrophoresis, extracted from the gel, and introduced by electrotransformation into recipient strain NZ131 (Fig. 1C). Chromosomal DNA from one of the erythromycin-resistant transformants (strain OK50) was isolated, and the presence of the insert containing the 65-bp- and 37-bp-long deletions of Ex DNA encompassing a stretch of DNA involved in recombination with the slo DNA was confirmed by sequencing; the chromosomal DNA harboring the insert was amplified with flanking primers P21 and P36, and the fragment obtained was cloned into pT7 and sequenced (Fig. 1D).
FIG. 3.
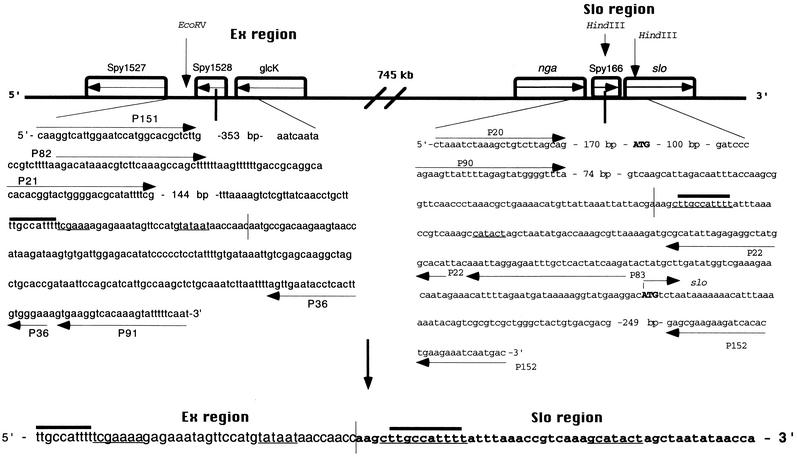
Primary structure of the Ex and slo loci around the sites involved in mutual recombination and the product of their recombination. Note an extra C at the junction of the recombinant molecule. The arrowed lines represent PCR primers, while thin plain lines signify the slo promoter and the putative Ex promoter. The arrowed lines inside the boxes mark the orientations of the genes. Vertical lines denote the recombination site between the Ex and slo DNAs. The nucleotide decamers are depicted by thick lines above the sequence. nga is the genetic symbol for the gene coding for NAD-glycohydrolase, slo denotes the gene for streptolysin O, while Spy1527 and glcK represent genes highly homologous to ylaG, the Bacillus subtilis gene coding for elongation factor G and the gene coding for glucose kinase, respectively (6). The open reading frame Spy1528 represents a gene coding for a conserved hypothetical protein showing a low degree of identity to a cell division protein from B. subtilis; it could also easily be a stretch of noncoding DNA (6). Similarly, Spy166 is a very short open reading frame with no similarity to any known gene (6, 25; W. M. McShan et al., unpublished data). Initiation codon of the slo gene and stop codon of the nga gene are given in bold capital letters. The figure is not drawn to scale.
FIG. 1.
Construction of 102-bp-long deletion in Ex region of strain NZ131. The deleted stretch of the chromosome (the sequence at the top) is the site in the Ex region that is involved in recombination with the slo DNA. The thin vertical line in the chromosome and in the sequence marks the recombination site. Horizontal arrows depict primers used for PCR. The figure is not drawn to scale. For details, see Materials and Methods.
Construction of fusions with erm as a reporter gene.
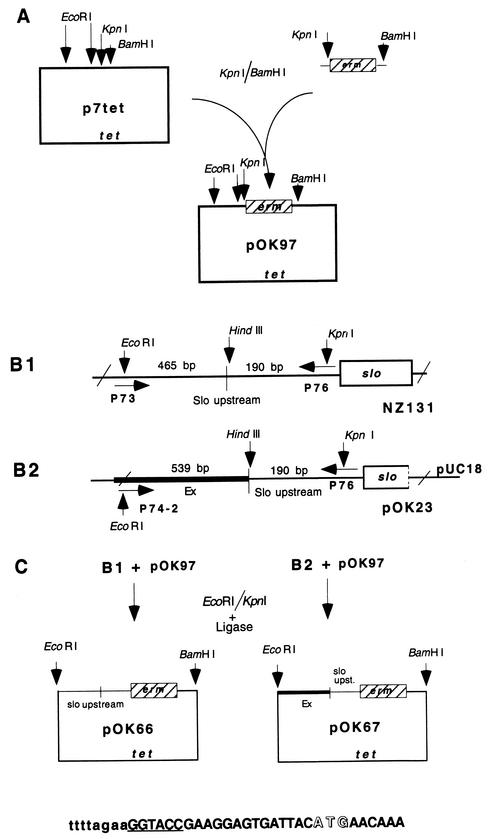
The erm gene was first amplified by PCR from plasmid p7erm and cloned into the KpnI and BamHI sites of the insertion vector p7tet, creating plasmid pOK97 (see Fig. 2A). In the following step, the native upstream region of the slo gene and the recombinant Ex/slo fragment were amplified by using as a substrate chromosomal DNA from strain NZ131 (Fig. 2B1 ) and plasmid pOK23 carrying the recombined Ex/slo fragment (Fig. 2B2), respectively, and cloned into the EcoRI and KpnI sites of the pOK97 vector, resulting in plasmids pOK66 and pOK67, respectively. The recombinant Ex/slo fragment in pOK23 was obtained by PCR on chromosomal DNA from strain NZ131 with primers 151 and 152 (Fig. 3). As presented (Fig. 2), EcoRI and KpnI restriction sites were introduced into the amplified fragments by mutagenic oligonucleotides containing these restriction sites. The correctness of the nucleotide sequence was confirmed by DNA sequencing (Fig. 2C).
FIG. 2.
