Abstract
In 1954, Cohen and Barner discovered that a thymine auxotrophic (thyA) mutant of Escherichia coli undergoes cell death in response to thymine starvation. This phenomenon, called thymineless death (TLD), has also been found in many other organisms, including prokaryotes and eukaryotes. Though TLD has been studied intensively, its molecular mechanism has not yet been explained. Previously we reported on the E. coli mazEF system, a regulatable chromosomal suicide module that can be triggered by various stress conditions. MazF is a stable toxin, and MazE is an unstable antitoxin. Here, we show that cell death that is mediated by the mazEF module can also be activated by thymine starvation. We found that TLD depends on E. coli mazEF and that under thymine starvation, the activity of the mazEF promoter P2 is significantly reduced. Our results, which describe thymine starvation as a trigger for a built-in death program, have implications for programmed cell death in both prokaryotes and eukaryotes.
As early as 1954, Cohen and Barner discovered that a thymine auxotrophic (thyA) mutant of Escherichia coli undergoes cell death in response to thymine starvation (10). This phenomenon, called thymineless death (TLD) (7), has also been found in many other organisms, including prokaryotes and eukaryotes (for a recent review, see reference 2). TLD is a unique effect, since starvation of bacteria for the other bases (adenine, guanine, or cytosine) or other nutrients, like amino acids or vitamins, causes growth to stop (i.e., is bacteriostatic) but does not cause cell death (i.e., is not bactericidal) (6). However, despite intensive research on TLD, its molecular mechanism has not yet been explained. Though many molecular events have been implicated in TLD, no compelling direct relationship has been found among them, and thus, no definite hypothesis has been formulated (2).
Most of the studies of TLD have been made in E. coli, where the first step in dTTP synthesis is the conversion of dUMP to dTMP. This conversion is catalyzed by thymidylate synthetase, which is encoded by the thyA gene (5), and involves the transfer of a C-1 unit from N5,N10-methylene tetrahydrofolate to the 5 position of dUMP (2); tetrahydrofolate is consumed in this reaction (15). Thus, thymine starvation in E. coli can be induced in several ways that inhibit the synthesis of thymidylate either directly or indirectly by interfering with folate metabolism (2).
We asked whether thymine starvation is a trigger for a built-in death program. In E. coli, programmed cell death (PCD) is mediated through special genetic elements called addiction modules (for reviews, see references 11, 13, 16, and 31). These modules consist of two genes, where the second gene encodes a stable toxin and the first gene encodes a labile antitoxin. Addiction modules were first discovered in a number of extrachromosomal elements, where they were found to be responsible for the postsegregational killing effect, that is, the death of cells from which these extrachromosomal elements have been lost. Thus, the cells are “addicted” to the short-lived antitoxin product, since its de novo synthesis is essential for cell survival (11, 13, 16, 31).
Pairs of genes homologous to some of these extrachromosomal addiction modules have been found on the E. coli chromosome (3, 16, 17, 21-23). The E. coli mazEF system, the first known regulatable prokaryotic chromosomal addiction module, has been reported (3). The mazEF module consists of two adjacent genes, mazE and mazF, located in the rel operon downstream from the relA gene (23). In the study (3), it was found that mazEF has the properties required for an addiction module: (i) MazF is toxic, and MazE is antitoxic; (ii) MazF is long-lived, while MazE is a labile protein degraded in vivo by the ATP-dependent ClpPA serine protease; (iii) MazE and MazF interact; (iv) MazE and MazF are coexpressed; and (v) The two mazEF promoters (P2 and P3) are strongly negatively autoregulated by the combined action of MazE and MazF (20). Another significant characteristic of the mazEF system is that its transcription from the P2 promoter is inhibited by high concentrations of guanosine 3′,5′-bispyrophosphate (ppGpp) (3), which is synthesized by the RelA protein under conditions of extreme amino acid starvation (9). Based on these properties of mazEF, and on the requirement for the continuous expression of MazE to prevent cell death, a model for PCD under conditions of nutritional stress (3) or under conditions in which the synthesis of RNA and/or protein are generally inhibited (19, 27) was presented. The model was supported by the results of previous experiments showing that mazEF-mediated cell death can be triggered by (i) the artificial overproduction of ppGpp (3, 14), (ii) several antibiotics (rifampin, chloramphenicol, and spectinomycin) that are general inhibitors of transcription and/or translation (12, 27), or (iii) the Doc protein, the toxic product of the addiction module phd-doc of the plasmid prophage P1, which is a general inhibitor of translation (19).
