Abstract
We found in the environmental strain Nitrosomonas europaea a chromosomal integron-like structure with an integrase gene, intINeu. We have tested the capacity of the IntINeu integrase to excise and integrate several resistance gene cassettes. The results allow us to consider IntINeu a new functional integron integrase.
Integrons are a gene capture system in which gene cassettes are mobile elements. They consist of an integrase gene (intI) (18), a member of the tyrosine recombinase family, followed by a nonpalindromic attI site specific to each integrase, and one or more integrase-dependent mobile cassettes expressed from a common promoter upstream of this site (4, 13). Excised cassettes in their free circular form consist of a structural gene and a palindromic attC site and are unable to replicate (5). Cassettes are preferentially integrated at the attI site, containing the GTTRRRY core site.
Four classes of multiresistance integrons have been identified to date. The first three classes are usually located on plasmids, while the fourth is chromosomal, in the SXT(ET) constin of Vibrio cholerae. Several new classes of integrons have been found to be chromosomally located in environmental bacteria, and their cassettes code for proteins with as yet unidentified functions (17, 23, 25).
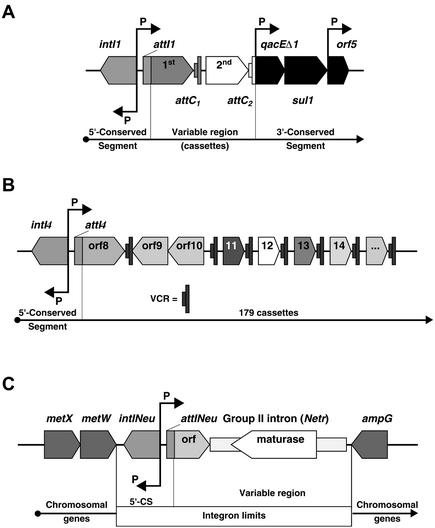
Class 1 integrons (Fig. 1A), which constitute the most widespread and studied class in the resistant bacterial population, are composed of two conserved segments and one variable region. The 5′-conserved segment (5′-CS) contains the integrase gene (intI1), a promoter region, and the IntI1-specific integration site attI1. The 3′-conserved segment (3′-CS) contains a quaternary ammonium resistance gene (qacEΔ1), and most segments also contain a sulfonamide resistance gene (sul1) (9, 19, 20) and an open reading frame (ORF5). Between the two conserved segments, the variable region (14) can contain from zero to eight cassettes.
FIG. 1.
Structures of integrons. (A) General structure of class 1 integrons, composed of two conserved segments flanking a variable region where cassettes containing an attC site are integrated at the attI1 site by the integron integrase by a site-specific recombination mechanism. P, promoters; intI1, integrase gene; qacEΔ1, antiseptic resistance gene; sul1, sulfonamide resistance gene; orf5, gene of unknown function. (B) Structure of the class 4 integron (superintegron of V. cholerae). P, promoters; intI4, integrase gene; attI4, integration site; orf, genes of unknown functions; VCR, V. cholerae repeat (attC site). (C) Structure of N. europaea chromosomal integron. P, promoters; intINeu, integrase gene; attINeu, integration site; orf, gene coding for a 123-amino-acid qacE-like protein; metX and metW, chromosomal genes involved in methionine biosynthetic pathway; ampG, chromosomal permease gene involved in β-lactamase induction and recycling of peptidoglycan.
Chromosomal integrons, named superintegrons (15, 23, 24), have been found in V. cholerae isolates from the 19th century and clearly predate the antibiotic era (Fig. 1B). This discovery indicates that the first function of integrons was probably a general, but not essential, gene capture system for bacterial genome evolution, which encodes adaptive rather than indispensable functions. Superintegrons differ from antibiotic resistance integrons by several characteristics: most of their cassettes encode unknown functions, their attC sites are more uniform, some cassettes possess their own promoters, and some carry ORFs in the orientation inverse to that of their recombination sequence.