Construction of transcription fusion strains with erm fused to fragments of different composition upstream of the slo gene. The sequence at the bottom of section C illustrates the fusion between the upstream region of the slo gene and the erm gene. The slo sequence is presented in lowercase letters, and the erm sequence is given in capital letters, the KpnI site is underlined, and the ATG initiation codon of the erm gene is presented in open letters. The picture is not drawn to scale. For more details, see Materials and Methods.
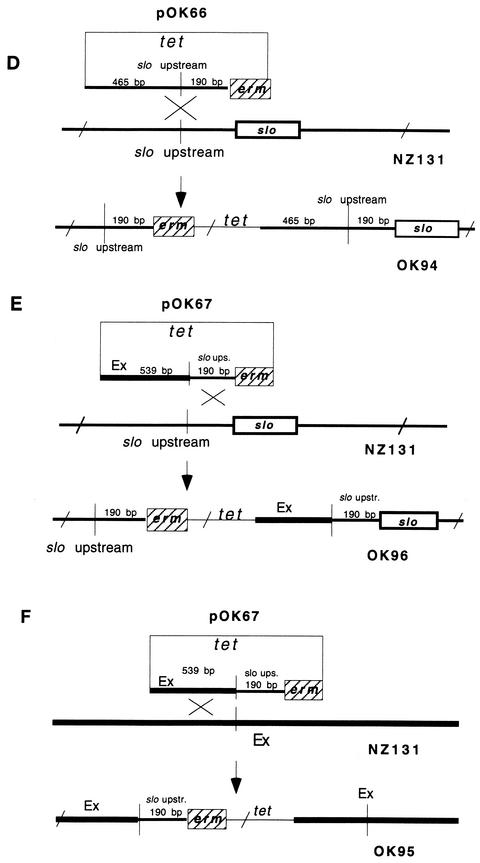
In the final step, plasmids pOK66 and pOK67 were introduced by electrotransformation into the competent recipient NZ131. Plasmid pOK66 can integrate into the chromosome in only one way, via the slo region (Fig. 2D). On the other hand, plasmid OK67 can produce two genetic constellations, depending on whether it has been integrated into the chromosome via slo (Fig. 2E) or via the Ex region (Fig. 2F). The genetic constellation of the final constructs upon insertion of recombinant plasmids into the NZ131 chromosome was confirmed by PCR with the appropriate primers. Finally, growing the constructs under nonselective conditions in THY broth without erythromycin demonstrated that the segregation of the erm marker was lower than 0.05%.
β-Galactosidase assay.
β-Galactosidase was determined as described before (7) by a modification of the original procedure (15). Overnight cultures of S. pyogenes were diluted approximately 20 times in fresh THY medium and then grown to mid-logarithmic phase (approximately 0.2 at an optical density of 600 nm). Cells were harvested by centrifugation, resuspended, and made permeable to O-nitrophenyl-β-d-galactopyranoside (ONPG) by adding 20 μl of chloroform per sample and vortexing for 30 s. β-Galactosidase activity was determined at a temperature of 37°C by measuring ΔE at 420 nm to detect O-nitrophenyl released from ONPG and was calculated as Miller units, proportional to enzyme activity per cell.
Selected oligonucleotides used in PCRs.
The following oligonucleotides (shown in the 5′-3′ direction) were used as PCR primers: P21, CACACGGTACTGGGGACGCATATTTTCG; P36, TTCCCACAAGTGAGGTATTCAACTA; P56, GTAAACAACTGGGAATTCCAATTGC; P57, CGCCGGTACCTTTTAAAGACGTGATGCGTC; P58, ATGCTCTAGACATATCCCCC TCCTATTTTG; P59, CGACAAGCTTCCATCAAACATCGCTTAGAC; P62. CTCTCACAAATACAGAAAAAAGC; P68, AAGCTTGCCATTTTATTTAAACCG; P69, TTTATATCAGTTAAGCTTGCC; P70, TTGGTTATTATACATGGAACTATTTC; P71, GTTGGTTATTATACATGGAACTAT; P73, TCAAAGAATTCTCTAAAGCTGTCTTAGCAG; P74′, TTCTTGAATT- CCTTAAGTTAGTCATTTGTTCC; P76, AAAAGGGTACCTTCTAAAATGTTTCTATTG; P82, TTAAGACATAAACGTCTTCAAAGCCAGC; P83, TCTTGATAGTGAGCAAATTCTCCTAATTTG; P90, AGAAGTTATTTTAGAGTATGGGGTTTA; P91, TATTGAAAAATACTTTGTGACCTTCAC; P97, ATGACTCTGCAGTAACTGGTCGCTTCAAAATTAAG; P98, GGACACTGCAGCAAGTCTGGATACAGCAAAAAAAG; P99, GGCGCCGGATCCTTTTTATTAGACATGTCCTTCATACC; P125, CAAAGAGTGGCGGAGCTTCATTGCTGACAC; P126, CAAATGAGCAACCAAAGCCAGAAAGTAGTGAG; P144, TGTTTTTCCGTGGTCAACGTGG; P145, TTTTCTGCAGATTCTAGTGGC; P151, CAAGGTCATTGGAATCCATGGCACG; and P152, GTCATTGATTTCTTCAGTGTGATCTTCTTCGCTC.
RESULTS
Evidence of chromosomal rearrangement.