Here, we asked whether mazEF-mediated cell death can be triggered in E. coli under conditions of thymine starvation. We generated the condition of thymine starvation in three ways: (i) we caused the inhibition of folate metabolism by adding trimethoprim to inhibit dihydrofolate reductase (4), (ii) we caused the inhibition of folate metabolism by adding sulfonamides to block folate synthesis (28), and (iii) we grew a thymine auxotrophic (thyA) mutant in a medium lacking thymine (1, 10). We found that in E. coli thymine starvation does trigger mazEF-mediated cell death. Furthermore, under thymine starvation, the activity of the mazEF promoter P2 is significantly reduced. Thus, the results of our work showing that thymine starvation is a trigger for a built-in death program provide a new insight into an old enigma.
MATERIALS AND METHODS
Materials and media.
We obtained [6-3H]thymidine (23.0 Ci/mmol) and [5,6-3H]uracil (45 Ci/mmol) from Amersham Pharmacia Biotech (Little Chalfont, United Kingdom) and [35S]methionine (1,175 Ci/mmol) from NEN (Boston, Mass.). Trimethoprim, sulfamethoxazole (SMZ) and inosine were obtained from Sigma (St. Louis, Mo.). We obtained avian myeloblastosis virus reverse transcriptase from Promega (Madison, Wis.), we obtained polynucleotide kinase from U.S. Biochemical Corp. (Cleveland, Ohio), and we obtained [γ-32P]ATP (3,000 Ci/mmol) from Amersham Pharmacia Biotech. The bacteria were grown either in M9 liquid medium with 1% glucose and a mixture of amino acids (20 μg/ml each) or in Luria-Bertani (LB) liquid medium and/or agar plates (24).
Bacterial strains and plasmids.
We used the following E. coli strains: MC4100relA+ [araD139 Δ(argF-lac)205 flb5301 pstF25 rpsL150 deoC1) and its derivative MC4100ΔmazEFrelA+ (mazEF::kan) (14). In addition, we used KL742 (λ− thyA748::Tn10 rph-1 deo-77) (from the E. coli Genetic Stock Center) (K. B. Low, unpublished strains). Its ΔmazEF derivative KL742mazEF::kan and its ΔclpP derivative KL742clpP::cat were constructed by P1 transduction from strains MC4100mazEF::kan and MC4100clpP::cat (3), respectively.
Plasmid pKK223mazEF (pmazEF) is a pKK223 derivative carrying mazEF (19). Plasmids pSK10Δ6-pef and pIB13 are derivatives of plasmid pSK10Δ6, which bears the lac′Z gene lacking its first eight codons. pSK10Δ6-pef carries the end of the relA gene, the mazEF promoter, and the first 17 codons of mazE fused to lac′Z (20). pIB13 carries the trpR promoter and the first 78 codons of trpR fused to lac′Z (8).
Thymine starvation induced by the addition of trimethoprim or SMZ.
Bacteria were grown in M9 minimal medium (with glucose and a mixture of amino acids) with shaking (160 rpm) at 37°C. After overnight growth, the cultures were (i) diluted to 106 cells/ml in M9 medium supplemented with trimethoprim (0.1 μg/ml) and inosine (50 μg/ml) or (ii) diluted in M9 medium supplemented with SMZ (10 μg/ml) and inosine (50 μg/ml). Inosine was added as a purine source (28). While these diluted cultures were growing at 37°C, samples were withdrawn at 30-min intervals, washed once with saline, and plated on LB plates that were incubated at 37°C for 18 h for viable-cell counts.
Thymine starvation of a thymine auxotrophic (thyA) mutant.
The E. coli thyA mutant strain KL742 was grown in M9 minimal medium supplemented with thymine (50 μg/ml) plus glucose and a mixture of amino acids with shaking (160 rpm) at 37°C. After overnight growth, the cultures were diluted 20-fold in the same medium and allowed to continue growing with shaking at 37°C. When the culture reached the exponential phase (108 cells/ml), 1.0-ml samples were withdrawn, centrifuged, washed two times with saline at 4°C, and resuspended in 2.0 ml of fresh M9 medium without thymine. These prepared samples were incubated at 37°C with aeration (shaking at 160 rpm). At 30-min intervals, 0.1-ml samples were withdrawn, diluted appropriately, and plated on LB nutrient agar plates supplemented with thymine (50 μg/ml).
