Abstract
CS31A produced by septicemic and diarrheic Escherichia coli belongs to the Pap-regulatory family of adhesive factors, which are under methylation-dependent transcriptional regulation. Common features of operons encoding members of this family include two conserved GATC sites in the upstream regulatory region, and transcriptional regulators homologue to the PapB and PapI proteins. Methylation protection of GATC sites was previously shown to be dependent on the leucine-responsive regulatory protein (Lrp). Lrp and ClpB, the PapB equivalent, repressed clp basal transcription. A PapI homologue (AfaF) was required together with Lrp to establish the phase variation control, which gave rise to phase-ON cells that expressed CS31A and phase-OFF cells that did not express CS31A. In phase-OFF cells, the GATCdist site was methylated and the GATCprox site was protected from methylation, whereas in phase-ON cells, the inverse situation was found. Unlike Pap fimbriae, CS31A synthesis was dramatically reduced in media containing l-alanine or l-leucine. l-Alanine prevented the OFF-to-ON switch, locking clp expression in the OFF phase, whereas l-leucine repressed transcription without obvious effect on the switch frequency of phase variation. In phase-variable cells, leucine and alanine promoted methylation of GATCdist and methylation protection of GATCprox, increasing the methylation pattern characteristic of repressed cells. Furthermore, alanine prevented the AfaF-dependent methylation protection of GATCdist and thus the appearance of phase-ON cells. In addition, analysis of clp expression in a Lrp-negative background indicated that alanine and leucine also repressed clp transcription by a methylation-independent mechanism.
Lrp (leucine-responsive regulatory protein) is a global transcriptional regulator of metabolism in Escherichia coli. It affects the expression of a number of operons involved in amino acid biosynthesis and degradation, nutrient transport, and formation of fimbriae. Many of these operons are also subject to control by leucine and alanine. All the possible regulatory patterns in response to Lrp and leucine have been demonstrated. Lrp positively regulates some operons and negatively regulates others. In each category, leucine antagonizes the action of Lrp on certain operons and potentiates it on other operons. Some other positively or negatively regulated operons are insensitive to leucine. In a number of cases, alanine has been shown to have a similar effect to leucine (for reviews, see references 7 and 31). It has been shown that leucine antagonizes the activating action of Lrp on ilvIH and gltBDF promoters and the repressive action of Lrp on the lysU promoter by reducing the affinity of Lrp for its binding sites (13, 25, 37). To explain why some operons are insensitive to leucine even when the cell contains enough leucine to bind to Lrp, Ernsting et al. suggested that the effect of leucine on positively regulated genes depends on the effective in vivo concentration of Lrp (13). At high Lrp concentrations, Lrp binds to the promoters regardless of the presence or absence of leucine, whereas at low Lrp concentrations, the activities of these promoters is leucine sensitive. If Lrp binds to different promoters with different affinities, the target genes with the lowest affinity for Lrp are the most sensitive to leucine. However, such a model does not explain how leucine can potentiate the action of Lrp.
Regulation of expression of fimbriae belonging to the P-regulatory family involves Lrp, which directly interacts with another regulatory protein encoded by the fimbrial operons, a PapI homologue. Expression of all the operons so far studied belonging to this family is insensitive to leucine, except for the clp and foo operons encoding CS31A and F1651 fimbriae, respectively (19, 28). Fimbrial operons belonging to the P-regulatory family share important regulatory features. They are subject to phase variation control (except fae, encoding K88), defined as the ability of cells to switch in a reversible manner between fimbriated, phase-ON cells and afimbriated, phase-OFF cells. Lrp and a PapI homologue are required for the switch to occur (20, 26). The Lrp target regulatory region contains two GATC sites that are spaced 102 or 103 bp apart. The differential methylation status of these sequences determines the binding of Lrp, because Lrp binds only nonmethylated or hemimethylated GATC sites (34). PapI or its homologues have the capacity to displace Lrp from one binding site to another (24, 33). The activity of the promoter depends on the binding of Lrp, since Lrp activates expression when bound to certain sites and represses expression when bound to others (33, 38, 40). All operons also encode a PapB homologue that appears to play a dual role in the control of pap expression. Low levels of PapB activate papI transcription, whereas high levels suppress papBA expression (3, 18, 42). Distinct regulatory features can also be observed between members of the P-regulatory family. Null mutations in the lrp gene reduce the transcription of some operons, such as pap, pef, sfa, and daa (4, 32, 39), and increase the transcription of others, such as clp or fae (22, 28). PapI homologues activate the transcription of pap, sfa, daa, and foo (33, 39) and repress the transcription of pef, fae, and clp (22, 28, 32).
CS31A, encoded by the clp operon is produced by septicemic and enterotoxigenic bovine (9, 16) and human (23) E. coli strains. In E. coli reference strain 31A, CS31A is encoded by a high-molecular-weight plasmid called p31A. CS31A mediates adhesion to Caco-2 and Int-407 cells (12, 23), is required for full bacterial virulence in the rat model, and protects bacteria from phagocytosis by bovine polymorphonuclear neutrophils (J. P. Girardeau, personal communication). In the absence of phase variation, i.e., in the absence of a PapI homologue in the cells, Lrp and the specific regulator ClpB negatively control the basal clp expression. Under these conditions, GATCdist is methylated whereas GATCprox is protected from methylation. Lrp, but not ClpB, is required for methylation protection of GATCprox. Methylation is required for repression (28). Leucine and alanine dramatically reduce CS31A production (16, 29). We have previously shown that alanine represses basal clp transcription twofold in the absence of a PapI homologue and inhibits phase variation in its presence (28).