The first suggestion or indication of a recombination event between two unlinked chromosomal loci was obtained inadvertently during construction of a local restriction map upstream of the slo gene of S. pyogenes strain NZ131 (M49). A PCR experiment with primers P21 and P22 was designed to amplify and clone a fragment from chromosomal DNA spanning the region immediately upstream of the slo gene and the DNA we thought was contiguous to the slo-carrying fragment. The reaction produced a fragment of the expected size that contained a HindIII site found previously at the end of the known sequence of the slo gene (Fig. 4A, lane 4) (10). However, Southern blotting and hybridization demonstrated that the DNA beyond the HindIII site must be at least 35 kb away from the slo locus (results not presented) and certainly beyond the range for standard PCR. Furthermore, hybridization of the slo DNA and this unknown DNA isolated from strains NZ131 (M49) and SF370 (M1) with pulsed-field gel electrophoresis, chromosomal fragments used in the construction of the S. pyogenes genome physical map (29) suggested that the two loci in these strains may be as much as one third of the chromosome away from each other (A. Suvorov, personal communication). This estimate is in accord with the data obtained recently from the genome sequence of stains SF370 (M1), MGAS8232 (M18), and NZ131 (M49) (6, 25; W. M. McShan et al., unpublished data); the distance between the two loci on the chromosome of about 1.852 Mb turned out to be approximately 746 kb. The product of the PCR amplification with primers P21 and P22 was cloned into the pT7Blue vector and sequenced.
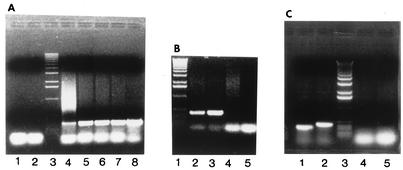
FIG. 4.
Electrophoretic profile of PCR products with primers pairs P21 × P22 and P82 × P83 on chromosomal DNA from different strains. (A) Products of PCR with primer pair P21 × P22 on chromosomal DNA from strains NZ131 (lane 4), NZ131 Rec− (lane 5), M1 (strain 370) (lane 6), M3 (strain 364) (lane 7), and M5 (strain 455) (lane 8). Lanes 1 and 2 are controls without chromosomal DNA, and lane 3 is molecular size standard (1-kb standard; Promega). (B) Lane 1, molecular size standards; lane 2, P82 × P83 on NZ131; lane 3, P82 × P83 on NZ131 Rec−; lane 4, P82 × P83 on OK50; lane 5, control with no DNA. (C) Lane 1, P21 × P22 on NZ131; lane 2, P82 × P83 on NZ131; lane 3, molecular size control; lanes 4 and 5, controls with no DNA.
The primary structure of the recombinant molecule showed that the chromosomal locus, temporarily named element X (Ex), is spliced to the upstream region of the slo gene a few nucleotides upstream of the slo native promoter and 189 bp upstream from the beginning of the slo gene (Fig. 3) (10, 22). Interestingly, an extra C was found at the recombination site (Fig. 3). Another feature of the recombinant fragment is that eight nucleotides upstream of the junction, the Ex DNA carries a promoter-like structure consisting of an E. coli −10 TATAAT consensus sequence and, 17 nucleotides upstream, a TCGAAA sequence identical to one of the −35 sequences found in the oriC region of Bacillus subtilis (18). It is also interesting that two TTGCCATTTT decamers, found 18 times in the chromosomes of strains SF370 (M1) (6), NZ131 (M49) (W. M. McShan et al., unpublished data), and MGAS8232 (M18) (25), are brought together by recombination into a proximity of only 50 nucleotides.
The nucleotide sequence that we determined for both the Ex region and the slo regions corresponded well to the sequence of the slo region of streptococcal strain Richards (M3) (10, 11) and to the nucleotide sequence of the slo and Ex regions from strains SF370 (M1), MGAS8232, and NZ131 (M49) (6, 25; W. M. McShan et al., unpublished data). The results of these analyses positioned the recombination site 189 bp upstream of the slo gene and 326 bp downstream from nga, the gene encoding NAD glycohydrolase, and between glcK and Spy1527, two genes showing similarity to glucose kinase and GTP binding protein, respectively, in the Ex region (Fig. 3) (6).
Figure 3 shows that the recombination site in the Ex region maps within the short open reading frame (Spy1528) which represents a gene encoding a conserved hypothetical protein with a low degree of identity to a cell division protein from B. subtilis. Given the size of the gene (387 bp) as well as its low identity value (E value = 8e−13 [1]), Spy1528 could easily be a stretch of noncoding DNA between Spy1527 and glcK. Similarly, Spy166 is a very short open reading frame (354 nucleotides) with no similarity to any known gene (6, 25; W. M. McShan et al., unpublished data). The fact that the slo promoter lies within this sequence (22) suggests that this open reading frame may also be a stretch of noncoding DNA.
The requirement for Ex DNA in the recombination event with slo DNA was verified in an experiment with an Ex deletion derivative of strain NZ131. The deletion, covering 102 bp around the junction point of Ex DNA, was constructed in strain NZ131 as described in Materials and Methods and illustrated in Fig. 1. Chromosomal DNA from this construct (OK50) was used as a substrate in PCR with a pair of primers, P82 and P83 (see below). No characteristic fragment like the one found with DNA from strain NZ131 could be observed. (Fig. 4B).
Recombination event is RecA independent and seems to be ubiquitous among GAS strains.
In order to further characterize the recombination event that takes place in the slo region, chromosomal DNA isolated from a recA mutant of strain NZ131 (30) was used as a substrate in PCR assays. Chromosomal DNAs isolated from representatives of three streptococcal M types, M1 (strain SF370), M3 (strain 364), and M5 (strain 455), were also included in this analysis to investigate whether the observed phenomenon is specific for M49 or is also present among other M types of GAS. The results of PCR experiments with the P21 and P22 primers (Fig. 4A) show that the recombination event creating the Ex/slo hybrid DNA is RecA independent and that it is widely present, possibly omnipresent, among GAS. The nature of the fragments obtained was also verified by sequencing. The nucleotide sequence obtained from the Ex/slo splicing region as well as the rest of the amplified fragments was virtually identical to the sequence obtained previously for strain NZ131, including an extra C at the recombination site (results not presented).