Chromosomal superintegrons have been found in other Vibrionaceae species (Vibrio mimicus [3], Vibrio salmonicida [H. Sørum, K. Dommarsnes, K. Sandersen, L. Sundström, M. Gullberg, and A. Solberg, unpublished data {GenBank accession number AJ277063}], and Listonella pelagia [23]) as well as in Pseudomonas alcaligenes (25) and uncultivated environmental bacteria (17). The integrases of V. cholerae (12) and Shewanella oneidensis (7) have been shown to be functional. Other chromosomal integrases from environmental bacteria, like IntIGsu from Geobacter sulfurreducens and IntITde from Treponema denticola, have been identified from genome projects (17).
Nitrosomonas europaea is an environmental gram-negative β-proteobacterium that plays a central role in the availability of nitrogen to plants. Its 2.81-Mbp genome has been completely sequenced by the U.S. Department of Energy Joint Genome Institute (JGI) (http://www.jgi.doe.gov/JGI_microbial/html/index.html) in order to explore the role of microorganisms in global carbon sequestration. Using the intI1 and the additional domain sequences characteristic to integron integrases (16), we identified in the genome of N. europaea two integrase-like genes. One lacks an identifiable initiation codon, and the second, named intINeu, is associated with a chromosomally located integron. In this study, we have tested the ability of the intINeu integrase to excise gene cassettes—resistance genes cloned into pACYC184—and integrate them by site-specific recombination into the attINeu site. We showed that in the presence of IntINeu most of the cassettes tested in this study, preceded by either attI or attC sites, could be excised from pACYC184 and integrated into the attINeu site cloned in pTrc99A. These results were presented at the 102nd Annual Meeting of the American Society for Microbiology (G. Leon and P. H. Roy, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. H-92, 2002).
Sequence environment of the N. europaea integron.
The integron of N. europaea is flanked by typical chromosomal genes. The metX and metW genes are found on the left-hand side as represented in Fig. 1C, while on the right-hand side an ampG gene is present. The metX and metW genes are involved in the biosynthetic pathway of methionine from homoserine and are homologous to Escherichia coli chromosomal genes. The ampG gene is also homologous to an E. coli chromosomal gene and codes for a permease essential to β-lactamase regulation and recycling of peptidoglycan. The integrase gene is adjacent to an attI site that is followed by an ORF. This integron possesses only one ORF coding for a 123-amino-acid protein, possibly related to qacE, inserted into the attINeu site and a group II intron (Netr), coded in the same orientation as the integrase gene. The group II intron (Netr) is integrated in the inverted core site of the single cassette's attC. However, the major part of the attC site was not present, as expected, on the right-hand extremity of the intron (Fig. 1C) but was found adjacent to a second copy of Netr 92 kb away. The cassette-intron and intron-attC assemblies may represent intermediates in cassette formation (2). Netr is a 2.0-kb element, with ribozyme activity and an ORF coding for a putative maturase/reverse transcriptase. A similar group II intron has been observed in Serratia marcescens, also integrated at the attC inverse core site in an integron cassette (2).
Construction of plasmids.