Assays for the effects of thymine starvation on the synthesis of DNA, RNA, and protein.
Synthesis of DNA, RNA, and protein was determined by measuring the incorporation of [6-3H]thymidine (0.2 μCi/ml), [5,6-3H]uracil (0.1 μCi/ml), or [35S]methionine (24 μCi/ml), respectively, into a trichloroacetic acid (TCA)-insoluble fraction. We added this radioactive material to the cultures at the beginning of the period of thymine starvation, induced by growing the cells in M9 medium in the presence of trimethoprim and inosine as described above. At various times during thymine starvation, the reactions were stopped by adding TCA to 1-ml samples to a final concentration of 10%, and the mixtures were incubated on ice for 30 to 60 min. The precipitates were filtered on Millipore (Bedford, Mass.) filters and washed with TCA (5%). The TCA solutions contained 100 μg of nonradioactive thymidine or nonradioactive uracil/ml. The filters were allowed to dry, and the TCA-insoluble counts were determined by using a scintillation counter.
Assay for the effect of thymine starvation on the activity of the mazEF promoter P2 and the trpR promoter.
E. coli strain MC4100relA+ harboring either plasmid pSK10Δ6-pef or plasmid pIB13 was grown in LB medium to 108 cells/ml, and trimethoprim (5 μg/ml) was added. At various times, RNA was extracted, and primer extension was carried out with avian myeloblastosis virus reverse transcriptase as described previously (20). The oligonucleotide primer used for the pSK10Δ6-pef construct was from positions −40 to −23 of the lacZ gene toward the transcription start site of mazEF (20). The primer used for pIB13 was from positions 82 to 59 of the trpR gene toward the promoter region of trpR (18). The primers were end labeled by polynucleotide kinase with [γ-32P]ATP. The reaction products were resolved on a 6% sequencing gel, and DNA-sequencing reactions were performed with the same primers and were run in parallel with the primer extension reaction as described previously (20). To quantify the RNA levels, the gels were analyzed and the bands were quantified using the Fujix BAS100 phosphorimager.
RESULTS
Thymine starvation by trimethoprim induces cell death through the mazEF system.
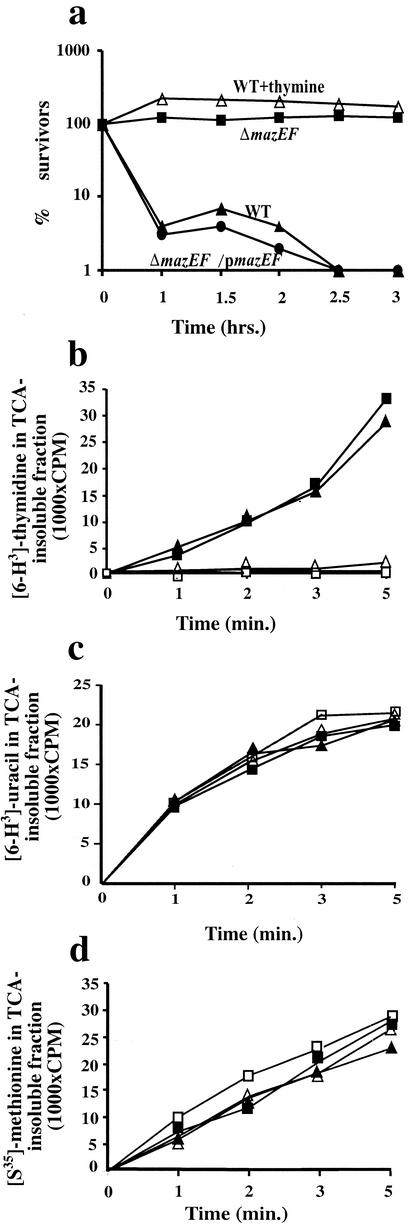
First, we studied the effect on mazEF-mediated cell death of thymine starvation induced by the addition of trimethoprim. Specifically, we compared the viability of the wild-type E. coli strain MC4100relA+ to that of its ΔmazEF derivative after exposure to trimethoprim. Growing the wild-type cells in the presence of trimethoprim caused 96% of the cells to die by 1.0 h of exposure and caused 99% of the cells to die by 2.5 h of exposure (Fig. 1a). Since the addition of thymine completely reversed this phenomenon, these data represent a TLD curve. In contrast to the wild-type strain, the ΔmazEF derivative did not die in the presence of trimethoprim, and close to 100% of these cells were viable even after 3.0 h (Fig. 1a). We also tested the effect of trimethoprim in ΔmazEF cells in which the mazEF gene module was present, on plasmid pKK223mazEF rather than on the chromosome. Here again, we observed TLD: 97% of the cells died after 1.0 h. Thus, we found that TLD induced by trimethoprim requires the mazEF system.
FIG. 1.
(a) Thymine starvation by trimethoprim in E. coli induces mazEF-mediated cell death. Cells were grown at 37°C in M9 liquid minimal medium plus 0.1 μg of trimethoprim/ml as described in Materials and Methods. Viability (% survivors) is plotted against the time of exposure to trimethoprim of E. coli MC4100relA+ (WT), MC4100ΔmazEFrelA+ (ΔmazEF), and MC4100ΔmazEFrelA+/pmazEF (ΔmazEF/pmazEF). MC4100relA+ was also grown in thepresence of 0.1 μg of trimethoprim/ml plus 50 μg of thymine/ml (WT+thymine). We calculated cell survival by comparing the colony-forming ability of trimethoprim-treated cells to that of untreated cells at zero time. The numbers represent the results of one out of five similar experiments. (b to d) Effect of thymine starvation on the synthesis of DNA (b), RNA (c), and protein (d) in MC4100relA+ and its ΔmazEF derivative. Cells were grown at 37°C in M9 liquid medium as described in Materials and Methods. After overnight growth, the cultures were diluted to 107 cells/ml and allowed to grow for 1 h in M9 medium plus inosine (50 μg/ml). Then, trimethroprim (2 μg/ml) was added; the cells were immediately labeled with either [6-3H]thymidine (b), [5,6-3H]uracil (c), or [35S]methionine (d); and their incorporation into the acid-insoluble fraction was determined as described in Materials and Methods. As controls, we used MC4100relA+ (▴) and its ΔmazEF derivative (▪) without the addition of trimethoprim. ▵, trimethoprim-treated MC4100relA+; □, ΔmazEF derivative.
In addition, we showed that the dependence of TLD on the function of the mazEF system has no connection with changes in the synthesis of macromolecules, since we found that DNA synthesis was inhibited in both the wild type and the ΔmazEF derivative mutant (Fig. 1b), while RNA and protein synthesis were unaffected (Fig. 1c and d).
Thymine starvation by a sulfonamide induces cell death through the mazEF system.
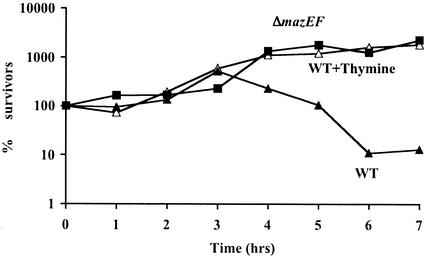
We then studied the effect of the induction of thymine starvation by the sulfonamide SMZ on mazEF-mediated cell death. We compared the viability of the wild-type E. coli MC4100relA+ to the viability of its ΔmazEF derivative after exposing each of the strains to SMZ (Fig. 2). Death of the wild-type strain occurred later in the presence of SMZ (Fig. 2) than in the presence of trimethoprim (Fig. 1a). In particular, 90% of the wild-type strain died after 6 h of exposure to SMZ (Fig. 2). In contrast, under identical conditions, we observed no killing of the ΔmazEF derivative at all (Fig. 2); in fact, this ΔmazEF strain continued growing at the same rate as the wild-type strain when thymine was added to reverse the effect of SMZ (Fig. 2). Thus, as when we induced thymine starvation by the addition of trimethoprim (Fig. 1a), TLD was also clearly mazEF dependent in the presence of SMZ (Fig. 2).
FIG. 2.