Given the extreme sensitivity of PCR, special precautions were taken during isolation of chromosomal DNA from strains other than RecA+ NZ131 for possible contamination with the previously obtained fragment P21 × P22. First, all DNAs, including DNA from strain NZ131, were used as substrates in PCRs in which a new pair of primers, P82 × P83, slightly outflanking the segment of DNA amplified by the pair P21 × P22, was used (Fig. 3). In this way it was ensured that neither possible contaminating P21 × P22 fragment nor the same fragment cloned into vectors pT7Blue or P7erm could be amplified. The PCR products obtained in all reactions, when analyzed by gel electrophoresis, demonstrated the expected increase in molecular size of approximately 80 nucleotides relative to the fragment obtained with primers 21 and 22 (Fig. 4C). An additional PCR (P82 × P152; Fig. 3) was also performed, producing a fragment of the expected size (results not presented).
DNA sequence analyses of the fragments obtained in both PCRs (P82 × P83 and P82 × P152) was the same (again, including the extra C at the Ex/slo junction) compared to the sequence obtained previously with DNA fragments from P21 × P22 PCRs. Finally, a test for possible contamination with P21 × P22 fragments cloned into the pT7Blue and p7Erm vectors was tested with primers which flank multiple cloning sites of these vectors (primers T7 and M13 Forward for pT7Blue and T7 and SP6 for p7Erm). None of the chromosomal DNAs produced any detectable fragment in these PCRs (results not presented). We conclude that the amplified P21 × P22 fragments are authentic and that, similar to the hybrid fragment obtained in the original experiment with strain NZ131 (M49), they illustrate an in vivo recombination event in the GAS M types 1, 3, and 5 as well as in the Rec− derivative of NZ131 (M9).
Detection of recombinant fragments between the slo and Ex DNAs is not the result of a coincidental in vitro hybridization of these two chromosomal segments.
The possibility was considered that the detected recombined DNA did not reflect an in vivo recombination event, but rather might have resulted from an in vitro hybridization of the slo and Ex chromosomal fragments in the course of PCR amplification. Provided that these two fragments harbor shorter or longer homologous DNA stretches, hybrid molecules could have been established either between single-stranded homologous DNA stretches at the recessed ends of the double-stranded fragments or between partial overlaps between single-stranded molecules. Double-stranded hybrids could readily serve as a substrate for the PCR amplification, while partially overlapping single-stranded molecules would require an earlier conversion into a fully double-stranded form by the extension of the internal 3′OH ends.
Although homologous sequences that would provide substrates for such reactions have not been found in the recombined section of Ex and slo DNAs (Fig. 3), control experiments were performed in the following way. Fragments of Ex and slo DNA of approximately 1 kb carrying the presumptive point of recombination were obtained from plasmid clones carrying these DNAs. The fragments obtained were separated by gel electrophoresis and purified from the gel as described above. The fragments were mixed at a 1:1 molar ratio and used as substrates for PCR amplification at 1, 10, 50, and 100 ng of DNA per reaction. No product was observed with primer pairs 21 × 22 and 82 × 83 (results not presented). We conclude that the observed recombinant Ex/slo fragments are not the result of a coincidental in vitro hybridization of these two chromosomal segments, but are rather the consequence of their prior in vivo recombinational fusion.
Quantification of recombination event.
The frequency of the described recombination event was determined by PCR. Original preparations of DNA isolated from strains NZ131 (M49) and SF370 (M1) were adjusted to an approximate concentration of 200 to 300 ng/μl and subsequently diluted up to 1010-fold by serial dilution in TE (Tris-EDTA) buffer. The first PCR, used as a positive control and reference point, was done with a pair of primers (P155 × P125) flanking a stretch of genetically stable DNA of an approximately 0.5-kb-long structural part of the slo gene (10). The highest dilution of both DNAs at which fragments were detected on the gel after PCR amplification was 108; PCR performed with DNA diluted 5 × 108 times did not produce a visible fragment (results not presented).
As expected, PCR amplification with primers P82 and P83, used in this study to detect the recombination event between Ex and slo DNA (Fig. 4), produced visible fragments at lower dilution values. Interestingly, the values were not the same for the two DNAs: the highest dilution of NZ131 DNA at which the amplified fragment was visible was 103, while the highest dilution of SF370 DNA that allowed synthesis of a detectable amount of the fragment was 104 (results not presented). We conclude that the frequency of rearrangement in strain NZ131 was 10−5 in the population, while the frequency of the rearrangement in strain SF370 was higher (10−4). The observed lower frequency values may have resulted from DNA shearing leading to the separation of Ex and slo loci in the subpopulation of chromosomal fragments. However, such shearing of 0.5-kb-long standard DNA is not to be expected. The numbers obtained in this type of experiment are always an approximation; it is difficult to expect that two pairs of primers will bind to their respective targets with the same affinity.
Search for the other terminus.
To understand the nature of the recombination event, an attempt was made to identify the other end of the detected rearrangement. In this pursuit, it was first hypothesized that the rearrangement might be an intragenomic transposition. Although contraindicated by the generation of an extra C at the splicing region that does not fit any of the mechanical models of transposition as well as by the lack of any transposase genes in the vicinity of the recombining DNAs, this belief was based on the fact that the two recombining loci were found to map far apart on the chromosome.