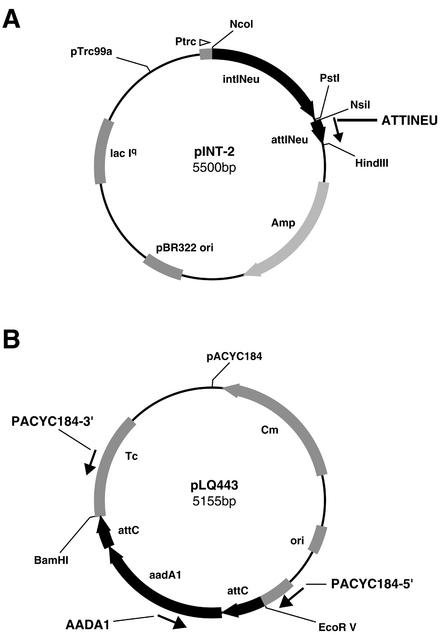
N. europaea was obtained from the ATCC collection (ATCC 25978) and cultured under aerobic conditions at 26°C with agitation, in a special Nitrosomonas medium (ATCC medium no. 221 broth). Total DNA isolation was done by using a phenol-chloroform purification method (1). The integrase gene intINeu was amplified by PCR on the genomic DNA with Herculase polymerase (New England BioLabs, Beverly, Mass.) and two primers, one of which is mutagenic and creates an NcoI site (Table 1) designed by using the OLIGO software package (version 4.1; National Biosciences, Plymouth, Minn.). The conditions for the PCR with mutagenic primers were as follows: 10 min at 95°C; 3 cycles consisting of 1 min at 95°C, 1 min at 42°C, and 1 min per kb at 72°C; 30 cycles consisting of 1 min at 95°C, 1 min at the appropriate annealing temperature (Table 1), and 1 min per kb at 72°C; and a final elongation step of 10 min at 72°C. For conventional primers, PCR conditions were the same except that the three cycles at a lower annealing temperature (42°C) were omitted. The amplicon was then digested with NcoI and PstI and cloned into the expression vector pTrc99A (Amersham-Pharmacia) digested with the same enzymes to yield clone pINT-1. The attINeu site was also amplified by PCR from genomic DNA by using the same protocol with mutagenic oligonucleotides (Table 1) containing the restriction sites NsiI (upper primer) and HindIII (lower primer), and then it was cloned into pINT-1 digested with PstI and HindIII to yield clone pINT-2 (Fig. 2A).
TABLE 1.
PCR primersa
| Primer pair | Nucleotide sequence (5′ to 3′) | Annealing temp. used (°C) | Use |
|---|---|---|---|
| INTINEU-5′ | TAT CAA CTT TCC ATG GGA AAT AC | 60 | intINeu cloning (NcoI-PstI) |
| INTINEU-3′ | CGT CCA CAA CTT TCT GAA TAC GG | 60 | |
| ATTINEU-5′ | TTG ATA CAG ATG CAT AGG CGT GAT | 57 | attINeu cloning (NsiI-HindIII) |
| ATTINEU-3′ | GAC AAC AAA GCT TGC GCT AGA GAC | 57 | |
| PACYC184-5′ | TGT AGC ACC TGA AGT CAG CC | 55 | Excision tests |
| PACYC184-3′ | ATA CCC ACG CCG AAA CAA G | 55 | |
| ATTINEUb | AGG CGT GAT AGA TTC TC | 48 | |
| AADA1c | TCG ATG ACG CCA ACT AC | 48 | |
| BLAIMPc | TGC GGT AGC AAT GCT GC | 48 | Integration tests |
| AACA1ac | TAA TTG CTG CAT TCC TCC GC | 48 | |
| PSE-1c | CGG ATG GTA TTA AAA GC | 44 |
For the first two pairs, mutagenic primers (restriction sites underlined) were used to clone the integrase intINeu and its recombination site attINeu. Excision of several cassettes by the integrase was verified by using the primer pair PACYC184-5′ and PACYC184-3′ for PCR amplifications. Integration of the excised cassettes was detected by using ATTINEU and a cassette-specific primer.
Upper primer.
Lower primer.
FIG. 2.
Vector constructions. (A) Clone pINT-2 used for the integration test, which differs from the pINT-1 clone by addition of the attINeu site. IntINeu, integrase gene cloned as an NcoI-PstI fragment; attINeu, recombination site for integration of cassettes cloned as an NsiI-HindIII fragment; amp, ampicillin resistance gene; pBR322 ori, replication origin; lacIq, constitutive repressor; Ptrc, inducible promoter. (B) Clone pLQ443 is the donor clone containing the aadA1 cassette used for excision and integration tests. Ori, replication origin; Cm, chloramphenicol resistance gene; attC-aadA1-attC, streptomycin-spectinomycin resistance cassette cloned as an EcoRV-BamHI fragment into the tetracycline resistance gene (Tc) of pACYC184.
Excision and excision-integration assays.
Nineteen clones containing resistance gene cassettes integrated in different recombination sites were used to test the activity of IntINeu. The integrase was induced in the presence of one or two excisable cassettes cloned in pACYC184 (F. Gagnon and P. H. Roy, unpublished results). Each resistance gene, bounded by two recombination sequences—the ORF-associated attC site downstream and a second recombination element (attI or attC) upstream—was susceptible to excision as a circular intermediate by the integrase and then to integration by a site-specific event into the attINeu site of pINT-2 (Table 2).
TABLE 2.
In vivo site-specific excision and integration by IntINeua
| pLQ clone | Left-hand neighbor | Gene cassette and associated attC | Right-hand neighbor
|
Excisionb | Integration | |
|---|---|---|---|---|---|---|
| Crossover site sequence | Gene | |||||
| 423 | attI1 | GTTAAAC + aadA1 | GTTAGAT | qacEΔ1 | − | No |
| 424 | attI2 | GTTAACC + dfrA1 | GTTAGGC | orfI | − | ND |
| 425 | attI3 | GTTAGAA + blaimp | GTTAGGC | aacA4 | − | ND |
| 427 | attI1 | GTTAACC + dfrA1 | GTTAAAC | aadA1 | − | ND |
| 429 | attI1 | GTTAGAC + aadA2 | GTTAGGG | orf1 | − | ND |
| 439 | attI1 | GTTAGAA + blaimp | GTTAGGC | orf1 | − | ND |
| 440 | attI1 | GTTAGGG + accA1a-orfG + orfH | GTTAGGC | orf1 | + | Yes |
| 441 | attI2 | GTTAGGG + accA1a-orfG + orfH | GTTAGGC | orf1 | + | Yes |
| 442 | attI3 | GTTAGGG + accA1a-orfG + orfH | GTTAGGC | orf1 | + | Yes |
| 443 | attCaadA2 | GTTAAAC + aadA1 | GTTAGAT | qacEΔ1 | +++ | Yes |
| 428 | attCaadA2 | GTTAGGG + accA1a-orfG + orfH | GTTAGGC | orf1 | ++ | Yes |
| 437 | attCaadA2 | GTTAACC + dfrA1 | GTTAAAC | aadA1 | − | No |
| 438 | attCaadA2 | GTTAGAA + blaimp | GTTAGGC | orf1 | +++ | Yes |
| 426 | attCdfrA1 | GTTAAAC + aadA1 | GTTAGAT | qacEΔ1 | − | No |
| 444 | attCdfrA1 | GTTAGGG + accA1a-orfG + orfH | GTTAGGC | orf1 | +++ | Yes |
| 430 | attCdfrA1 | GTTAGGC + sat | GTTAAAC | aadA1 | − | ND |
| 445 | attCaacA1a-orfG | GTTAAAC + aadA1 | GTTAGAT | qacEΔ1 | +++ | Yes |
| 446 | attCblaimp | GTTAGGG + accA1a-orfG + orfH | GTTAGGC | orf1 | + | Yes |
| 431 | attCaacA4 | GTTAGCC + pse1 | GTTAGAC | aadA2 | + | Yes |
Gene cassettes and their associated attC sites (core site in bold type) were cloned into pACYC184 and contained the GTTRRRY recombination consensus sequences of various attI or attC sites. One cassette, aacA1a-orfG, was always cloned in tandem with orfH. Right-hand neighbors (qacEΔ1, orfI, aacA4, aadA1, aadA2) were not complete cassettes and were not excised by the integrase.
−, no excision; +, weak excision (<20%); ++, moderate excision (20 to 75%); +++, strong excision (>75%); ND, not determined.
In vivo tests were done by double transformation of pINT-1 (for the excision test) or pINT-2 (for the excision-integration test) into E. coli DH5α [supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] with one of the cassette-containing pLQ clones (Table 2). Double transformants were grown overnight at 37°C in a modified Luria-Bertani medium (10 g of Bacto Tryptone, 5 g of yeast extract, 5 g of NaCl, and 2 g of glucose) with selective antibiotics (50 μg each of ampicillin and chloramphenicol per ml). Then, 15 ml of a new medium (without antibiotic) was inoculated with 2% of the overnight culture and grown until the optical density at 600 nm reached 0.6 at 37°C before adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM for an overnight induction. Plasmids were purified from 5 ml of the overnight culture with the Miniprep kit (Qiagen) and used for detection by PCR of excision or integration by using the appropriate primer pair (Table 1). For each positive integration, the attINeu site was sequenced to determine the exact point of integration.
The results of the excision tests using PACYC184-5′ and PACYC184-3′ primers for PCR amplifications showed that IntINeu is unable to excise cassettes integrated into a class 1, 2, or 3 attI site, except for the clones containing the aacA1a-orfG and orfH cassettes, from clones pLQ440, 441, and 442.
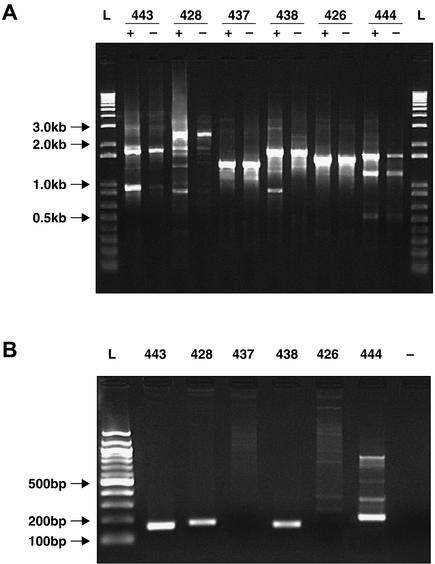
In clones pLQ443, 428, 437, and 438, cassettes downstream of the aadA2 attC site showed good levels of excision by IntINeu integrase (Fig. 3A), except for the trimethoprim resistance cassette dfrA1 (pLQ437). This cassette has previously been shown to be less frequently excised by IntI1 than the aadA1 cassette (10). For pLQ443, excision of the aadA1 cassette yields a PCR amplicon of 900 bp whereas the amplicon is 1,800 bp when the cassette is not excised. In each test, excision by IntINeu was not totally efficient and there were still unexcised cassettes in each induced reaction. We observed in the induced excision of the pLQ428 cassettes (Fig. 3A) PCR products of 889, 1,341, and 2,047 bp, corresponding to the excision of both cassettes (aacA1a-orfG+orfH) and of either of the two cassettes independently (aacA1a-orfG and orfH), respectively. Excision of orfH alone had not been previously observed with the IntI1 integrase (8). Excision results were confirmed by excision-integration tests, and the amplicons obtained by using ATTINEU and several cassette-specific primers (Table 1) were around the expected 200 bp in length (Fig. 3B).
FIG. 3.
PCR amplification results after induced excision or integration of cassettes by the IntINeu integrase. (A) Example of results obtained from the excision tests. Six clones were tested with the IntINeu integrase induced (+) or not induced (−) (see Table 2 for description). A positive excision gives, in the induced sample, one or more bands that are smaller than in the noninduced sample. L, DNA molecular weight marker (1kb plus; Gibco BRL). (B) The same clones from the excision test used in integration tests. Integration was confirmed by a positive amplification. L, DNA molecular weight marker (100 bp; Gibco BRL); −, negative PCR control (see Table 1 for the primer pairs used in these two tests).
Clones pLQ426, 444, and 430 possess cassettes downstream of the dfrA1 attC site. The dfrA1 attC site is an unusual element (95 bp) and is not in one of the usual three groups (22). When this attC site was the left-hand neighbor, excisions of the aminoglycoside resistance gene aadA1 (pLQ426) and the streptothricin acetyltransferase gene sat (pLQ430) (result not shown in Fig. 3A) were not observed. Only the aacA1a-orfG and orfH cassettes in pLQ444 were excised. Finally, clones pLQ445, 446, and 431 have different cassettes, each with a different left-hand neighbor: a 117-bp attCaacA1a-orfG site for pLQ445, a group 3 attCblaimp site for pLQ446 (127 bp), and a group 2 attCaacA4 site for pLQ431 (70 bp) (21). In all cases, excision of the cassettes was observed but a greater efficiency was obtained for the pLQ445 cassette.
These results have also been confirmed by subsequent integrations of the excised cassettes into the attINeu site. Sequencing amplicons from some of the integration tests and comparison with pINT-2 and the donor clones confirm that the excised cassettes were integrated specifically into the G/TTRRRY consensus site of the attINeu site (Fig. 4).
FIG. 4.
Sequence alignment confirming site-specific integration of the aacA1a-orfG cassette from pLQ428 into the consensus attINeu site GTTRRRY. A sequence from the amplicon obtained from the integration test is compared to the sequences of the attINeu site (pINT-2) and the donor clone (containing the cassettes).
IntINeu is able, like IntI1, to excise cassettes integrated in a heterologous attI site, but this ability is limited to easily excisable cassettes such as aacA1a-orfG and orfH. In accordance with the excision results from IntISon (7) and with the first three classes of integrases (6, 11), our data confirmed that the efficiency of excision depends not only on the cassette's own attC site but also on the neighboring left-hand site (attC or attI). Certain attC sites have a negative effect on excision when in either position. The attCdfrA1 to the left of several cassettes (in pLQ426, 444, and 430) and as part of its own dfrA1 cassette (clone pLQ437) is an example of this effect, as only one clone, with the aacA1a-orfG+orfH cassettes bounded by this attCdfrA1, was excised. Excision of the dfrA1 cassette itself was not observed. The first few bases of the right-hand neighbor of the cassette may also influence the level of excision. These bases correspond to the last few bases of the core site (TTRRRY) of the attC site of the excisable cassette and vary according to the right-hand neighbor (Gagnon and Roy, unpublished).
Sequence comparison with other tyrosine recombinases.
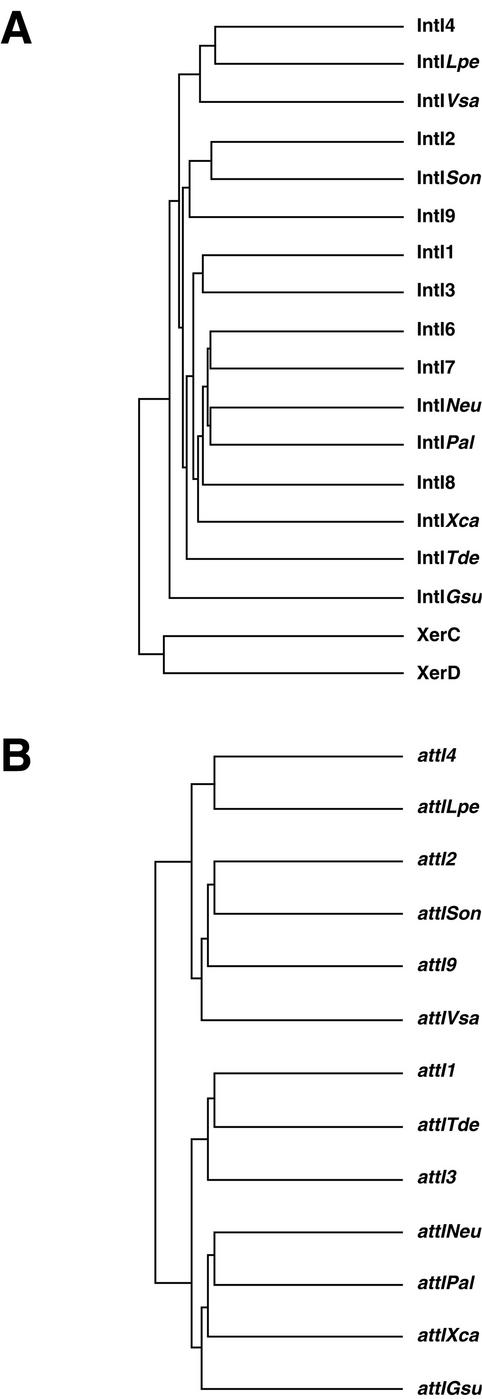
A characteristic of integron integrases, permitting them to form a specific clade within the tyrosine recombinase family, is an extra domain, implicated in the recognition of the attI and attC sites, near Patch III of the family (16). This domain is absent from the XerC and XerD recombinases of E. coli and from other tyrosine recombinases. Comparison with 16 integron integrase sequences identified to date allowed us to construct a dendrogram (Fig. 5A). IntINeu is not closely related to chromosomal integron integrases such as IntI4 (from the superintegron of V. cholerae), IntISon (from S. oneidensis), IntIGsu (from G. sulfurreducens), or IntITde (from T. denticola). However, several superintegron integrases, like IntIPal (from P. alcaligenes) (25), IntIXca (from Xanthomonas campestris pathovar campestris) (23), and IntI6, IntI7, and IntI8 (from uncultured bacteria) (17), are the closest relatives of IntINeu, with up to 60% amino acid identity. Also, other integron integrases form separate branches within the integron integrase clade.
FIG. 5.
Phylogenetic relationships between integron integrase genes (A) and between attI sites (B) among the proteobacteria. Dendrograms are based on known intI gene and attI site sequences. IntI1 and attI1, class 1 integrase and attI site; IntI2 and attI2, class 2 integrase and attI site; IntI3 and attI3, class 3 integrase and attI site; IntI4 and attI4, class 4 integrase and attI site; IntI6, 7, and 8, class 6, 7, and 8 integrases (uncultured bacteria); IntINeu and attINeu, integrase of N. europaea and attI site; IntISon and attISon, integrase of S. oneidensis and attI site; IntIGsu and attIGsu, integrase of G. sulfurreducens and attI site; IntITde and attITde, integrase of T. denticola and attI site; IntIXca and attIXca, integrase of X. campestris pv. campestris and attI site; IntIPal and attIPal, integrase of P. alcaligenes and attI site; IntIVsa and attIVsa, integrase of V. salmonicida and attI site; IntILpe and attILpe, integrase of L. pelagia and attI site; XerC and XerD, recombinases from E. coli.
Excision tests on IntI1, IntI2, and IntI3 (Gagnon and Roy, unpublished) on IntISon (7) and on IntINeu show similar patterns of excision levels for IntINeu, IntI1, and IntI3. A distinct pattern was observed for IntI2 and IntISon, which are closely related in sequence, and which also possess similar attI sites (7).
The IntINeu sequence is closest to IntIPal, which is associated with the superintegron of P. alcaligenes. This superintegron possesses a cluster of ORFs, but each ORF is associated with smaller recombination elements (P. alcaligenes repeats [PAR] of 75 to 89 bp in length) than V. cholerae repeats (25).
Sequence comparison with other attI recombination sites.
For excision and integration of cassettes, integrases act preferentially at specific sequences designated attI attachment sites. The sequences of attI sites are not closely related to each other. A multiple sequence alignment of the attI sequences identified to date, using the 70 bp up to the consensus site, including the GTT, allowed us to construct a dendrogram of 13 attI sequences (Fig. 5B). Interestingly, the results obtained with the attI sites yield a dendrogram comparable to that of the tyrosine recombinases, except that with the former there are three major groups formed. The IntINeu recombination site (attINeu) is most closely related to the IntIPal recombination site (attIPal), with which it forms a separate group within the chromosomal integron attI sites (attIPal, attIXca, and attIGsu). A distinct group is formed by the attI sites of integrons and superintegrons of Vibrio and related species (attI4, attILpe, attI9, and attIVsa). As with the IntI2 and IntISon integrases, their respective recombination sites, attI2 and attISon, are very similar, correlating with the similar excision-integration results obtained with these two integrases (7).
The finding of superintegrons in environmental strains such as V. cholerae (15), P. alcaligenes (25), and S. oneidensis (7) supports the hypothesis of an environmental origin of multiresistant integrons (23) and is consistent with the hypothesis that integrons are a common feature of bacterial populations. However, small chromosomal integrons, such as those of N. europaea, S. oneidensis, and G. sulfurreducens, rather than superintegrons may have been a source of the integrases and attI sites of multiresistance integrons.
Acknowledgments
We thank the Joint Genome Institute (JGI) for the genome sequence of N. europaea. For the phylogenetic analysis, the integrase and the attI sequences were obtained from EMBL or GenBank, except for G. sulfurreducens, S. oneidensis, and T. denticola (obtained from The Institute for Genomic Research) and N. europaea (obtained from JGI). The sequences were compared by using the Genetics Computer Group program Pileup.
This work was supported by grant MT-13564 from the Canadian Institutes of Health Research (CIHR) to P. H. Roy.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology, vol. 1, p. 2.4.1-2.4.2. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 2.Centrón, D., and P. H. Roy. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, C. A., L. Purins, P. Kaewrakon, T. Focareta, and P. A. Manning. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146:2605-2612. [DOI] [PubMed] [Google Scholar]
- 4.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis, C. M., and R. M. Hall. 1992. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6:2875-2885. [DOI] [PubMed] [Google Scholar]
- 6.Collis, C. M., M. J. Kim, S. R. Partridge, H. W. Stokes, and R. M. Hall. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J. Bacteriol. 184:3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drouin, F., J. Melançon, and P. H. Roy. 2002. The IntI-like tyrosine recombinase of Shewanella oneidensis is active as an integron integrase. J. Bacteriol. 184:1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravel, A., N. Messier, and P. H. Roy. 1998. Point mutations in the integron integrase IntI1 that affect recombination and/or substrate recognition. J. Bacteriol. 180:5437-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansson, K., O. Sköld, and L. Sundström. 1997. Non-palindromic attI sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol. Microbiol. 26:441-453. [DOI] [PubMed] [Google Scholar]
- 11.Hansson, K., L. Sundström, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lévesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 14.Lévesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 16.Messier, N., and P. H. Roy. 2001. Integron integrases possess a unique additional domain necessary for activity. J. Bacteriol. 183:6699-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 18.Ouellette, M., and P. H. Roy. 1987. Homology of ORFs from Tn2603 and from R46 to site-specific recombinases. Nucleic Acids Res. 15:10055.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen, I. T., T. G. Littlejohn, P. Radström, L. Sundström, O. Sköld, G. Swedberg, and R. A. Skurray. 1993. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 37:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rådström, P., G. Swedberg, and O. Sköld. 1991. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 35:1840-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 22.Recchia, G. D., and R. M. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5:389-394. [DOI] [PubMed] [Google Scholar]
- 23.Rowe-Magnus, D. A., A. M. Guerout, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. USA 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe-Magnus, D. A., and D. Mazel. 1999. Resistance gene capture. Curr. Opin. Microbiol. 2:483-488. [DOI] [PubMed] [Google Scholar]
- 25.Vaisvila, R., R. D. Morgan, J. Posfai, and E. A. Raleigh. 2001. Discovery and distribution of super-integrons among Pseudomonads. Mol. Microbiol. 42:587-601. [DOI] [PubMed] [Google Scholar]