Thymine starvation by SMZ in E. coli induces mazEF-mediated cell death. Viability (% survivors) is plotted against time of exposure to SMZ (10 μg/ml) of E. coli MC4100relA+ (WT) and its ΔmazEF derivative (ΔmazEF) in M9 liquid medium at 37°C. MC4100relA+ was also grown in the presence of 10 μg of SMZ/ml plus 50 μg of thymine/ml (WT+Thymine). Cell survival was determined as described in the legend to Fig. 1a.
Thymine starvation of a thyA mutant induces cell death through the mazEF system.
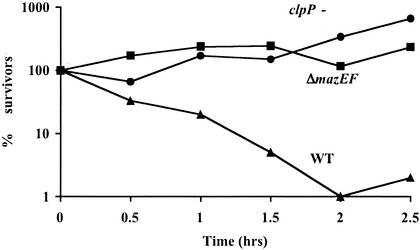
Finally, we studied the involvement of the mazEF module in TLD induced by the lack of thymine in a thymine auxotrophic (thyA) mutant. For this pur pose, we used E. coli strain KL742, which carries the mutation thyA748::Tn10. Here, we observed 95% cell death after 1.5 h and 99% cell death after 2.0 h of growth in the absence of thymine. However, we observed almost no killing during this period in either ΔmazEF or ΔclpP derivatives of E. coli KL742 thyA (Fig. 3). Thus, TLD induced by thymine starvation in a thymine auxotrophic mutant is both mazEF and clpP dependent.
FIG. 3.
Thymine starvation in an E. coli thyA mutant induces mazEF-mediated cell death. Viability (% survivors) is plotted against times of growth of E. coli strains KL742thyA748::Tn10 (WT), KL742thyA748::Tn10ΔmazEF (ΔmazEF), and KL742thyA748::Tn10ΔclpP (clpP −) at 37°C in M9 liquid medium lacking thymine. Cell survival was determined as described in the legend to Fig. 1a.
Thymine starvation in E. coli reduces transcription from the mazEF promoter P2.
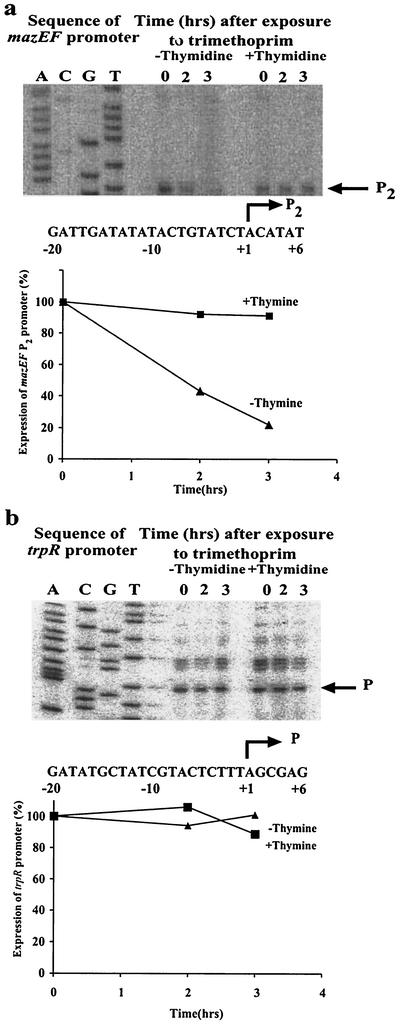
We asked whether thymine starvation affects transcription from the mazEF promoter. To test the mazEF promoter activity, we used plasmid pSK10Δ6-pef, which was previously constructed for this purpose (20). It carries a gene fusion in which the beginning of mazE is fused to the eighth codon of lacZ (see Materials and Methods). We transformed this plasmid into E. coli MC4100 relA+. The cells harboring the plasmid were starved for thymine with trimethoprim, and during starvation, the activity of the mazEF promoter was determined by primer extension. As shown, the level of transcription from P2 was reduced to ∼25% after 2 h of thymine starvation compared to unstarved cells (Fig. 4a). We also found that similar conditions of thymine starvation by trimethoprim do not affect the activity of the E. coli trpR promoter (Fig. 4b).
FIG. 4.
(a) Thymine starvation by trimethoprim in E. coli MC4100relA+ reduces transcription from the mazEF promoter P2. E. coli strain MC4100relA+ harboring plasmid pSK10Δ6-pef was treated with trimethoprim; RNA was extracted at the indicated times, primer
DISCUSSION
In summary, we have shown that TLD induced in E. coli K-12 strains is mediated through the mazEF clpP-dependent system (3, 12-14, 19, 20, 27). We used three different procedures to induce thymine starvation: treatment with trimethoprim or with a sulfonamide antibiotic or thymine starvation of a thyA auxotroph. Moreover, we found that thymine starvation by trimethoprim drastically reduces transcription from the mazEF promoter P2 (Fig. 4a), which was previously described as mainly responsible for transcription and regulation of E. coli mazEF (3, 20). mazEF has an additional promoter, P3, located 13 nucleotides downstream from P2, that has only 1/10 of the P2 activity (20). Here, we could hardly detect the activity of P3.
Based on these results, we suggest the following model for the induction of mazEF-mediated cell death by thymine starvation. Previous research has shown that thymine starvation provokes DNA damage that involves a unique breaking or twisting of the chromosome into a configuration that defies all the repair/protective mechanisms of the cell (references 2 and 26 and references therein). We assume that such substantial damage to the DNA is responsible for the reduction of transcription from the mazEF promoter P2 (Fig. 4a). The effect of thymine starvation on the activity of this promoter may result from the direct starvation effect on P2. Note that the promoter region of P2 is particularly A:T rich: 20 nucleotides out of 26 (−20 to +6) are A or T (Fig. 4a). However, the interference with transcription from P2 by thymine starvation may also be caused indirectly by the induction of ppGpp synthesis, known to inhibit the mazEF P2 promoter (3), and/or by some specific protein(s) that could sense the damage to the DNA. Our experiments indicate that ppGpp might be involved. In these experiments, we compared the mazEF-dependent killing caused by trimethoprim in MC4100, which carries a defective relA gene (relA1) (3) and therefore does not synthesize ppGpp (9), to that of the relA+ strain. We found that mazEF-dependent death appears in the relA+ strain at 0.1 μg of trimethoprim/106 cells, while in the relA1 strain it appears only at 0.3 μg of trimethoprim/106 cells (data not shown). Altogether, as a consequence of thymine starvation, prevention of the continuous expression of the labile antitoxin MazE results in cell death. Although we have shown that thymine starvation does not affect the activity of the E. coli trpR promoter (Fig. 4b), other promoters may be affected; these may include other genes involved in mazEF-mediated death.
Our results showing that thymine starvation can trigger a built-in death program provide new insight into an old enigma and may have implications for PCD in both prokaryotes and eukaryotes. (i) Until now, mazEF-directed PCD has been studied only in E. coli. However, this system is not unique to E. coli and is found on the chromosomes of other bacteria (12, 25). Nevertheless, TLD has been described in a wide range of microorganisms (2), suggesting the existence of analogous suicide modules and PCD not only in E. coli but in other bacteria as well. (ii) Our present results also increase the repertoire of antibiotics that operate in various bacteria by triggering a chromosomal suicide module. In previous work, it was shown that the mazEF suicide system in E. coli can be triggered by antibiotics like rifampin, chloramphenicol, and spectinomycin that inhibit transcription and/or translation (12, 27). Here, we extend this list to include trimethoprim and sulfonamides that inhibit folic acid metabolism and thereby cause TLD. (iii) Furthermore, our results may have implications for both mammalian TLD and cancer research. For decades, clinicians have used thymine starvation, leading to mammalian cell death, as a major method of cancer therapy. TLD is induced by treatment with the analogue 5-fluorouracil and with the folic acid antagonist methotrexate (29, 30). Thus, the knowledge gained from TLD as a trigger of bacterial PCD may be relevant to apoptosis in mammals and to cancer therapy.
Acknowledgments
We thank F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript.
The research described here was supported by grant no. 215/99-2 from the Israel Science Foundation administered by the Israel Academy of Science and Humanities.
REFERENCES
- 1.Ahmad, S. I. 1980. Thymineless death in recombination deficient mutants of Escherichia coli K12. Z. Naturforsch. Teil C 35:279-283. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 52:591-625. [DOI] [PubMed] [Google Scholar]
- 3.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angehrn, P., and R. Then. 1973. Nature of trimethoprim-induced death in Escherichia coli. Arzneimittelforschung 23:447-451. [PubMed] [Google Scholar]
- 5.Bachmann, B. J. 1990. Linkage map of Escherichia coli K-12, edition 8. Microbiol. Rev. 54:130-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barner, H. D., and S. S. Cohen. 1957. The isolation and properties of amino acids requirement mutants of thymineless bacterium. J. Bacteriol. 74:350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazill, G. W. 1967. Lethal unbalanced growth in bacteria. Nature 216:346-349. [DOI] [PubMed] [Google Scholar]
- 8.Benhar, I., and H. Engelberg-Kulka. 1993. Frameshifting in the expression of the E. coli trpR gene occurs by bypassing of a segment of its coding sequence. Cell 72:121-130. [DOI] [PubMed] [Google Scholar]
- 9.Cashel, M., D. R. Gentry, V. Z. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtis III, J. L. Ingrahm, E. C. C. Lin, K. B. M. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 10.Cohen, S. S., and H. D. Barner. 1954. Studies on the unbalanced growth in Escherichia coli. Proc. Natl. Acad. Sci. USA 40:885-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couturier, M., E. M. Bahassi, and L. Van Melderen. 1998. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 12.Engelberg-Kulka, H., B. Sat, and R. Hazan. 2001. Bacterial programmed cell death and antibiotics. ASM News 67:617-624. [Google Scholar]
- 13.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka, H., M. Reches, S. Narasimhan, R. Schoulaker-Schwarz, Y. Klemes, E. Aizenman, and G. Glaser. 1998. rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. USA 95:15481-15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedkin, M., and A. Kornberg. 1957. The enzymatic conversion of deoxyridylic acid to thymidine acid and the participation of tetrahydrofolic acid, p. 609. In W. D. McElroy and B. Glass (ed.), A symposium on the chemical basis of heredity. Johns Hopkins University Press, Baltimore, Md.
- 16.Gerdes, K., A. P. Gultyaev, T. Franch, K. Pederson, and N. D. Milkkelsen. 1997. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 19:49-61. [DOI] [PubMed] [Google Scholar]
- 17.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 18.Gunsalus, R. P., and C. Yanofsky. 1980. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc. Natl. Acad. Sci. USA 77:7117-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marianovsky, I., E. Aizenman, H. Engelberg-Kulka, and G. Glaser. 2001. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. J. Biol. Chem. 276:5975-5984. [DOI] [PubMed] [Google Scholar]
- 21.Masuda, Y., and E. Ohtsubo. 1994. Mapping and disruption of the chpB locus in Escherichia coli. J. Bacteriol. 176:5861-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzger, S., I. B. Dror, E. Aizenman, G. Schreiber, M. Toone, J. D. Friesen, M. Cashel, and G. Glaser. 1988. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J. Biol. Chem. 263:15699-15704. [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics, p. 205-210. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 25.Mittenhuber, G. 1999. Occurrence of MazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1:295-302. [PubMed] [Google Scholar]
- 26.Nakayama, K., K. Kusano, N. Irino, and H. Nakayama. 1994. Thymine starvation-induced structural changes in Escherichia coli DNA. Detection by pulse field gel electrophoresis and evidence for involvement of homologous recombination. J. Mol. Biol. 243:611-620. [DOI] [PubMed] [Google Scholar]
- 27.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Then, R., and P. Angehrn. 1973. Sulphonamide-induced ‘thymineless death’ in Escherichia coli. J. Gen. Microbiol. 76:255-263. [DOI] [PubMed] [Google Scholar]
- 29.Walder, S., R. Horowitz, X. Mao, and E. L. Schwartz. 1996. Effect of interferon on 5-fluorouracil-induced perturbations in pools of deoxynucleotide triphosphates and DNA strand breaks. Can. Chemother. Pharmacol. 38:529-535. [DOI] [PubMed] [Google Scholar]
- 30.Wei, X., H. L. McLeod, J. McMurrough, F. J. Gonzalez, and P. Fernandez-Salguero. 1996. Molecular basis of the human dihydropyrimidine dehydrogenase deficiency and 5-fluorouracil toxicity. J. Clin. Investig. 98:610-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarmolinsky, M. B. 1995. Programmed cell death in bacterial populations. Science 267:836-837. [DOI] [PubMed] [Google Scholar]