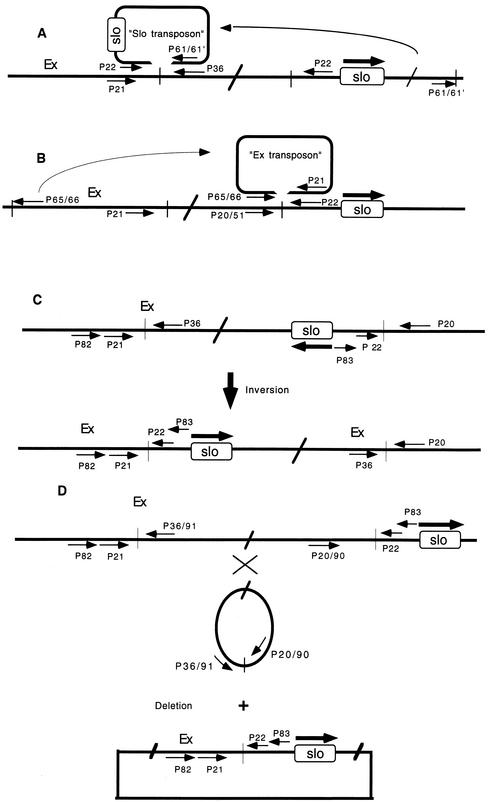
The first model tested was that the slo gene might be part of a transposon that had been inserted into the target sequence within the Ex DNA (Fig. 5A). The alternative hypothesis was that the Ex “transposon” was inserting itself into the target sequence upstream of the slo gene (Fig. 5B). To test these hypotheses, primers were designed for PCR based on the assumption that the termini of both “transposons” were composed of either inverted or direct repeats. The known nucleotide sequences at one end of the hypothesized transposons, i.e., DNA stretches immediately flanking the splicing point of the Ex/slo recombinant DNA, served as a reference for designing these primers. To minimize difficulties due to the possible incomplete identity of the known ends and the projected ends, a pair of partially overlapping primers differing in their 3′OH ends were designed for each end of the putative Ex and slo transposons. The reactions, performed under various experimental conditions, produced either no PCR products or bands which differed vastly from the expected sizes (results not presented). After several additional unsuccessful attempts, it was concluded that the observed rearrangement possibly was not due to a standard bacterial transposition event.
FIG. 5.
Hypothetical models of chromosomal rearrangement between the slo and Ex chromosomal loci by a transposition of the “slo transposon” (A), by a transposition of the “Ex transposon” (B), by an inversion event (C), and by deletion (D). Thin arrows denote oligonucleotide primers used in PCR, while thick arrows indicate the transcriptional polarity of the slo gene. The primers 68, 69, 70 and 71 (not presented) are inverse complements of primers P61, P61′, P65, and P66, respectively (see Materials and Methods); they were used to test the possibility that the hypothetical transposons are flanked by direct repeats. The figure is not drawn to scale.
Although most Rec-independent inversion or deletion-generating events are characterized by shorter or longer repeats around recombination sites (28), the possibility that the rearrangement might be a consequence of either inversion or deletion of the chromosomal segment between the Ex and slo loci was also considered (Fig. 5C and D). At the time we performed these experiments, the relative orientations of the slo and Ex loci on the chromosome were not known and the distance between them was not firmly established, so both possibilities had to be taken into account.
The experimental strategy employed to ascertain this possibility was basically the same as that exercised in a search for transposons. In the case of deletion, the PCR primers were designed to detect an excised, circular chromosomal fragment (Fig. 5D). Again, no PCR fragments were detected to verify the occurrence of inversion or deletion.
Rearrangement represses slo gene expression.
To investigate the possible effect of recombination on the activity of the nearby slo gene, sets of fusion strains with erm and lacZ reporter genes were constructed. Figure 2 illustrates the construction of the first set of strains with different slo upstream regions fused to an erm reporter gene. Resistance to erythromycin in these strains was tested on THY plates containing erythromycin by inspecting growth after overnight incubation at 37°C and also by measuring growth in THY broth supplemented with erythromycin. Strains OK94 and OK96, in which the erm gene is fused to the natural upstream region of the slo gene (Fig. 2D and E), produced individual colonies on plates containing 0.1 μg of erythromycin per ml. Surprisingly, strain OK95 harboring the recombinant Ex/slo fragment with the putative Ex promoter upstream of the erm gene (Fig. 2F) was sensitive to erythromycin at 0.1 μg/ml (results not presented). The same pattern of strain sensitivity to erythromycin was reproduced in an experiment in which bacterial growth was monitored in THY broth supplemented with 0.1 μg of erythromycin per ml (results not presented).
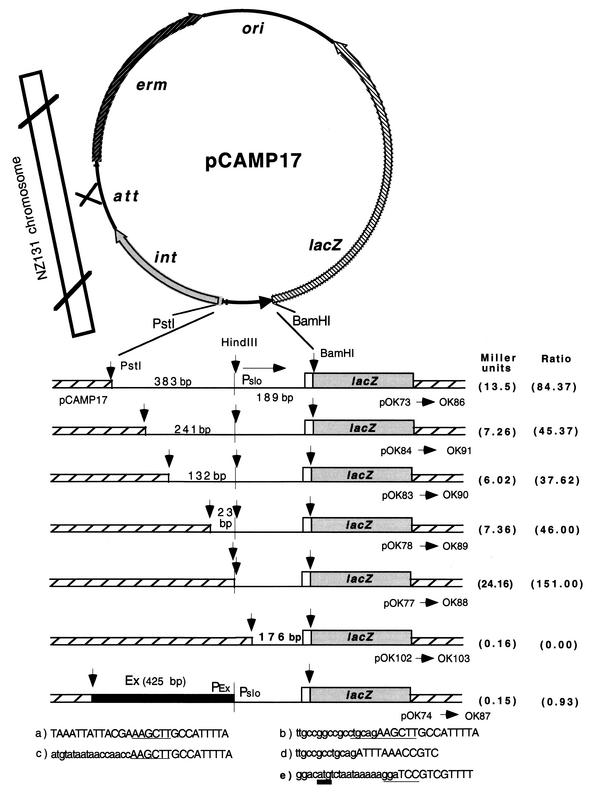
In a parallel experiment, a set of strains containing a protein fusion with lacZ as a reporter gene were constructed. Similar to previous constructions, the fragments fused to the lacZ gene were obtained by PCR amplification with mutagenic oligonucleotides containing PstI and BamHI restriction sites. The fragments amplified from strain NZ131 chromosomal DNA contained the proximal part of the slo gene and different lengths of the slo upstream region (Fig. 6). The fragment amplified from pOK23-1 contains the Ex/slo recombinant fragment (Fig. 6). pOK23-1 is a derivative of pOK23 (Fig. 2) that carries a mutated form (ACTG) of the slo gene initiation codon (not shown). A wild-type initiation codon was recreated by PCR from pOK23 with the mutagenic primer P99. Primer P99 also introduces a flanking BamHI restriction site, while the other primer, P98, creates a PstI restriction site.