In this study, CS31A phase variation was examined using AfaF, a PapI equivalent. The influence of AfaF and Lrp on the methylation patterns and their role in phase variation were investigated. The influence of leucine and alanine on clp expression and on the methylation pattern was examined. As emphasized by Blomfield, changes in phase variation as opposed to steady-state control could have different outcomes on fimbriae production (1a). Thus, we tried to distinguish the effect of leucine and alanine on each process by studying the steady-state expression (i.e., the basal transcription) and the phase variation in the absence and presence of AfaF.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli strains MC4100.λ6, MC4100.λ15, DL844.λ6, and DL844.λ15/1 contain a single chromosomal copy of a clp-lacZYA fusion (see Fig. 1). The λ6 lysogens contain an entire functional clpB gene, whereas clpB is truncated in the λ15 lysogens (28). DL844.λ15/1 was constructed as previously described to replace DL844.λ15 (28). The PCR assay described by Powell et al. (36) was used to verify that the four strains contained single lysogens. By this method, the DL844.λ15 strain used in a previous study (28) was shown to harbor more than one copy of the prophage, which was not detected by the λ terminase test (2) that was used previously. DL844 is an Lrp-negative isogenic MC4100 strain (5). ptrf1 and pCLB99 are multicopy recombinant plasmids derived from ptrc99A (Pharmacia Biotech) and express afaF and clpB, respectively, under the control of the trc promoter (28). Luria-Bertani broth, Luria-Bertani agar, M9 minimal broth, and M9 minimal agar were prepared by the method of Miller (30). Glycerol as carbon source was added at 0.2% (wt/vol). Supplements, when appropriate, were used at the following concentrations: 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 40 μg ml−1; isopropyl-β-d-thiogalactopyranoside (IPTG), 1 mM; l-leucine or l-alanine, 5 mM; ampicillin, 50 μg ml−1; and kanamycin, 50 μg ml−1. Isoleucine (0.4 mM) and valine (0.6 mM) were added to media containing 5 mM leucine. Cultures of strains harboring ptrf1 were done without IPTG to avoid too high an expression of afaF.
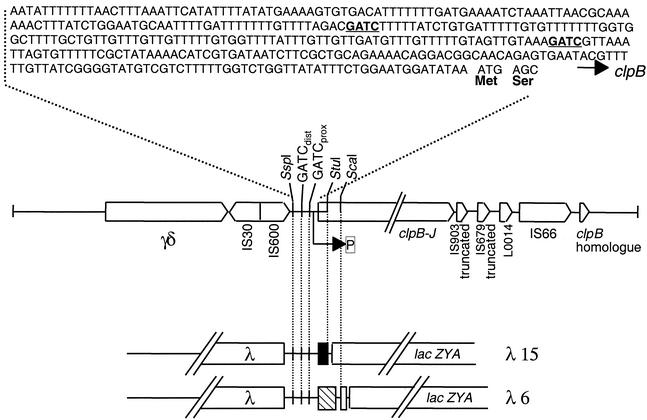
FIG. 1.
The clp regulatory region, sequences surrounding the clp operon, and operon fusion constructs. The nucleotide sequence of the clp regulatory region is shown at the top, with the GATC sites in bold. The GenBank accession number for this sequence is L48184. The organization of DNA surrounding the clp operon is shown below the regulatory region. Boxes with arrowheads indicate the positions of sequences with high homologies to sequences present in data banks as found using the BLASTn program (National Center for Biotechnology Information). The operon fusion constructs are shown in expanded scale at the bottom. The clpB gene is truncated in λ15 (black box) and is entire in λ6 (hatched box). P indicates the putative clp promoter.
Assay of β-galactosidase activity.
A single bacterial colony picked on M9-glycerol agar plates containing X-Gal was used to inoculate an overnight culture in M9-glycerol medium. This culture was diluted 50-fold, grown to logarithmic phase (optical density at 600 nm, 0.5 to 0.8) in M9-glycerol medium supplemented with amino acids as indicated. Cultures were assayed for β-galactosidase activity as described previously (30). For strains showing phase variation, ON (blue) or OFF (white) colonies were picked from M9-glycerol-X-Gal plates as starting material for assays of activity of ON and OFF cultures, respectively. Each experiment was done in duplicate at least five times.
Calculation of the switch frequency.
Lysogen strains were inoculated onto M9- glycerol-X-Gal plates supplemented with l-leucine or l-alanine or left unsupplemented. Colonies showing a uniform phenotype were resuspended in M9 salts. Appropriate dilutions were spread onto the same medium as the parent colony. After growth, the colonies were scored for a light blue or dark blue phenotype. The switch frequencies were calculated using the formula (M/N)/g, where M is the number of cells that underwent phase transition, N is the total number of cells evaluated, and g is the total number of generations, estimated to be 25, that gave rise to the colony.
DNA sequencing.
For sequencing of DNA regions adjacent to the clp operon, the genome walker kit from Clontech Laboratories was used. p31A DNA was digested by restriction endonucleases that generate blunt ends and ligated to the adaptors provided in the kit as specified by the manufacturer. PCR amplification using the adaptor primer and a clp-specific primer allowed sequencing of unknown DNA fragments adjacent to clp without previous cloning. Templates for DNA sequencing were prepared using a PE Applied Biosystem dye terminator cycle-sequencing kit. Products were analyzed on an ABI Prism 310 DNA sequencer. Oligonucleotides were from Eurogentec or MWG Biotech.
DNA hybridizations.
For chromosomal MC4100.λ6 DNA purification, a single blue or a single white colony was picked from M9-glycerol-X-Gal agar, resuspended in M9 salts, and used to inoculate three flasks containing the appropriate medium. Isoleucine (0.4 mM) and valine (0.6 mM) were added to the medium containing 5 mM leucine. Chromosomal DNA was isolated from overnight cultures using the DNeasy tissue kit (Qiagen). Aliquots of the cultures used for DNA purification were plated on the corresponding medium to calculate the percentage of ON cells. Digestions with restriction endonucleases MboI and DpnI were performed as specified by the manufacturer. DNA restriction fragments were separated by electrophoresis on 1.5% agarose in TBE buffer and transferred to a nylon membrane (Hybond N+; Amersham Biotech) in 20× SSC (0.3 M sodium citrate, 0.33 M NaCl [pH 7.0]) using a vacuum blotter (Appligene). Transfer was carried out at 4.5 kPa for 60 min. DNA fragments were cross-linked to the filter with UV light by using a Spectrolinker (Spectronics Corp.). The filter was probed with digoxigenin-labeled DNA (240 bp) generated by PCR using the PCR DIG probe synthesis kit (Roche) and primers RU (5′-CCTGTTTGATTTCGTGC-3′) and RD (5′-CTGCAGCGAAGATTATC-3′). Hybridization was detected by chemiluminescence using CDP-Star (NEN) as a substrate for alkaline phosphatase. Quantification was performed by densitometry analysis using the Diversity DataBase software from Bio-Rad.
RESULTS
CS31A phase variation.
To analyze CS31A phase variation, we attempted to clone the clp papI homologue. By analogy to other fimbrial operons belonging to the P regulatory family, we supposed that the papI homologue would be present upstream or downstream next to the clp operon. However, nucleotide sequencing revealed that diverse insertion sequences and transposons flank the clp operon up to 7.8 kb upstream and 4.6 kb downstream. An open reading frame presenting 44% identity to the functional clpB gene is present 3 kb downstream of the clp operon but is isolated and also flanked by insertion sequences or unknown sequences (Fig. 1). Furthermore, our numerous efforts to clone the papI homologue from plasmid p31A were unsuccessful. PCR assays using afaF- or papI-derived primers failed to amplify a DNA fragment from p31A. A p31A DNA library was constructed and hybridized at high stringency with a 400-bp afaF probe (28). Hybridization was found only with a 65-bp sequence derived from an insertion sequence. No more hybridization was found after deletion of this sequence in the probe, even at low stringency. Transformation of p31A in MC4100.λ6 or MC4100.λ15 failed to restore phase variation. Either this gene indeed exists on p31A and we were not able to clone it, or CS31A phase variation is controlled by a PapI homologue encoded to a related chromosomally located operon. We therefore decided to analyze CS31A phase variation by using in trans AfaF, the PapI homologue belonging to the afa-3 gene cluster. Afa-3 is a nonfimbrial adhesin (15) with a structural organization more closely related to CS31A than to P-fimbriae. AfaF was chosen because the ClpB and AfaA regulators (PapB homologues) have 60.6% amino acid sequence identity (28). Furthermore, we have shown previously that plasmid ptrf1, harboring afaF under the control of the trc promoter, promotes phase variation when introduced into E. coli strains harboring clp-lacZYA transcriptional fusions (28). To study transcriptional clp regulation, we used different strains that harbor a single chromosomal clp-lacZYA transcriptional fusion, with (λ6 lysogens) or without (λ15 lysogens) an intact clpB gene (Fig. 1). The fusions were created either in MC4100 E. coli or in DL844 E. coli to provide Lrp+ or Lrp− backgrounds, respectively. As already demonstrated, uniform blue colonies arose by plating λ6 orλ15 lysogens on M9-glycerol plates containing X-Gal in the absence of AfaF (28). Table 1 shows that ClpB and Lrp regulated the clp basal transcription even in the absence of the phase variation control (i.e., in the absence of AfaF): comparison of DL844.λ6 (493 MU) with MC4100.λ6 (112 Miller units [MU]) showed that Lrp played a repressive role. Similarly, comparison of MC4100.λ15 (775 MU) with MC4100.λ6 (112 MU) showed that ClpB also played a repressive role, which was previously demonstrated in MC4100.λ15 complemented in trans by pCLB99, a multicopy plasmid expressing clpB under the control of the trc promoter (28). It is not clear why the transcription was not totally derepressed in the DL844.λ15/1 strain, which lacks Lrp and ClpB (see Discussion). After the plasmid encoding AfaF was introduced into the strains, phase variation occurred only in the Lrp+ backgrounds, with emergence of a majority of white colonies among the blue ones, indicating a global repressive effect. Measurements of the β-galactosidase activity of Lac+ (phase-ON) and Lac− (phase-OFF) colonies are reported in Table 1. Transcription was highly repressed in phase-OFF cells (18 and 38 MU in ClpB+ and ClpB− strains, respectively). When AfaF was overexpressed due to the addition of 1 mM IPTG to the growth medium, the colonies presented a uniform Lac− phenotype on M9-glycerol-X-Gal agar plates, indicating that transcription of clp-lacZ was completely inactive (data not shown). Without Lrp, transcription was locked in the ON phase [DL844.λ6(ptrf1), 338 MU]. Thus, Lrp was required for repression of the promoter and emergence of phase-OFF cells. However, when we took into account the fraction of phase ON cells in the MC4100.λ6(ptrf1) colonies, it appeared that the transcription level of phase-ON cells was higher than in the absence of AfaF (87 MU for 21% of phase ON-cells versus 112 MU for 100% of phase-ON cells). Since AfaF had no regulatory effect in an Lrp− background [Table 1, DL844.λ15/1 versus DL844.λ15/1(ptrf1)], our data suggest that AfaF modulated the regulatory activity of Lrp by increasing its repressive effect in a large fraction of cells (phase OFF) and by conferring to it an activating activity in the remaining fraction (phase ON). In contrast, ClpB was not required for the phase variation to occur. The ClpB-dependent repression was still effective in phase-ON cells [compare MC4100λ6(ptrf1) to MC4100.λ15(ptrf1)]. However, results obtained with MC4100.λ15(ptrf1) were difficult to analyze: while Lac− colonies were really white, Lac+ colonies presented all the gradations of blue, leading to a fraction of phase-ON cells in a Lac+ colony that varied greatly from one experiment to another [which was not the case for MC4100.λ6(ptrf1)], ranging from 6 to 78%. Such a variability has already been observed in Salmonella enterica serovar Typhimurium populations expressing Pef fimbriae (32), but it remains unexplained.
TABLE 1.
Transcription levels of clp-lacZYA in different media and different genetic backgrounds
| Strain | Genotype | Growth mediuma | Starting colony | Switch frequency
|
β-Galactosidase activity (MU)c | % of phase- ON cells | |
|---|---|---|---|---|---|---|---|
| ON to OFF | OFF to ON | ||||||
| MC4100.λ6 | clpB+lrp+afaF | M9 | Ub | 112 ± 9 | |||
| M9-alanine | U | 36 ± 3 | |||||
| M9-leucine | U | 53 ± 7 | |||||
| MC4100.λ6(ptrf1) | clpB+lrp+afaF+ | M9 | Phase ON | 3.16 × 10−2 | 87 ± 21 | 21.1 | |
| Phase OFF | 1.47 × 10−3 | 18 ± 5 | 3.68 | ||||
| M9-leucine | Phase ON | 3.24 × 10−2 | 10 ± 3 | 19.12 | |||
| Phase OFF | 1.81 × 10−3 | 8 ± 3 | 4.53 | ||||
| DL844.λ6 | clpB+lrp afaF | M9 | U | 493 ± 59 | |||
| M9-alanine | U | 197 ± 39 | |||||
| M9-leucine | U | 250 ± 3 | |||||
| DL844.λ6(ptrf1) | clpB+lrp afaF+ | M9 | U | 338 ± 57 | |||
| M9-alanine | U | 163 ± 33 | |||||
| M9-leucine | U | 200 ± 66 | |||||
| MC4100.λ15 | clpB lrp+afaF | M9 | U | 775 ± 60 | |||
| M9-alanine | U | 170 ± 20 | |||||
| M9-leucine | U | 238 ± 71 | |||||
| MC4100.λ15(ptrf1) | clpB lrp+ afaF+ | M9 | Phase ON | 2.37 × 10−2 | 336 ± 144 | 40.35 | |
| Phase OFF | 1.15 × 10−3 | 38 ± 11 | 2.96 | ||||
| M9-alanine | U | 11 ± 4 | |||||
| M9-leucine | Phase ON | 2.64 × 10−2 | 152 ± 30 | 33.6 | |||
| Phase OFF | 1.16 × 10−3 | 31 ± 4 | 2.91 | ||||
| DL844.λ15/1 | clpB lrp afaF | M9 | U | 276 ± 36 | |||
| M9-alanine | U | 202 ± 28 | |||||
| M9-leucine | U | 295 ± 47 | |||||
| DL844.λ15/1(ptrf1) | clpB lrp afaF+ | M9 | U | 278 ± 81 | |||
| M9-alanine | U | 221 ± 44 | |||||
| M9-leucine | U | 290 ± 26 | |||||
Glycerol is the carbon source. Media containing leucine are supplemented with isoleucine (0.4 mM) and valine (0.6 mM).
U, uniform phenotype of the colonies.
Results are the means±standard deviations of at least five independent experiments made in duplicate.
Influence of leucine and alanine on clp expression.
Because CS31A is one of the only two known P-related fimbriae sensitive to leucine and alanine, we analyzed in further detail the effect of these amino acids on clp transcription. Alanine abolished clp transcription, locking cells in phase OFF (Table 1). In contrast, leucine did not dramatically alter the switch frequency of phase variation. We measured the β-galactosidase activity in different strain backgrounds starting either from a Lac+ or from a Lac− colony grown on M9-glycerol-X-Gal with l-alanine or l-leucine (Table 1). Alanine and, to a lesser extent, leucine dramatically decreased clp transcription in MC4100.λ6 and MC4100.λ15 strains in the presence and absence of AfaF. Repression was also observed in the Lrp− ClpB+ (DL844.λ6) backgrounds with and without AfaF. However, in the simultaneous absence of ClpB and Lrp [DL844.λ15(ptrf1) and DL844.λ15], neither leucine nor alanine repressed clp transcription.
DNA methylation pattern analysis.
In the absence of AfaF (i.e., in the absence of phase variation), the GATCdist site was methylated and the GATCprox site was protected from methylation in about 50% of the DNA molecules whereas both GATCdist and GATCprox sites were methylated in the remaining 50% of DNA molecules. This methylation pattern was similar in ClpB+ and ClpB− strains. In an Lrp− background, both GATC sites were methylated in 100% of the molecules (28). By analogy to pap, it was concluded that Lrp was bound to the GATCprox site in 50% of the molecules and that the affinity of Lrp for GATCprox is stronger than for GATCdist. To determine whether AfaF, leucine, or alanine modifies the methylation pattern, we performed Southern analyses on MC4100.λ6 and MC4100λ6(ptrf1) DNA. Single colonies cultured on M9-glycerol-X-Gal were used to inoculate M9-glycerol supplemented with the amino acids or left unsupplemented. For MC4100λ6(ptrf1), inoculation was done with either a Lac+ or a Lac− colony. The corresponding percentage of phase-ON cells was calculated. MC4100.λ6 cells presented a uniform phenotype; therefore, a single colony was picked at random. DNA was digested with DpnI, which cuts only methylated GATC sites, or MboI, which cuts only nonmethylated GATC sites, and hybridized with a digoxigenin DNA probe that encompasses the two GATC sites (Fig. 2). The results obtained (Fig. 2) are schematically represented in Table 2.
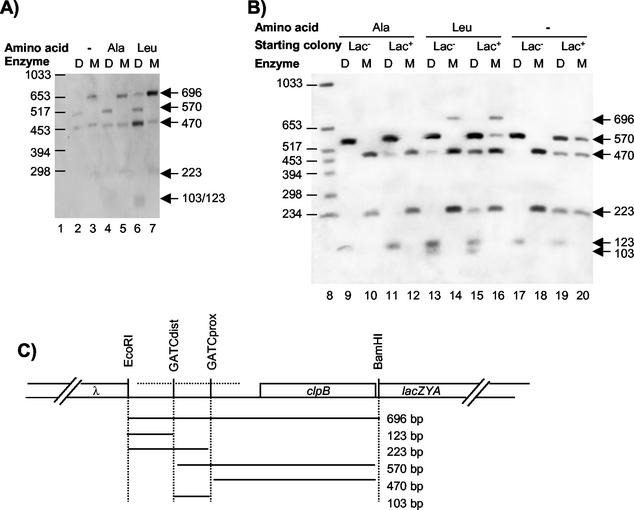
FIG. 2.
Analysis of the in vivo GATC methylation states of the clp regulatory region. A Southern blot of chromosomal DNA obtained from MC4100.λ6 (A) and MC4100.λ6(ptrf1) (B) grown in M9-glycerol (lanes 2, 3, and 17 to 20), in M9-glycerol plus alanine (lanes 4, 5, and 13 to 16), or in M9-glycerol plus leucine, isoleucine, valine (lanes 6, 7, and 9 to 12) is shown. For MC4100.λ6, a colony grown on M9-glycerol was chosen at random among uniform colonies as the starting material. For MC4100.λ6(ptrf1), a Lac+ and a Lac− colony were used as the starting material. The DNA was digested with BamHI plus EcoRI and either DpnI (D) or MboI (M). DNA hybridization was carried out with the digoxigenin-labeled probe indicated in panel C by a horizontal dotted line. The digoxigenin-labeled DNA molecular weight marker VI (Roche) was loaded in lanes 1 and 8. Its fragment sizes are indicated on the left. Size of DNA fragments that hybridized with the probe are shown on the right. (C) Schematic representation of DNA fragments that may hybridize with the probe.
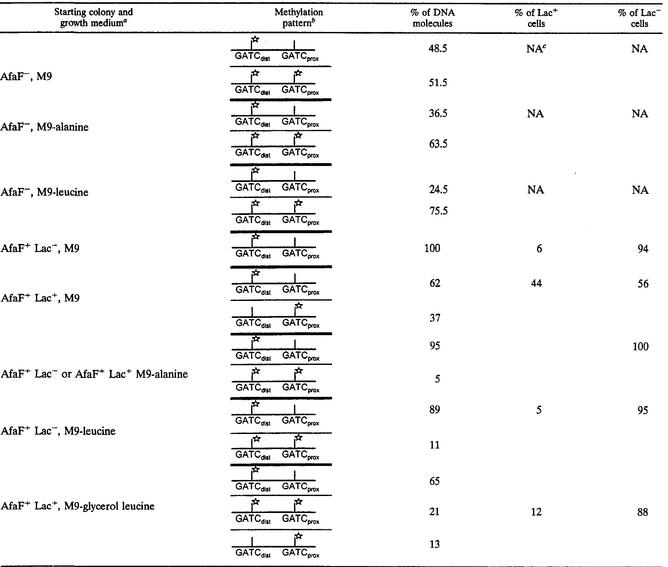
TABLE 2.
Methylation patterns of the clp regulatory region
The strain used is MC4100.λ6 (ClpB+ Lrp+) harboring or not harboring ptrf1 (AfaF+ and AfaF−, respectively). The carbon source is glycerol. Media containing leucine are supplemented with 0.4 mM isoleucine and 0.6 mM valine.
Stars indicate methylation of GATC.
NA, not applicable (cells present a uniform phenotype).
In the absence of AfaF and the presence of alanine, fewer than 40% of the DNA molecules presented methylation protection of GATCprox (Fig. 2; Table 2), suggesting that alanine slightly lowered the affinity of Lrp for its target GATCprox site. Leucine altered the methylation pattern in the same way, but its effect was more drastic since only 25% of the DNA molecules were protected from methylation on GATCprox.
In the presence of AfaF, the DNA molecules from a Lac− colony cultured in M9-glycerol-X-Gal showed methylated GATCdist sites whereas GATCprox sites were protected from methylation. Since 94% of the colonies presented a Lac− phenotype, we can assume that this methylation pattern is characteristic of phase-OFF cells and that the method was not sensitive enough to detect molecules corresponding to fewer than 6% of the cells. DNA molecules from a Lac+ colony presented two methylation patterns: 62% of the molecules showed the methylation pattern characteristic of phase-OFF cells, and 37% of the molecules showed the opposite pattern, with GATCdist protected from methylation and with methylated GATCprox sites. These patterns correlated well with the 56% Lac− colonies and 44% Lac+ colonies observed in the culture. Thus, we can assume that the characteristic pattern of phase-ON cells is GATCdist protected from methylation and GATCprox methylated. Comparison of the methylation patterns of the AfaF− and AfaF+ cells grown in M9-glycerol suggests that AfaF increased the affinity of Lrp for GATCprox, producing highly repressed phase-OFF cells, and that in a small proportion of cells, AfaF allowed Lrp binding to GATCdist, producing activated phase-ON cells. Together with measurement of β-galactosidase activities, our results indicate that maximal repression requires Lrp binding to GATCprox and maximal activation Lrp binding to GATCdist. When alanine was added to the growth medium, 100% of the colonies appeared Lac−. This phenotype correlated well with 95% of the DNA molecules presenting the methylation pattern characteristic of phase-OFF cells and 5% being fully methylated. Leucine had a less drastic effect on clp transcription since phase variation still occurred. The Lac− colony presented the same methylation patterns as the colonies cultured in alanine and was composed of 95% of Lac− cells. The Lac+ colony presented 12% Lac+ cells, and 13% of the DNA molecules showed the methylation pattern characteristic of phase-ON cells in addition to the two methylation patterns observed in the Lac− colony. Comparison of the methylation patterns with and without the amino acids shows that leucine decreased and alanine prevented the AfaF-dependent methylation protection of GATCdist. In consequence, alanine prevented the emergence of phase-ON cells. Furthermore, alanine increased the methylation protection of GATCprox since 95% of the DNA molecules from AfaF+ cells presented GATCprox protected from methylation in the presence of alanine instead of 62% in its absence.
DISCUSSION
CS31A, encoded by the clp operon, belongs to the P-regulatory family of fimbriae. Its expression is subject to a tight control at the transcriptional level, mediated by the global regulator Lrp, as well as by two specific regulators, ClpB and a PapI homologue, and is dramatically repressed by l-leucine and l-alanine. Leucine bound to Lrp modulates the expression of a number of operons by altering the affinity of Lrp for its target sites. In some cases, alanine plays the same role as leucine. The mode of action of leucine on operons subjected to phase variation control, involving an Lrp binding protein of the PapI family, has not been investigated until now. The intensively studied pap operon is insensitive to leucine, and CS31A and F1651 are the only two fimbriae of the P-regulatory family known to be responsive to leucine and alanine. In this study, we investigated the molecular mechanisms of the phase variation control and the influence of leucine and alanine on clp expression by analyzing their effects (i) on the clp promoter, which is repressed by Lrp and ClpB independently of phase variation, and (ii) on phase variation, which is mediated by Lrp and the PapI homologue AfaF.
AfaF was used in this study because our numerous efforts to clone the homologous PapI homologue from the p31A virulence plasmid were unsuccessful. These results suggest that p31A does not harbor a clp specific homologue to papI. The phase variation control observed in wild-type strains could result from a cross talk between clp and a chromosomal pap-related operon. Recently it has been shown that the reference strain 31A encodes P fimbriae (1). Whether PapI encoded by this pap operon indeed cross-regulates clp transcription has not be yet investigated. Functional complementation with heterologous PapI homologues has already been demonstrated: PefI expressed from a multicopy plasmid activates papBA transcription in E. coli whereas it represses pef transcription in S. enterica serovar Typhimurium (32), and PapI, which is a positive regulator of pap transcription, represses K88 production, as does the homologous FaeA regulator (22). SfaC and DaaF, controlling the production of S and F1845 fimbriae, respectively, are also functional PapI homologues (17, 39). Therefore, it was rational to assume that AfaF would regulate clp expression in the same manner as would the homologous Clp regulator.
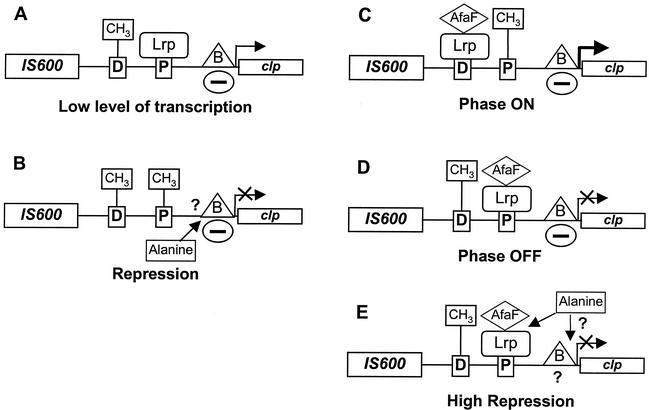
A regulatory model for clp expression may be proposed (Fig. 3). We have shown previously that both GATC sites are methylated in the absence of Lrp and that methylation protection of these sites requires Lrp but does not require ClpB (28). By analogy to Pap, we postulate that Lrp protects the GATC sites from methylation and cannot bind methylated GATC sites. Lrp presents a lower affinity for GATCdist than for GATCprox. The clp basal transcription level is low due to the repression exerted by ClpB and Lrp, which is bound to GATCprox (Fig. 3A). Leucine and alanine decrease the affinity of Lrp for GATCprox and repress clp basal transcription by a noncharacterized Lrp-independent mechanism (Fig. 3B). AfaF, the PapI homologue, is required for Lrp to bind GATCdist and increases the affinity of Lrp for GATCprox, so that phase variation can occur. The methylation pattern characteristic of phase-ON cells is GATCdist protected from methylation and GATCprox methylated (Fig. 3C), whereas the methylation pattern characteristic of phase-OFF cells is the opposite (GATCdist methylated and GATCprox protected) (Fig. 3D). Leucine lowers and alanine prevents Lrp binding to GATCdist. Leucine and alanine thus appear as antagonists of AfaF for Lrp binding to GATCdist. By contrast, alanine acts in conjunction with AfaF to increase Lrp binding to GATCprox, so that cells are locked in a highly repressed OFF phase independently of ClpB. Whether maximal repression by alanine also involves the Lrp-independent mechanism cannot be determined (Fig. 3E).
FIG. 3.
Model for control of clp expression by Lrp, AfaF, ClpB, and alanine. D and P in open squares represent GATCdist and GATCprox sites, respectively. B in a triangle represents ClpB. The minus sign in an oval indicates a repressive effect on clp basal transcription. (A) Low level of basal transcription in the absence of AfaF and alanine; GATCdist is methylated, whereas Lrp is bound to GATCprox; ClpB is bound near to the promoter region. (B) Alanine represses the basal transcription independently of Lrp by an unknown mechanism and decreases Lrp binding to GATCprox. (C and D) AfaF bound to Lrp allows Lrp binding to GATCdist in phase-ON cells (C) and increases Lrp binding to GATCprox in phase-OFF cells (D). (E) Alanine prevents the Lrp-AfaF complex from binding to GATCdist and increases its binding to GATCprox, locking cells in the OFF phase.
According to this model, a high level of transcription should be expected in the simultaneous absence of Lrp, ClpB, and AfaF (DL844.λ15/1), but this was not the case (276 MU). It has been shown that pap transcription is highly repressed by H-NS in the absence of Lrp (38). We could hypothesize that clp transcription is moderately repressed by H-NS or another unidentified regulatory protein in the absence of Lrp and ClpB. This repression could be alleviated by Lrp or ClpB individually but amplified by the simultaneous action of Lrp and ClpB.
Without AfaF, about 50% of the DNA molecules were methylated on both GATC sites whereas in the other 50%, GATCprox was protected from methylation. Thus, under these conditions all GATCdist sites were methylated. A hypothesis to explain this result is that Lrp by itself has no affinity for GATCdist and that Dam and Lrp bind equally to GATCprox, so that at each replication cycle, Lrp binds to half of the GATCprox sites. This pattern was not observed in the presence of AfaF. AfaF by itself had no direct effect on clp transcription. Rather, it modified the affinity of Lrp for the GATC sites. In phase-OFF cells, AfaF increased Lrp binding to GATCprox, so that this site was protected from methylation in 100% of the DNA molecules, leading to a highly repressed state. In phase-ON cells, AfaF allows Lrp binding to GATCdist. Thus, Lrp plays dual roles as both an AfaF-dependent activator (when bound to GATCdist) and a repressor (when bound to GATCprox) of clp transcription, similar to its role with the pap operon (38). GATCdist and GATCprox could be considered activator and repressor sites, respectively. However, pap expression is totally abrogated in the absence of PapI (41) whereas clp transcription is active in the absence of AfaF. High levels of PapI increase pap transcription (34, 38), whereas high levels of AfaF abrogate clp expression. Similarly, transcription of pef (Pef pili in S. enterica serovar Typhimurium) is repressed by PefI, the PapI/AfaF counterpart (32). To explain these differences, Hernday et al. (21) suggested that the binding of Lrp to pef sites around GATCdist would occur with the highest affinity, with PefI facilitating the transition to the OFF state by movement of Lrp to sites around GATCprox. Clearly, this is not the case for clp, as demonstrated by our results. The differences between clp and pap expression could, instead, be due to different Lrp-binding sites, to divergent nucleotide sequences surrounding the GATC sites, and to the different action of the other regulatory proteins on the promoter region. Indeed, ClpB is a repressor of clp expression whereas PapB at physiological levels has no direct influence on the papBA promoter. Transcription of pap but not of clp requires Lrp binding to GATCdist (6, 38) and is highly repressed by H-NS in the absence of Lrp (38). In contrast, the clp promoter is moderately active in the absence of Lrp, ClpB, and AfaF, suggesting that H-NS or another unidentified regulatory protein acts as a repressor of clp expression less efficiently than does Lrp or ClpB.
Among fimbriae belonging to the P-regulatory family, only CS31A and F1651 respond to leucine and alanine levels (10, 19, 28). The synthesis of K99 and type 1 fimbriae is also sensitive to leucine and alanine (11, 14). However, expression of these fimbriae is controlled by a mechanism involving Lrp but is distinct from that of members of the P-regulatory family, and the mode of action of leucine and alanine is not well understood. Two mechanisms by which leucine and alanine repress clp transcription were evidenced in this study. First, alanine and leucine repressed the basal transcription about twofold in the absence of Lrp, and this repression required ClpB (Table 1). Second, alanine and leucine altered the methylation patterns in the presence of Lrp, suggesting that they modified the affinity of Lrp for its target sites: in the absence of AfaF, alanine and leucine increased methylation of GATCprox (Table 2). This leads to an increase in the amount of fully methylated DNA. However, fully methylated DNA is probably not associated with the repression of clp expression, since clp transcription is active in an Lrp-negative background (Table 1). This indicates that the twofold repression observed in the absence of AfaF is due mainly to the Lrp-independent mechanism. In the presence of AfaF, alanine prevented and leucine decreased the AfaF-dependent methylation protection of GATCdist, acting as antagonists of AfaF. In addition, alanine acted in conjunction with AfaF to increase methylation protection of GATCprox. These results suggest that alanine and leucine decreased Lrp binding to the site with the lowest affinity (GATCdist) and increased or had no effect, respectively, on Lrp binding to the site with the highest affinity (GATCprox).
It has been shown that leucine antagonizes the activating action of Lrp on ilvIH and gltBDF promoters and its repressive effect on the lysU promoter by reducing the affinity of Lrp for its binding sites (13,25,37). Zhi et al. (43) have identified Lrp-binding sites in the dadAX promoter region. Lrp sites that overlap the dad promoters are involved in repression, whereas sites upstream of the promoters are required for activation. Leucine and alanine, which activate dad expression, decrease the affinity of Lrp to repressor sites while having little or no effect on the binding of Lrp to activator sites. The inverse situation is found in clp expression: leucine or alanine, which represse clp expression, increase the methylation protection of the repressor site (GATCprox) and dramatically reduce the methylation protection of the activator site (GATCdist). Since methylation protection is very probably due to Lrp binding, in both cases leucine or alanine decreases the binding of Lrp to low-affinity sites. However, in the case of clp, an extra protein (AfaF) is required to increase the affinity of Lrp for the repressor site, and leucine and alanine appear as agonists of AfaF. Direct interaction between leucine and Lrp has been demonstrated (27), and Lrp mutations which cause Lrp to be insensitive to leucine have been identified (35). It has been shown that Lrp self-associates in vitro to form a hexadecamer and an octamer and that leucine induces the dissociation of the Lrp hexadecamer to an octamer (8). The authors proposed a model for operons for which the effects of Lrp are potentiated by leucine: the Lrp octamer would bind with moderate affinity, and leucine would potentiate the effect of Lrp by increasing the amounts of leucine-bound octamers. However, the Lrp-mediated regulation of clp expression is even more complex, since Lrp binding requires an extra protein such as AfaF.
REFERENCES
- 1.Bertin, Y., J. P. Girardeau, A. Darfeuille-Michaud, and C. Martin. 2000. Epidemiological study of pap genes among diarrheagenic Escherichia coli strains producing CS31A and F17 adhesins, and characterization of the Pap31A fimbriae. J. Clin. Microbiol. 38:1502-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Blomfield, I. C. 2001. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 45:1-49. [DOI] [PubMed] [Google Scholar]
- 2.Blyn, L. B., B. A. Braaten, and D. A. Low. 1990. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9:4045-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blyn, L. B., B. A. Braaten, C. A. White-Ziegler, D. H. Rolfson, and D. A. Low. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braaten, B. A., L. B. Blyn, B. S. Skinner, and D. A. Low. 1991. Evidence for a methylation-blocking factor (mbf) locus involved in pap pilus expression and phase variation in Escherichia coli. J. Bacteriol. 173:1789-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braaten, B. A., X. Nou, L. S. Kaltenbach, and D. A. Low. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577-588. [DOI] [PubMed] [Google Scholar]
- 6.Braaten, B. A., J. V. Platko, M. W. van der Woude, B. H. Simons, F. K. de Graaf, J. M. Calvo, and D. A. Low. 1992. Leucine-responsive regulatory protein controls the expression of both the pap and fan pili operons in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:4250-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo, J. M., and R. G. Matthews. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S., M. H. Rosner, and J. M. Calvo. 2001. Leucine-regulated self-association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 312:625-635. [DOI] [PubMed] [Google Scholar]
- 9.Contrepois, M., H. C. Dubourguier, A. L. Parodi, J. P. Girardeau, and J. L. Ollier. 1976. Septicemic Escherichia coli and experimental infections of calves. Vet. Microbiol. 12:109-118. [DOI] [PubMed] [Google Scholar]
- 10.Daigle, F., C. Forget, C. Martin, M. Drolet, M. C. Tessier, H. Dezfulian, and J. Harel. 2000. Effects of global regulatory proteins and environmental conditions on fimbrial gene expression of F165(1) and F165(2) produced by Escherichia coli causing septicaemia in pigs. Res. Microbiol. 151:563-574. [DOI] [PubMed] [Google Scholar]
- 11.de Graaf, F. K., P. Klaasen-Boor, and J. E. van Hees. 1980. Biosynthesis of the K99 surface antigen is repressed by alanine. Infect. Immun. 30:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Martino, P., Y. Bertin, J. P. Girardeau, V. Livrelli, B. Joly, and A. Darfeuille-Michaud. 1995. Molecular characterization and adhesive properties of CF29K, an adhesin of Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 63:4336-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernsting, B. R., J. W. Denninger, R. M. Blumenthal, and R. G. Matthews. 1993. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J. Bacteriol. 175:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia, M. I., P. Gounon, P. Courcoux, A. Labigne, and C. Le Bouguenec. 1996. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol. Microbiol. 19:683-693. [DOI] [PubMed] [Google Scholar]
- 16.Girardeau, J. P., M. Der Vartanian, J. L. Ollier, and M. Contrepois. 1988. CS31A, a new K88-related fimbrial antigen on bovine enterotoxigenic and septicemic Escherichia coli strains. Infect. Immun. 56:2180-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goransson, M., K. Forsman, and B. E. Uhlin. 1988. Functional and structural homology among regulatory cistrons of pili-adhesin determinants in Escherichia coli. Mol. Gen. Genet. 212:412-417. [DOI] [PubMed] [Google Scholar]
- 18.Goransson, M., P. Forsman, P. Nilsson, and B. E. Uhlin. 1989. Upstream activating sequences that are shared by two divergently transcribed operons mediate cAMP-CRP regulation of pilus-adhesin in Escherichia coli. Mol. Microbiol. 3:1557-1565. [DOI] [PubMed] [Google Scholar]
- 19.Harel, J., F. Daigle, C. Forget, M. C. Tessier, C. Crost, and C. Martin. 2000. Phase variation of F165(1) (Prs-like) fimbriae from Escherichia coli causing septicaemia in animals. Can. J. Microbiol. 46:1101-1107. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 21.Hernday, A., M. Krabbe, B. Braaten, and D. Low. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16470-16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huisman, T. T., D. Bakker, P. Klaasen, and F. K. de Graaf. 1994. Leucine-responsive regulatory protein, IS1 insertions, and the negative regulator FaeA control the expression of the fae (K88) operon in Escherichia coli. Mol. Microbiol. 11:525-536. [DOI] [PubMed] [Google Scholar]
- 23.Jallat, C., A. Darfeuille-Michaud, J. P. Girardeau, C. Rich, and B. Joly. 1994. Self-transmissible R plasmids encoding CS31A among human Escherichia coli strains isolated from diarrheal stools. Infect. Immun. 62:2865-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaltenbach, L. S., B. A. Braaten, and D. A. Low. 1995. Specific binding of PapI to Lrp-pap DNA complexes. J. Bacteriol. 177:6449-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, R., B. Ernsting, I. N. Hirshfield, R. G. Matthews, F. C. Neidhardt, R. L. Clark, and E. B. Newman. 1992. The lrp gene product regulates expression of lysU in Escherichia coli K-12. J. Bacteriol. 174:2779-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low, D. A., B. A. Braaten, and M. van der Woude. 1996. Fimbriae, p. 146-157. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 27.Marasco, R., M. Varcamonti, F. La Cara, E. Ricca, M. De Felice, and M. Sacco. 1994. In vivo footprinting analysis of Lrp binding to the ilvIH promoter region of Escherichia coli. J. Bacteriol. 176:5197-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, C. 1996. The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and l-alanine at the transcriptional level. Mol. Microbiol. 21:281-292. [DOI] [PubMed] [Google Scholar]
- 29.Martin, C., C. Boeuf, and F. Bousquet. 1991. Escherichia coli CS31A fimbriae: molecular cloning, expression and homology with the K88 determinant. Microb. Pathog. 10:429-442. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Newman, E. B., and R. Lin. 1995. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu. Rev. Microbiol. 49:747-775. [DOI] [PubMed] [Google Scholar]
- 32.Nicholson, B., and D. Low. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35:728-742. [DOI] [PubMed] [Google Scholar]
- 33.Nou, X., B. Braaten, L. Kaltenbach, and D. A. Low. 1995. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14:5785-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nou, X., B. Skinner, B. Braaten, L. Blyn, D. Hirsch, and D. Low. 1993. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol. Microbiol. 7:545-553. [DOI] [PubMed] [Google Scholar]
- 35.Platko, J. V., and J. M. Calvo. 1993. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J. Bacteriol. 175:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, M. P. Rivas, and C. L. Turnbough. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricca, E., D. A. Aker, and J. M. Calvo. 1989. A protein that binds to the regulatory region of the Escherichia coli ilvIH operon. J. Bacteriol. 171:1658-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Woude, M. W., L. S. Kaltenbach, and D. A. Low. 1995. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol. Microbiol. 17:303-312. [DOI] [PubMed] [Google Scholar]
- 39.van der Woude, M. W., and D. A. Low. 1994. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in Escherichia coli. Mol. Microbiol. 11:605-618. [DOI] [PubMed] [Google Scholar]
- 40.Weyand, N. J., and D. A. Low. 2000. Regulation of pap phase variation—Lrp is sufficient for the establishment of the phase off pap DNA methylation pattern and repression of pap transcription in vitro. J. Biol. Chem. 275:3192-3200. [DOI] [PubMed] [Google Scholar]
- 41.White-Ziegler, C. A., M. L. Angus Hill, B. A. Braaten, M. W. van der Woude, and D. A. Low. 1998. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 28:1121-1137. [DOI] [PubMed] [Google Scholar]
- 42.Xia, Y., K. Forsman, J. Jass, and B. E. Uhlin. 1998. Oligomeric interaction of the PapB transcriptional regulator with the upstream activating region of pili adhesin gene promoters in Escherichia coli. Mol. Microbiol. 30:513-523. [DOI] [PubMed] [Google Scholar]
- 43.Zhi, J., E. Mathew, and M. Freundlich. 1999. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol. Microbiol. 32:29-40. [DOI] [PubMed] [Google Scholar]