FIG. 6.
Plasmid constructs with lacZ reporter gene fused to the proximal part of the slo gene and fragments harboring different constructs of the slo upstream region. The vertical line represents the recombination point between Ex and slo DNAs. Pslo and Pex denote the native slo promoter (22) and the hypothetical promoter situated on Ex DNA, respectively. The correctness of the nucleotide sequence in all constructs was verified by DNA sequencing. Strains OK86 to OK103 were generated by introducing plasmids pOK73 to pOK102, respectively, into the recipient strain NZ131. The genetic configuration of the plasmids with fusion upon their integration into the chromosome of NZ131 was verified by PCR with appropriate primers. The nucleotide sequences at the bottom denote the fusion point between the slo upstream region and pCAM17 DNA or Ex DNA in the presented constructs. (a) Strains OK86, OK91, OK90, and OK89; (b) strain OK88; (c) strain OK87; (d) strain 103; (e) nucleotide sequence at the junction (BamHI site) between the proximal part of the slo gene (first 17 bp; open box) and the lacZ gene (shaded box) shared by all fusions. HindIII, PstI, and BamHI restriction sites are underlined. The translation initiation codon of the slo gene is underlined with a thick line. The values in the last column demonstrate the ratio between the β-galactosidase activity presented in the first column and the β-galactosidase activity obtained in strain OK103 with a deleted slo promoter (22).
Upon treatment with PstI and BamHI enzymes, the fragments were cloned into the PstI- and BamHI-treated vector pCAMP17. As shown (Fig. 6), the BamHI restriction site is located inside both the slo and lacZ genes, so that the products obtained at the splicing region between the lacZ gene and the slo gene included the Shine-Dalgarno sequence, ATG initiation codon, and the first four codons of the slo gene. A special advantage of the pCAMP17 vector is that besides the lacZ reporter gene and the erm selective marker, it also harbors the S. pyogenes phage T12 genes for integrase and its specific attachment site (8). These characteristics enable pCAMP17 to simulate a temperate pathway of phage T12, i.e., to integrate into a specific attachment site in the S. pyogenes chromosome (8, 14). Finally, integration of pCAMP17 into the chromosome is irreversible because excision of phage T12 also requires the enzyme excisionase, which is lacking in this system (14).
Preliminary measurements with strain OK86 showed that slo, i.e., the lacZ gene, is expressed practically at the same level from the early log to the stationary phase in THY broth, so the samples for measuring β-galactosidase activity were taken at the end of the log phase. The strains carrying increasing sections of the slo upstream region demonstrated low but reproducible expression of the reporter gene (Fig. 6). The negative control strain NZ131 with no plasmid (results not presented) as well as strain OK103 with a deletion of the −35 sequence of the slo promoter (22) demonstrated no β-galactosidase activity. Similar to strain OK95 with the erm reporter gene (Fig. 2F), the strain carrying the recombinant Ex/slo upstream region (OK87) showed no β-galactosidase activity. Interestingly, strain OK88, in which the natural slo upstream sequence terminates at practically the same nucleotide as in strain OK87 but is spliced at the breakpoint not with Ex DNA but with the vector pCAMP17 DNA, demonstrated a higher level of β-galactosidase activity than strain OK86, harboring the longest segment (573 bp) of the natural slo upstream region (Fig. 6).
DISCUSSION
In this study we describe the detection and partial characterization of a rearrangement involving two S. pyogenes chromosomal loci separated by 745 kb on the 1.85-Mb-long chromosome. By its nature, the event is RecA independent and seems to be ubiquitous among GAS. The phenomenon was detected by PCR, and its functional consequence on expression of the slo gene was monitored by fusing reconstructed recombinant molecule with the erm and lacZ reporter genes.
The structural aspect of this rearrangement remains obscure for the moment. The experimental strategies applied in the quest for the other terminus, i.e., design of appropriate primers for PCR, were grounded in structural models of genomic rearrangements such as transpositions, inversions, and deletions (2, 4). With this method, we found neither inversion nor deletion as a basis for the rearrangement. The failure to identify an inversion is not surprising because the same genetic polarity of the recombining loci was found postfestum during genome sequencing of strains M370, MGAS8232, and NZ131 (6, 25; W. M. McShan et al., unpublished data).
Constant generation of a G/C pair at the Ex/slo junction only seemingly indicates a classical transposition event almost always characterized by duplication of the target sequence. However, this extra G/C does not fit into any known mechanism of target duplication following transposition (2). We also did not succeed in describing a mechanism by which an extra G/C pair would be generated via the conjugative transposon insertion pathway, known to generate not only base substitutions but also frameshift mutations in the course of homogenization of mismatched staggered ends (4). The fact that the chromosomal regions in the neighborhood of both slo and the Ex recombination sites do not demonstrate any resemblance to transposons (6, 25; W. M. McShan et al., unpublished data) also speaks against a transposon model. The failure to demonstrate the transposon nature of the rearrangement might have been caused by larger than expected divergence in homology of the postulated transposon termini. Designing more alternative primers for PCR experiments, particularly various nucleotides at the 3′OH end, might help in further testing of this possibility. Finally, there is a possibility that this rearrangement might be related to transposons which diverge widely from the common models. The most extreme example is the reported case of transposition without transposase, in which a block of donor DNA is inserted into the target sequence without the involvement of any known transposase. In addition, the described transposable element lacks typical transposon ends, and insertion of a transposon is not associated with characteristic duplication of the target sequence (19).
Aside from transposition models, the structural requirements in genomic rearrangements such as direct or inverted repeats or monotonous nucleotide stretches seem not to be universally required. The recent report on capsular phase variation in Streptococcus pneumoniae clearly shows that the event is due to an apparently random formation of duplications and their subsequent precise excision within a heterogeneous nucleotide sequence of the cap3A gene (31).
The reproducibility of the recombination event and the characteristics of the sequences around the junction between the Ex and slo DNAs argue that this rearrangement is not a stochastic, pathological event, but a chromosomal rearrangement of some biological significance. Our failure to detect the relevant PCR fragments in the search for the other terminus and the failure to observe amplified fragments in several PCR experiments with primers located in either the slo or Ex locus paired with primers around the chromosome (results not presented) indicate that background recombination between distant chromosomal loci, as opposed to the Ex/slo recombination event, is a rare event. Finally, positioning two decamers into a proximity of only 50 nucleotides (Fig. 3) argues in favor of the rearrangement as a nonstochastic event. The TTGCCATTTT decamer appears 18 times in all three sequenced strains SF370, MGAS8232, and NZ131 (6, 25; W. M. McShan et al., unpublished data). The average distance between decamers in these strains is about 103 kb, while minimal and maximal distances between two decamers are approximately 4.2 kb and 280 kb, respectively. As computed from the chromosomal sequence of strain SF370, the TTGCCATTTT sequence is statistically expected only 3.4 times (17), which also argues against the random nature of this decamer and its coincidental positioning of two copies in the recombinant.
The results obtained with the erm (Fig. 2) and lacZ (Fig. 6) reporter genes for monitoring the effect of the rearrangement on the expression of the slo gene in a certain way confirmed the assumption about the nonrandom nature of the Ex/slo rearrangement. The rearrangement did produce an effect on expression of the slo gene, although the effect was quite opposite to what was expected, that positioning of the putative Ex promoter upstream of the slo gene would boost the expression of that gene in the recombinants (strain OK95, Fig. 2; strain OK87, Fig. 6). In both reporter systems, joining of the putative Ex DNA promoter to the slo gene upstream region resulted in a complete lack of expression of the reporter genes. The fact that the clone containing the identical segment of the slo gene upstream region joined not to the Ex DNA but to the pCAMP17 vector DNA (Fig. 6, strain OK88) showed significant expression of the reporter gene argues against the possibility that recombination immediately upstream of the slo gene native promoter (22) was detrimental to the function of that promoter. The lack of β-galactosidase activity in the strain with the deleted −35 sequence of the native slo promoter (Fig. 6) (22) shows that the activity of the reporter gene in strain OK88 is not a consequence of some spurious transcription coming from the pCAMP17 vector.
One of the possible answers for the difference in expression of the lacZ gene in strains OK87 and OK88 is that the native slo promoter might be a superhelicity-sensitive promoter, its activity being affected in different ways by the nearness of Ex DNA (OK87) and pCAMP17 DNA (OK88). A similar explanation is that the applied experimental system is unsuitable, i.e., that in strains OK87 and OK95, both native and putative Ex promoters are inhibited by structural constraints induced by inappropriate superhelicity of the chromosomal region in which they integrate. However, plasmid pOK67 generated strain OK95 by integrating itself into the Ex region (Fig. 2F), in contrast to pOK74, which integrated at a completely different site on the chromosome, the gene for serine-tRNA (Fig. 6) (14). It is hard to envision that only in the strains having Ex DNA joined to the slo DNA (OK87 and OK95), as opposed to other strains (e.g., OK96 and OK88), the local topology is affected so as to impede functioning of the promoters. In addition, the activity of promoters of the cfa gene (8) and the speA gene (J. Liu and J. J. Ferretti, unpublished data) monitored by the same system demonstrated normal β-galactosidase activities.
A recent important report on a type III-like secretion system in S. pyogenes may offer an alternative explanation for the biological meaning of the Ex/slo rearrangement (12). In that work, a model is presented in which the effector NAD-glycohydrolase, coded by nga (spn), the gene immediately upstream of the slo gene, is transported through the Slo pore into the target cell. It also suggests that the slo gene is a part of an nga-slo operon transcribed from a promoter immediately upstream of the nga gene, implying their common regulation of transcription (12). Consequently, the Slo protein is always physiologically connected to NAD-glycohydrolase, providing the secretion function. However, in our independent study (22), we showed that the slo gene, although being mostly transcribed from the strong nga promoter, also possesses its own promoter. This promoter shows weak activity, at least under in vitro experimental conditions (growth in THY), which explains the low values obtained with the lacZ reporter gene clones not containing the upstream nga promoter (Fig. 6) (22). We also found that the slo promoter is negatively regulated by the 19-kb-distant gene sloR (alias Spy0146 [6, 22]). These data suggest that the regulatory network of the nga-slo operon is more complex and that the products of the nga and the slo genes may also have other, independent roles in pathogenesis. The Ex/slo recombination that splits the nga-slo operon and physiologically separates NAD-glycohydrolase from Slo introduces a new variable into this genetic system.
The frequency of rearrangement, as measured in vitro by PCR, is low (10−4 in strain SF370 and 10−5 in strain NZ131). The detected frequency of recombination in these experiments may not be too low, since a quantitative study by Simpson and Cleary (24) showed that the gene for M12 protein reversibly switches between on and off states at a “high” frequency, from 10−3 to 10−4, which is close to the frequency of Ex/slo rearrangement found in strain SF370 (10−4). Thus, the observed rearrangement may belong to a plethora of still undefined genetic switches and rearrangements that cause instability of GAS strains. In addition, the Ex/slo rearrangement might be advantageous under certain physiological conditions, and mutants harboring the rearrangement could quickly become a significant or dominant part of the population.
We still do not know the extent of the chromosomal perturbation caused by the rearrangement. More genes may be involved, repression of the slo gene being only part of a more complex picture. Further experiments, including immortalization of the described rearrangement as a priority, are necessary for complete understanding of this phenomenon.
Acknowledgments
We are grateful to D. Clewell and K. Dybvig for critical reading of the manuscript and helpful suggestions. We also thank D. Ajdic, R. E. McLaughlin, W. M. McShan, and A. Suvorov for generous assistance during this project and C. Primeaux and G. Savic for excellent technical service.
This study was supported by NIH grant C1105902 to J.J.F.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, X. Schäffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped blast and PSI-BLAST: a new generation of protein data base search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, D. E., and M. M. Howe (ed.). 1989. Mobile DNA. American Society for Microbiology, Washington, D.C.
- 3.Borst, P., and D. R. Greaves. 1987. Programmed gene rearrangements altering gene expression. Science 235:658-667. [DOI] [PubMed] [Google Scholar]
- 4.Clewell, D. B., and S. E. Flannagan. 1993. The conjugative transposons of Gram-positive bacteria, p. 369-393. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 5.Dybvig, K. 1993. DNA rearrangement and phenotypic switching in prokaryotes. Mol. Microbiol. 10:465-471. [DOI] [PubMed] [Google Scholar]
- 6.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gase, K., T. Ellinger, and H. Malke. 1995. Complex transcriptional control of the streptokinase gene of Streptococcus equisimilis H46A. Mol. Gen. Genet. 247:749-758. [DOI] [PubMed] [Google Scholar]
- 8.Gase, K., J. J. Ferretti, C. Primeaux, and W. M. McShan. 1999. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect. Immun. 67:4725-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haselkorn, R. 1992. Developmentally regulated gene rearrangements in prokaryotes. Annu. Rev. Genet. 26:113-130. [DOI] [PubMed] [Google Scholar]
- 10.Kehoe, M. A., L. Miller, J. Walker, and G. J. Boulnois. 1987. Nucleotide sequence of the streptolysin O (SLO) gene: structural homologies between Slo and other membrane-damaging thiol-activated toxins. Infect. Immun. 55:3228-3232., [DOI] [PMC free article] [PubMed]
- 11.Kolstø, A. B. 1997. Dynamic bacterial genome organization. Mol. Microbiol. 24:241-246. [DOI] [PubMed] [Google Scholar]
- 12.Madden, J. M., N. Ruiz, and M. Caparon. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in Gram-positive bacteria. Cell 104:143-152. [DOI] [PubMed] [Google Scholar]
- 13.McLaughlin, R. E., and J. J. Ferretti. 1998. Electrotransformation of streptococci. Methods Enzymol. 47:185-193. [DOI] [PubMed] [Google Scholar]
- 14.McShan, M. W., R. W. McLaughlin, A. Nordstrand, and J. J. Ferretti. 1998. Vectors containing streptococcal bacteriophage integrases for site-specific gene insertion. Methods Cell Sci. 20:51-57. [Google Scholar]
- 15.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 16.Musser, J. M., V. Kapur, J. Szeb, X. Pan, D. S. Swanson, and D. R. Martin. 1995. Genetic diversity and relationships among serotype M1 strains of Streptococcus pyogenes. Dev. Biol. Stand. 85:209-213. [PubMed] [Google Scholar]
- 17.Nei, M., and W. S. Li. 1979. Mathematical model for studying genetic variations in terms of restriction nucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogasawa, N., S. Moriya, and H. Yoshikawa. 1985. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase and other open reading frames. Nucleic Acid Res. 13:2267-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rappleye, C. A., and J. R. Roth. 1997. Transposition without transposase. J. Bacteriol. 179:2047-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain- terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savic, D. J., W. M. McShan, and J. J. Ferretti. 2002. Autonomous expression of the slo gene of the bicistronic nga-slo operon of Streptococcus pyogenes. Infect. Immun. 70:2730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon, D., and J. J. Ferretti. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 82:219-224. [DOI] [PubMed] [Google Scholar]
- 24.Simpson, W. J., and P. P. Cleary. 1987. Expression of M type 12 protein by a group A streptococcus exhibits phase-like variation: evidence for coregulation of colony opacity determinants and M protein. Infect. Immun. 55:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smoot, J. C., K. D. Barbian, J. J. Van Gompel. L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. F. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stivens, F. A., and R. A. Dochez. 1924. Studies on the biology of the streptococcus. III. Agglutination and absorption of agglutinin with Streptococcus scarlatinae. J. Exp. Med. 49:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stragier, P., B. Kunkel, L. Kroos, and R. Losick. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507-512. [DOI] [PubMed] [Google Scholar]
- 28.Suvorov, A. N., and J. J. Ferretti. 1994. Heterogeneity of group A streptococcal M1 chromosomal restriction patterns as determined by PFGE, p. 21-23. In A. Totolian (ed.), Pathogenic streptococci: present and future. Lancer Publications, St. Petersburg, Fla.
- 29.Suvorov, A. N., and J. J. Ferretti. 1996. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J. Bacteriol. 178:5546-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao, L., S. K. Hollingshead, A. N. Suvorov, J. J. Ferretti, and W. M. McShan. 1995. Construction of a Streptococcus pyogenes recA mutant via insertional inactivation, and cloning and sequencing of the complete recA gene. Gene 162:59-62. [DOI] [PubMed] [Google Scholar]
- 31.Waite, R. D., J. K Struthers, and C. G. Dowson. 2001. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high-frequency phase variations. Mol. Microbiol. 42:1223-1232. [DOI] [PubMed] [Google Scholar]
- 32.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed]