Abstract
We investigated the effect of antisense RNA (asRNA) structural properties on the downregulation efficacy of enzymes in the acetone-formation pathway (acetoacetate decarboxylase [AADC] and coenzyme A-transferase [CoAT]) of Clostridium acetobutylicum strain ATCC 824. First, we generated three strains, C. acetobutylicum ATCC 824 (pADC38AS), 824(pADC68AS), and 824(pADC100AS), which contain plasmids that produce asRNAs of various lengths against the AADC (adc) transcript. Western analysis showed that all three strains exhibit low levels of AADC compared to the plasmid control [ATCC 824(pSOS95del)]. By using computational algorithms, the three different asRNAs directed toward AADC, along with previously reported clostridial asRNAs, were examined for structural features (free nucleotides and components). When the normalized metrics of these structural features were plotted against percent downregulation, only the component/nucleotide ratio correlated well with in vivo asRNA effectiveness. Despite the significant downregulation of AADC in these strains, there were no concomitant effects on acetone formation. These findings suggest that AADC does not limit acetone formation and, thus, we targeted next the CoAT. Using the component/nucleotide ratio as a selection parameter, we developed three strains [ATCC 824 (pCTFA2AS), 824(pCTFB1AS), and 824(pCOAT11AS)] which express asRNAs to downregulate either or both of the CoAT subunits. Compared to the plasmid control strain, these strains produced substantially low levels of acetone and butanol and Western blot analyses showed significantly low levels of both CoAT subunits. These results show that CoAT is the rate-limiting enzyme in acetone formation and strengthen the hypothesis that the component/nucleotide ratio is a predictive indicator of asRNA effectiveness.
Clostridium acetobutylicum strain ATCC 824 is a gram-positive, spore-forming, obligate anaerobe able to ferment various sugars into the commercial solvents acetone and butanol. Recent developments in the genetics (29), genomics (20), and metabolic engineering of solventogenic clostridia have revived interest in this fermentation process. Applications in bioremediation (15) and gene therapy (28) are also actively being pursued.
Although not widely exploited in prokaryotic systems, antisense RNA (asRNA) is a potent and flexible tool for manipulating microbial metabolism. Desai and Papoutsakis first demonstrated the effectiveness of asRNA strategies for the metabolic engineering of C. acetobutylicum (5). They developed asRNA molecules against two genes involved in the primary metabolic network of C. acetobutylicum. One was directed toward the mRNA of the butyrate kinase (buk) gene, while the other one was directed toward the mRNA of the phosphotransbutyrylase (ptb) gene. However, no rules or patterns have been examined regarding the effectiveness of different asRNA designs and the generality of these asRNA findings in downregulating other C. acetobutylicum proteins and pathways remains to be tested. The present study was undertaken in an effort to address these two significant issues.
Most methods for designing effective asRNA molecules are based on the concept that, by increasing the association rate between asRNA and target mRNA, inhibition of target gene expression can also be increased (13, 14, 23, 26). One method that was evaluated for designing asRNAs with higher annealing rates to the target mRNA is to incorporate stem-loop structures that were designed specifically for interacting with stem-loop structures in the target mRNA (6). Experimental data, however, suggest that this approach does not necessarily work well (i.e., only 50% downregulation of the target was achieved). Another method examined for generating asRNA candidates with high annealing rates to target mRNAs is the in vitro selection of asRNA (21). This technique involves using only asRNA candidates that bind with target mRNA with the highest association rate constants as determined from nondenaturing polyacrylamide gel electrophoresis (23, 26). However, only ca. 75% overall downregulation of the target protein was achieved by this method (21). The disadvantage of using in vitro selection is that in vitro conditions might not represent in vivo conditions accurately and thus may lead to inaccurate prediction of the ideal asRNA candidates. Finally, using theoretical analyses and computer algorithms, which can provide information on structural elements that could be important for RNA duplex formation, a technique for developing asRNAs based on structural properties has been investigated (22). By examining several different structural features (including terminal unpaired nucleotides, components, and loop degree) of many different asRNAs designed to inhibit human immunodeficiency virus type 1 replication in human cells, Patzel and Sczakiel (22) showed a correlation between asRNA effectiveness and the number of terminal unpaired nucleotides.
Since structural-feature-based asRNA design (22) has not been reported for prokaryotic systems, we decided to investigate the effect of structural properties on downregulation efficacy in C. acetobutylicum. For this study, we focused on the acetone formation pathway in order to examine the possibility of increasing the ratio of butanol/acetone formed. The acetone formation pathway consists of two enzymes, acetoacetyl-coenzyme A (CoA):acetate/butyrate:CoA-transferase (CoAT) and acetoacetate decarboxylase (AADC). The first enzyme in the pathway, CoAT, which is composed of 2 subunits (A and B) coded by ctfA and ctfB, respectively, catalyzes the formation of acetoacetate by transferring the CoA group of acetoacetyl-CoA to either acetate or butyrate to form acetyl-CoA or butyryl-CoA, respectively (32). AADC (coded by adc) then catalyzes the irreversible decarboxylation of acetoacetate, producing acetone and CO2 (9, 31). Sequence and cloning of adc (24, 25) and adc gene expression in C. acetobutylicum (10, 12) have been previously reported. Sequence and cloning of the CoAT genes ctfA and ctfB (3, 25) have also been previously reported. adc is part of a monocistronic operon (10), while the ctfA and ctfB genes are transcribed as part of a polycistronic operon (aad-ctfA-ctfB) that includes aad (or adhE1), which is the gene that encodes for the enzyme aldehyde/alcohol dehydrogenase (8, 19).
In this study, we first chose AADC as our target for asRNA downregulation because it is transcribed as a monocistronic operon, which ensures that potential effects on the other cistrons of a polycistronic operon are not an issue. Then, using computational algorithms that predict RNA secondary structures, the asRNAs directed against adc (adc-asRNAs) were examined for structural information that might explain differences in adc-asRNA effectiveness and thus provide potential rules for the design of future asRNA constructs. We found that, although dramatic AADC downregulation can be achieved with asRNA, the effects on acetone formation were not equally dramatic. Thus, we next targeted the CoAT enzyme, despite the complexity that derives from the fact that its 2 units are transcribed from a large tricistronic transcript.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strain | ||
| E. coli ER2275 | NEBb | |
| E. coli OneShot chemically competent TOP10 | Invitrogen | |
| C. acetobutylicum ATCC 824 | ATCCc | |
| M5 | AADC− CoAT− | 4 |
| Plasmid | ||
| pAN1 | Cmr; carries the φ3T I gene | 17 |
| pFNK6 | Ampr MLSr; ace operon | 18 |
| pFNK7 | Ampr MLSrctfA ctfB | 18 |
| pSOS95 | ace2 operon | 30 |
| pSOS95del | Ampr MLSr; thl promoter | This study |
| pADC38AS | Ampr MLSr; thl promoter; antisense adc38 | This study |
| pADC68AS | Ampr MLSr; thl promoter; antisense adc68 | This study |
| pADC100AS | Ampr MLSr; thl promoter; antisense adc100 | This study |
| pCTFA2AS | Ampr MLSr; thl promoter; antisense ctfa2 | This study |
| pCTFB1AS | Ampr MLSr; thl promoter, antisense ctfb1 | This study |
| pCOAT11AS | Ampr MLSr; thl promoter; combined antisense ctfb1 and antisense ctfa1 | This study |
adc, AADC gene; ctfA, CoAT subunit A gene; ctfB, CoAT subunit B gene; Cmr, chloramphenicol resistant; φ3T I, Bacillus subtilis phage φ3T I methyltransferase gene; Ampr, ampicillin resistant; MLSr, macrolide-lincosamide-streptogramin B resistant; ace operon, synthetic operon which contains the three homologous acetone formation genes (adc, ctfA, and ctfB) transcribed from the adc promoter; ctfA, gene encoding for the A subunit of CoAT; ctfB, gene encoding for the B subunit of CoAT; ace2 operon, synthetic operon which contains the three homologous acetone formation genes (adc, ctfA, and ctfB) transcribed from the thl promoter; thl promoter, promoter region for the thiolase gene (thl) of C. acetobutylicum strain ATCC 824; adc38, DNA fragment containing approximately 38% of the sequence of the adc transcript; adc68, DNA fragment containing approximately 68% of the sequence of the adc transcript; adc100, DNA fragment containing approximately 100% of the sequence of the adc transcript; ctfa2, DNA fragment containing approximately 79% of the ctfA structural gene; ctfb1, DNA fragment containing approximately 39% of the ctfB structural gene; ctfa1, DNA fragment containing approximately 53% of the ctfA structural gene.
NEB, New England Biolabs (Beverly, Mass.).
ATCC, American Type Culture Collection (Manassas, Va.).
Plasmid DNA isolation.
An alkaline lysis method was used for plasmid isolation from Escherichia coli strains containing plasmid pAN1 (16). For plasmid isolations from all other E. coli strains, the method of Berghammer and Auer (1) was used. For plasmid isolation from recombinant C. acetobutylicum strains, an alkaline lysis method was used (11).
DNA manipulation.
All commercial enzymes utilized in this study (restriction enzymes, T4 DNA ligase, calf intestinal alkaline phosphatase, T4 DNA polymerase, Klenow fragment of DNA polymerase, and vent DNA polymerase) were used under supplier-recommended conditions. DNA fragments were isolated from agarose gels with GFX PCR DNA and a gel band purification kit (Amersham Pharmacia Biotech, Piscataway, N.J.).
Cell transformation.
All plasmids were constructed in E. coli and then transformed into C. acetobutylicum. Transformation with ligation mixtures was done with E. coli TOP10 OneShot competent cells from Invitrogen Corporation (Carlsbad, Calif.) according to the manufacturer's instructions. E. coli(pAN1) and C. acetobutylicum were electrotransformed according to previously published methods (17, 27).
Construction of plasmids.
For adc-asRNA experiments, three plasmids (pADC38AS, pADC68AS, and pADC100AS) that contain various lengths of the DNA encoding for the adc transcript in an antisense orientation with respect to the strong, constitutive-like thiolase (thl) promoter (12, 30) of C. acetobutylicum strain ATCC 824 were developed. Using plasmid pFNK6 (18) as a template, we first PCR amplified three different-sized DNA fragments (adc38, adc68, and adc100) that were approximately 38, 68, and 100% of the length of the adc transcript, respectively. The PCR primers used for these reactions are listed in Table 2. adc38 and adc100 were modified by restriction enzyme digestion with BamHI and SalI, respectively. SalI-digested adc100 was further modified by filling in its 5′ overhang with the Klenow fragment of DNA polymerase. To form pADC38AS, BamHI-digested adc38 was ligated in an antisense orientation to BamHI- and EheI-digested pSOS95 (30), which contains the thl promoter. adc68 and the blunt-ended adc100 were then ligated separately in an antisense orientation into BamHI- and EheI-digested pSOS95, which was blunt ended with the Klenow fragment of DNA polymerase, to form pADC68AS and pADC100AS, respectively. The blunt-ended, BamHI- and EheI-digested pSOS95 was recircularized to form the control plasmid pSOS95del (it contains the thl promoter). The lengths of the asRNAs that were produced from pADC38AS, pADC68AS, and pADC100AS were 548, 799, and 1,068 nucleotides, respectively. The asRNA length was determined to be the number of nucleotides from the transcription start site of the thl promoter of each plasmid to the last nucleotide of the terminator sequence for each asRNA.
TABLE 2.
Oligonucleotides used for PCR
| Primer | Oligonucleotide sequencea (5′-3′) | Use |
|---|---|---|
| ADCUP | GTAAATATAAATAAATAGGACTAGAGGCGA | Upstream primer for adc38, adc68, and adc100 |
| ADC38DOWN | CTGTCCGCTTTCTGGATCCCAA | Downstream primer for adc38 |
| ADC68DOWN | GCTTCATTAGCATCTAAGGCT | Downstream primer for adc68 |
| ADC100DOWN | CACAGTTTAAAAAACCTCCAATCA | Downstream primer for adc100 |
| CTFAUP | ATCTTAAATACTCGAGTTAAGAATTT | Upstream primer for ctfa1 and ctfa2 |
| CTFADOWN1 | GATCCGCCTGCTCGAGTTGGATCCACTAGAG | Downstream primer for ctfa1 |
| CTFADOWN2 | ATAGAAGGTGGATCCGGCCTCGAGTACAAT | Downstream primer for ctfa2 |
| CTFBUP | AGCAATGACCCTCGAGTTCTTAT | Upstream primer for ctfb1 |
| CTFBDOWN1 | CGAAAAATGTGCCGTCTCGAGGATCCGTTGT | Downstream primer for ctfb1 |
Boldface letters in the oligonucleotide sequences indicate nucleotide substitutions used to generate useful restriction sites near the termini of the PCR products. Relevant restriction sites are underlined.
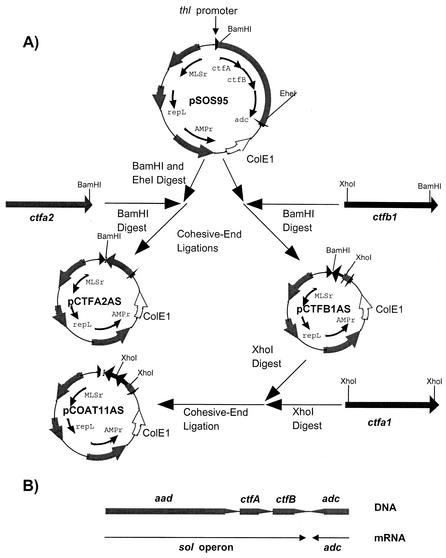
For experiments involving asRNA aimed at the downregulation of CoAT, two plasmids that targeted the downregulation of subunits A and B of CoAT separately (pCTFA2AS and pCTFB1AS, respectively) and one plasmid in which both subunits were targeted in unison (pCOAT11AS) were constructed. We first PCR amplified two DNA fragments of the ctfA transcript (ctfa1 and ctfa2) and one fragment of the ctfB transcript (ctfb1) with the primers shown in Table 2 and plasmid pFNK6 as template. After PCR amplification, all fragments were gel purified as described above. Both ctfa2 and ctfb1, which had BamHI sites engineered in their downstream primers, were then digested with BamHI and cohesive-end ligated separately to BamHI- and EheI-digested pSOS95 in an antisense orientation to form pCTFA2AS and pCTFB1AS, respectively, as shown in Fig. 1. ctfa1, which contains engineered XhoI sites in both its upstream and downstream primers, was digested with XhoI and cohesive-end ligated to XhoI-digested pCTFB1AS in an antisense orientation to form pCOAT11AS. Thus, pCOAT11AS is a plasmid that contains adjacent fragments on the asRNA construct of the naturally distant ctfB and ctfA structural genes in an antisense orientation (Fig. 1). The lengths of the asRNAs that were produced from pCTFA2AS, pCTFB1AS, and pCOAT11AS were 762, 522, and 873 nucleotides, respectively. The asRNA length was determined as described above.
FIG. 1.
Construction of plasmids that express asRNA directed toward the downregulation of CoAT. (A) Construction of pCTFA2AS, pCTFB1AS, and pCOAT11AS. For each plasmid, the locations and directions of transcription of the relevant genes are indicated (arrows). Relevant restriction sites are shown. Abbreviations: thl promoter, promoter region for the thiolase gene of C. acetobutylicum strain ATCC 824; ctfA, CoAT subunit A gene; ctfB, CoAT subunit B gene; adc, AADC gene; MLSr, macrolide-lincosamide-streptogramin B resistance gene; repL, pIM13 origin of replication; AMPr, ampicillin resistance gene; ColE1, ColE1 origin of replication. All genes and plasmids are not drawn to scale. (B) Genetic organization of the sol operon. Abbreviations: aad, alcohol/aldehyde dehydrogenase structural gene; sol operon, polycistronic message that includes the mRNA for aad, ctfA, and ctfB. Lines with arrows represent the location and direction of each transcript.
Static-flask experiments.
Maintenance and static-flask cultures of all strains were carried out as described by Tummala et al. (30) except for the following modifications: (i) static-flask-culture starting volumes were 400 ml rather than 750 ml, and (ii) flasks were inoculated with 4 ml of preculture, which came from a 10-ml tube culture rather than a 50-ml flask culture. For each sample, 1 to 1.5 ml of culture supernatant was collected in duplicate and stored at −20°C. For Western blot analysis, 8 to 10 ml of culture suspension with an A600 of <1.0, 4 to 8 ml for samples with 1.0<A600<4.0, and 2 ml for samples with an A600 of >4.0 were centrifuged at 16,000 × g for 2 min at room temperature and the pellets were then stored at −20°C.
Bioreactor experiments.
Large-scale, pH-controlled batch fermentations were performed in a 2.0-liter Biostat M (B. Braun, Allentown, Pa.) or a 5.0-liter BioFlo II (New Brunswick Scientific, Edison, N.J.) bioreactor with a working volume of 1.8 or 3.6 liters, respectively. All fermentations were performed as described by Desai and Papoutsakis (5) with two modifications. The initial sparge rate of N2 for the 2-liter reactor was 55 ml/min, but once the A600 of the culture reached 0.1 to 0.4, the sparge rate was reduced to 11 ml/min. Secondly, the agitation rate for the 2-liter reactor was set at 400 rpm. Culture supernatants and cell pellets for protein extracts were also collected as described above.
Product analysis.
Culture supernatants were analyzed for product concentrations as described previously (2) by using a Waters (Milford, Mass.) high-performance liquid chromatography system (model no. 717 plus autosampler, no. 1515 high-performance liquid chromatography pump, no. 2410 refractive index detector, and in-line vacuum degasser) and Waters' Breeze software. The standard error of the mean for product concentrations was determined from replicate analyses.
Western blots.
Cell extracts were prepared as described previously (27) except that sodium dodecyl sulfate gel-loading buffer without bromophenol blue was used for the resuspension of the pellet. This was done to avoid interference of the bromophenol blue in subsequent protein assays of crude extracts. Protein concentrations of crude extracts were determined by the RC DC protein assay (Bio-Rad Laboratories, Hercules, Calif.) and had a standard error of the mean of less than 10%. For electrophoresis of the proteins, 5 μg (for AADC immunoblots) or 10 μg (for CoAT immunoblots) of total protein for each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Ready Gels (12% Tris-HCl polyacrylamide, 4% resolving) from Bio-Rad Laboratories. After electrophoresis, proteins were transferred to 0.45-μm-pore-size Hybond-P polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). The membranes were then blocked with Tris-buffered saline-Tween 20 (TBST) plus 4% enhanced chemiluminescence (ECL) blocking agent (Amersham Pharmacia Biotech) and hybridized to either rabbit anti-AADC antiserum (1:3,000 dilution in TBST) (24) (for AADC immunoblots) or sheep anti-CoAT antiserum (1:10,000 dilution in TBST plus 4% ECL blocking reagent) (3) (for CoAT immunoblots). Goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (1:10,000 dilution in TBST) (Sigma, St. Louis, Mo.) (for AADC immunoblots) or donkey anti-sheep immunoglobulin G (Sigma) conjugated with horseradish peroxidase (1:10,000 dilution in TBST plus 4% ECL blocking reagent) (for CoAT immunoblots) was then used as a secondary antibody. Fluorescence on each membrane was generated according to the instructions of the ECL+ kit (Amersham Pharmacia Biotech) and detected with a Storm 860 imager (Molecular Dynamics, Sunnyvale, Calif.).
In Western blot experiments in which the nonspecific hybridization and background intensity in AADC blots were reduced, we modified the AADC Western blot protocol described above by adding antibodies in TBST plus ECL blocking reagent (4%) instead of in TBST buffer only. In a separate protocol, we also added 0.5 mg of protein from crude extracts of M5 strain exponential-phase samples to the same amount of primary antibody used in other blots. After bringing the final volume up to 1 ml with TBST buffer, we incubated the antibody and crude extract mixture for an hour at room temperature with gentle shaking. Then, the mixture was added to 25 ml of TBST plus 4% ECL blocking reagent (for a final dilution of 1:3,000 for the primary antibody) and incubated with the membrane as described above.
Quantitative analysis of Western blots.
ImageQuant version 5.0 (Molecular Dynamics) was used to view and quantify Western blots. Quantification of gel band intensities was initiated by drawing rectangles around desired gel bands from all Western blots in ImageQuant version 5.0. By using the method of local averages for background correction, the software reports band intensities for each rectangle. For AADC blots, the range of band intensities was ca. 0 to 93,000, while for CoAT blots, the range was ca. 25 to 47,000. The lower threshold value of detection was determined by visual inspection of Western blots. Gel bands with intensities below 500 (after background intensity was subtracted) were considered to not contain quantifiable amounts of AADC. For CoAT immunoblots, the threshold value was 100. Intensity values for gel bands that were below the threshold value were set equal to zero in all subsequent manipulations of band intensity data. The standard error of the mean for gel band intensities was determined from replicate (two to four) Western blots and represents the cumulative error for the total Western analysis assay.
asRNA structure analysis.
DNA sequences were manipulated and visualized using the Gene Construction Kit 2 (Textco, Inc., West Lebanon, N.H.). Analysis of the secondary structures of RNA molecules was carried out with the Mfold and Plotfold algorithms of the Wisconsin Package, version 9.1 (Genetics Computer Group, Madison, Wis.). For visualization of the secondary structures, GCGFigure 1.1 (Genetics Computer Group) was used as a metafile viewer and PICT file converter. For quantitation of structural features, only the most energetically favorable structure as predicted by Mfold was used.
Calculation of percent overall protein downregulation by asRNA constructs.
To compare the effectiveness of the various asRNA constructs of this study as well as of the asRNA molecules discovered or designed previously, we used the percentage of overall protein downregulation. For studies in which enzyme activities were used to evaluate asRNA effectiveness (5, 7), the percent overall protein downregulation was determined by averaging the target protein's enzyme activity for both control and asRNA-expressing strains throughout their respective cultures and then calculating the percent decrease in average enzyme activity for the asRNA-expressing strain. For the asRNA-producing strains used in this study, the percent overall protein downregulation was determined by averaging the ratio of the target protein's gel band intensities in the asRNA-expressing strains over those of the control strain for all phases of culture in which Western blot analysis was performed.
RESULTS
Differential AADC downregulation with three distinct asRNA constructs.
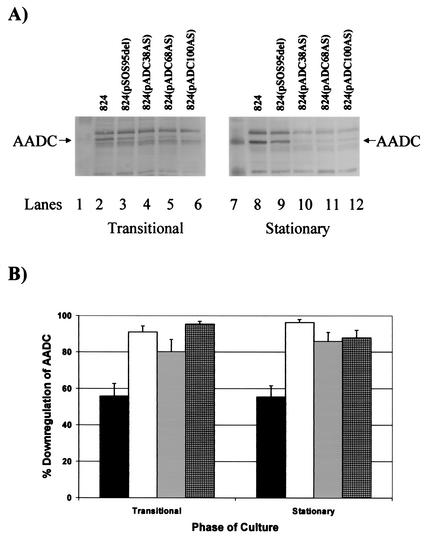
To examine the effect of asRNA length and structural properties on asRNA efficacy, we used three plasmids (pADC38AS, pADC68AS, and pADC100AS), which produce asRNAs of various lengths against the adc transcript. These plasmids, along with a plasmid control (pSOS95del), were then introduced into C. acetobutylicum to generate the four strains of C. acetobutylicum ATCC 824: 824(pADC38AS), 824(pADC68AS), 824(pADC100AS), and 824(pSOS95del). We carried out Western blot analyses of samples from the late exponential and stationary phases and the transitional point between the exponential and stationary phases (hereafter referred to as the transitional phase) of duplicate fermentations for each strain to assess the effectiveness of the various asRNA constructs. Since levels of AADC expression were very low in the late-exponential-phase samples, we were not able to reliably quantify band intensities for this culture stage. Typical Western blots from the transitional and stationary phases of fermentations of each strain are shown in Fig. 2A, and a quantitative summary of the results from these Western blots is shown in Fig. 2B. ATCC 824(pSOS95del) exhibited downregulation of AADC compared to the parental strain in both phases, suggesting that the plasmid itself has a significant effect on AADC levels. However, ATCC 824(pADC38AS), 824(pADC68AS), and 824(pADC100AS) had much greater downregulation of AADC than 824(pSOS95del), which indicates that downregulation of AADC due to adc-asRNA occurred in each adc-asRNA-expressing strain. ATCC 824(pADC38AS) showed the highest percentage of overall downregulation (Fig. 5) of AADC compared to the plasmid control (86% ± 6%), followed by 824(pADC100AS) (81% ± 7%) and then 824(pADC38AS) (62% ± 14%). Static-flask culture experiments were also performed for all these strains, and similar overall trends were observed (data not shown).
FIG. 2.
Downregulation of AADC in C. acetobutylicum adc-asRNA-expressing strains. (A) AADC Western blots from the transitional and stationary phases of fermentation. AADC bands as well as the closest marker bands are indicated for both blots. The culture phase from which the samples on each blot were taken is indicated below each blot. Lanes: 1, kaleidoscope-prestained marker from Bio-Rad Laboratories; 2, ATCC 824; 3, ATCC 824(pSOS95del); 4, ATCC 824(pADC38AS); 5, 824(pADC68AS); 6, 824(pADC100AS); 7, biotinylated protein marker from the horseradish peroxidase protein marker detection pack from New England Biolabs; 8, ATCC 824; 9, ATCC 824(pSOS95del); 10, ATCC 824(pADC38AS); 11, ATCC 824(pADC68AS); 12, ATCC 824(pADC100AS). (B) Percent downregulation of AADC in C. acetobutylicum adc-asRNA-expressing strains. The percent downregulation was calculated as the percent decrease of AADC gel band intensity in ATCC 824(pSOS95del) (▪), ATCC 824(pADC38AS) (□), ATCC 824(pADC68AS) (igwidth>), and ATCC 824(pADC100AS) (igwidth>) compared to that of the parental strain at the same culture phase in Western blots. The standard error of measurement was calculated from two to four different blots.
FIG. 5.
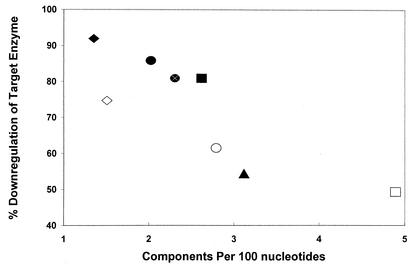
Relationship between the component/nucleotide ratio and the percentage overall protein downregulation for asRNA in solventogenic clostridia. The different asRNA are represented by the following symbols: ▴, glna-asRNA; □, ptb-asRNA; ▪, buk-asRNA; •, adc38-asRNA; ○, adc68-asRNA;  , adc100-asRNA; ♦, ctfb1-asRNA; (⋄), coat11-asRNA-b.
, adc100-asRNA; ♦, ctfb1-asRNA; (⋄), coat11-asRNA-b.
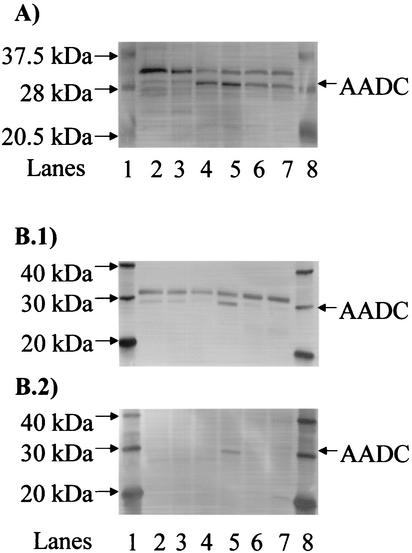
In all Western blots, a significant number of bands, other than AADC, were also detected. To ensure that these bands were not AADC, we performed a Western blot analysis (Fig. 3A) with samples from the early exponential, transitional, and stationary phases of parental (wild-type) fermentations and the stationary phase of a static-flask culture of strain M5. Since adc mRNA is not expressed until the late exponential phase of ATCC 824 fermentations (12) and M5 is a degenerate strain that does not contain the pSOL1 plasmid which carries the adc gene (4), only the transitional and stationary samples from the wild-type fermentations should have contained AADC. As expected, AADC was clearly seen in all of the ATCC 824 transitional and stationary samples at the appropriate size of ca. 28 kDa (24) while only very faint bands were seen in the early exponential phase of a wild-type fermentation and the stationary phase of a M5 static-flask culture (Fig. 2). To further support this conclusion, we subsequently performed another Western blot in which we modified our Western blot protocol by incubating membranes with antibodies in TBST buffer plus ECL blocking reagent (4%) as opposed to TBST buffer solely. This was done to further block any nonspecific hybridization and to decrease background intensity. These Western blots (Fig. 3B.1) showed much-reduced nonspecific hybridization and only two distinct bands, one at the appropriate size of ca. 28 kDa and one at ca. 33 kDa. The latter was present in all strains, including M5, and thus could not possibly be AADC. To further ensure that this larger unknown band was not AADC or a modified version of AADC, we performed another Western blot in which we pretreated the primary antibody, before incubation with the membrane, with crude extracts prepared from M5 protein samples. Since M5 does not contain AADC, this step should have further eliminated any nonspecific binding of our primary antibody, and this is indeed what the Western blots of Fig. 3B.2 show. The very faint bands seen in some M5 lanes may derive from possible persistent contamination of the M5 strain culture with the parent strain since we had no means (selective pressure) to ascertain that the M5 strain is 100% pure (4).
FIG. 3.
Verification of the AADC band in Western blots. (A) Western blot with negative and positive controls for AADC expression. AADC and several of the closest marker bands (20.5, 28, and 37.5 kDa) are indicated. Lanes: 1 and 8, biotinylated protein marker from the horseradish peroxidase protein marker detection pack (New England Biolabs); 2, ATCC 824 at early exponential phase (2-liter bioreactor); 3, strain M5 at stationary phase (static-flask culture); 4, ATCC 824 at stationary phase (5-liter bioreactor); 5, ATCC 824 at transitional phase (5-liter bioreactor); 6, ATCC 824 at stationary phase (2-liter bioreactor); 7, ATCC 824 at transitional phase (2-liter bioreactor). (B) Improved detection of AADC in Western blots. (B.1) Western blot in which antibodies were added in the presence of blocking reagent. (B.2) Western blot in which the primary antibody was pretreated with crude extracts of strain M5 prior to incubation with the membrane. AADC and several of the closest marker bands (20, 30, and 40 kDa) are indicated. Lanes: 1 and 8, MagicMark Western protein standard Invitrogen; 2, ATCC 824(pADC100AS) at transitional phase; 3, ATCC 824(pADC68AS) at transitional phase; 4, ATCC 824(pADC38AS) at transitional phase; 5, ATCC 824(pSOS95del) at transitional phase; 6, strain M5 at stationary phase; 7, strain M5 at exponential phase.
asRNA effectiveness correlates with an asRNA structural feature.
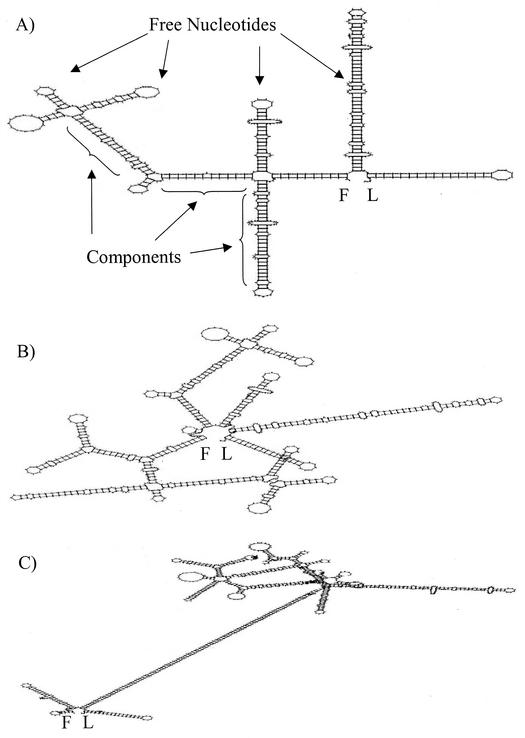
We used two criteria to assess asRNA structural differences: free nucleotides and components (Fig. 4A). Free nucleotides are defined as nucleotides in an asRNA molecule that are not involved in intramolecular bonding and thus are thought to provide potential sites with which the asRNA and target mRNA might interact (22). Components are structural features that contain regions of high complementarity within an asRNA molecule. The numbers of free nucleotides and components (normalized per 100 nucleotides of each asRNA molecule) were plotted separately for each asRNA against the percentage of overall protein downregulation of each asRNA's target enzyme. By using the data from the asRNA constructs developed in this study (pADC38AS, pADC68AS, and pADC100AS) as well as those of Desai and Papoutsakis (5) (pRD4 for buk-asRNA and pRD1 for ptb-asRNA) and the natural glnA-asRNA (7), only the normalized number of components correlated well with in vivo asRNA effectiveness, with a correlation coefficient of 0.87 for a second-order polynomial (Fig. 5). This value increased to 0.97 when only the adc-asRNAs developed in this study were considered. These results suggest that the (number of components)/(number of total nucleotides) ratio may be a useful predictor of asRNA effectiveness.
FIG. 4.
Predicted secondary structures of adc-asRNA. (A) adc38-asRNA; (B) adc68-asRNA; (C) adc100-asRNA. Examples of free nucleotides and components are shown for the asRNA of panel A. The first and last nucleotides of each asRNA molecule are designated F and L, respectively.
Downregulation of AADC does not result in concomitant effects on acetone production.
Solvent analysis and cell (optical) density measurements of all fermentations were performed for each strain to examine the possible effects of downregulation of AADC on cell metabolism (Table 3). Both ATCC 824(pADC38AS) and 824(pADC100AS) had slightly higher maximum A600 values (14 and 16%, respectively) than 824(pSOS95del). Although acetone and butanol concentrations appeared to vary among the strains, only a slight decrease for a normalized (based on the maximum A600) acetone concentration in ATCC 824(pADC38AS) (17%) and a slight increase for a normalized butanol concentration in 824(pADC100AS) (27%) were observed compared to 824(pSOS95del). Larger differences could be seen in the normalized final butyrate levels of ATCC 824(pADC38AS) and 824(pADC100AS) (47 and 32% lower, respectively) than in 824(pSOS95del), which indicates more butyrate uptake in 824(pADC38AS) and 824(pADC100AS) than in 824(pSOS95del). Finally, small increases in the butanol/acetone ratio were observed in only 824(pADC38AS) (23%) and 824(pADC68AS) (12%) compared to that for the plasmid control strain.
TABLE 3.
Fermentation characteristicsa of strains for the asRNA downregulation of AADC in pH-controlled bioreactor experimentsb
| Strain | No. of expts | Growth rate (h−1) | Maximum A600 | Acetone concn | Acetone concn/maximum A600c | Butanol concn | Butanol concn/maximum A600c | Peak acetate concn/maximum A600c | Final acetate concn/maximum A600c | Peak butyrate concn/maximum A600c | Final butyrate concn/maximum A600c | Butanol/acetate ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATCC 824 | 2 | 0.47 ± 0.07 | 7.63 ± 0.33 | 79 ± 4 | 10.4 ± 0.0 | 139 ± 9 | 18.2 ± 0.0 | 9.8 ± 0.0 | 9.1 ± 0.2 | 11.3 ± 0.4 | 6.4 ± 0.6 | 1.75 ± 0.03 |
| ATCC 824(SOS95del) | 6 | 0.45 ± 0.03 | 5.56 ± 0.42 | 49 ± 2 | 9.5 ± 1.0 | 93 ± 3 | 17.3 ± 1.6 | 15.9 ± 1.8 | 14.7 ± 1.2 | 13.4 ± 1.1 | 10.3 ± 0.7 | 1.91 ± 0.08 |
| ATCC 824(pADC38AS) | 2 | 0.43 ± 0.04 | 6.34 ± 0.04 | 50 ± 1 | 7.9 ± 0.2 | 118 ± 2 | 18.6 ± 1.8 | 17.2 ± 1.2 | 16.3 ± 0.9 | 11.9 ± 0.2 | 5.5 ± 1.8 | 2.34 ± 0.17 |
| ATCC 824(pADC68AS) | 2 | 0.42 ± 0.02 | 5.56 ± 0.19 | 47 ± 7 | 8.5 ± 0.9 | 101 ± 10 | 18.0 ± 1.1 | 17.4 ± 0.3 | 17.0 ± 0.5 | 14.2 ± 0.2 | 9.2 ± 0.7 | 2.14 ± 0.09 |
| ATCC 824(pADC100AS) | 2 | 0.44 ± 0.06 | 6.43 ± 0.42 | 73 ± 1 | 11.5 ± 0.9 | 141 ± 1 | 22.0 ± 1.6 | 15.9 ± 0.4 | 14.5 ± 0.4 | 12.5 ± 0.5 | 7.0 ± 0.2 | 1.92 ± 0.01 |
Mean ± standard error of the mean.
Concentrations are millimolar.
Ratios of product concentrations over the maximum cell density (A600) were calculated in order to normalize some key data for comparison purposes.
Although drastic differences in normalized product concentrations were not seen between the adc-asRNA-expressing strains and the plasmid control strain, large differences were observed between ATCC 824(pSOS95del) and the parental strain. First, the maximum A600 of ATCC 824(pSOS95del) was 27% lower than the maximum A600 for strain 824. In addition, normalized peak acetate, final acetate, and final butyrate concentrations were 62, 62, and 61% greater, respectively, in ATCC 824(pSOS95del) than in strain 824. These results further demonstrate that the plasmid itself has an impact on cell physiology.
AADC is necessary for acetone formation.
Despite the significant downregulation of AADC, acetone levels of ATCC 824(pADC38AS) and 824(pADC68AS) did not differ from the plasmid control as significantly as would be expected from the drastic downregulation of AADC. These unexpected results suggest that AADC is not rate limiting and perhaps is not even necessary for acetone production. To test the hypothesis that AADC is not necessary for acetone formation, we complemented a C. acetobutylicum mutant, M5, which does not produce acetone (it lacks all the solvent formation genes discussed above), with plasmid pFNK7, which contains the genes ctfA and ctfB, and also with plasmid pFNK6, which contains the genes encoding for the complete acetone formation pathway (adc, ctfA, and ctfB). Both of these plasmids were shown to produce functional proteins in C. acetobutylicum (18). Acetone (ca. 23 mM) was detected only in cultures of M5(pFNK6). We concluded that AADC is necessary for acetone formation, although AADC is not apparently limiting the rate of acetone formation, i.e., very low levels (e.g., 10% of those normally expressed [Fig. 4]) of AADC are sufficient for high rates of acetone formation.
CoA-transferase limits acetone formation.
For each CoAT subunit gene, three potential asRNA constructs were examined by using DNA representation software. All three included the DNA sequence of the ribosome binding site and a portion of the subunit's structural gene in an antisense orientation. Potential asRNA candidates were further selected by calculating a (number of components)/(number of nucleotides) ratio based on each asRNA's secondary structure. The DNA sequence for the asRNA candidate that produced the lowest component/nucleotide ratio was then cloned in the antisense orientation into BamHI- and EheI-digested pSOS95. The asRNAs, which were separately designed against each CoAT subunit, were then combined in several different combinations to create constructs that could potentially downregulate both subunits of CoAT simultaneously. The selection process for choosing the best asRNA candidate discussed above was also used in the screening process for these combined asRNA molecules.
These three asRNA constructs (Fig. 1) were introduced into C. acetobutylicum to generate strains [ATCC 824(pCTFA2AS), 824(pCTFB1AS), and 824(pCOAT11AS)] that were then testedin static-flask culture experiments (Table 4). All three of these strains had significantly lower maximum A600 values (22, 26, and 21%, respectively) than the plasmid control strain ATCC 824(pSOS95del). The most notable difference between the CoAT-asRNA-expressing strains and the plasmid control strain can be seen in the normalized (based on the maximum A600) acetone concentrations. ATCC 824(pCTFA2AS) and 824(pCOAT11AS) had normalized acetone concentrations approximately eightfold lower than the plasmid control levels, with 824(pCTFB1AS) showing no acetone production at all [although in pH-controlled fermentations of 824(pCTFB1AS), 6 mM acetone was produced (data not shown)]. Normalized butanol levels for ATCC 824(pCTFA2AS), 824(pCTFB1AS), and 824(pCOAT11AS) were also lower than the normalized butanol levels seen in the plasmid control (69, 82, and 70% lower, respectively). Normalized peak and final acetate levels were slightly higher in the asRNA-producing strains than the ATCC 824(pSOS95del) normalized peak and final acetate levels. Normalized peak butyrate levels were 86, 113, and 54% higher in ATCC 824(pCTFA2AS), 824(pCTFB1AS), and 824(pCOAT11AS), respectively, than in the plasmid control strain. More importantly, for the three asRNA-expressing strains, peak and final butyrate levels did not change (no butyrate uptake), whereas for ATCC 824(pSOS95del), final butyrate levels were 66% lower than its peak value (strong butyrate uptake).
TABLE 4.
Fermentation characteristicsa of strains for the asRNA downregulation of CoAT in static-flask culturesb
| Strain | No. of expts | Maximum A600 | Acetone concn/maximum A600c | Butanol concn/maximum A600c | Peak acetate concn/maximum A600c | Final acetate concn/maximum A600c | Peak butyrate concn/maximum A600c | Final butyrate concn/maximum A600c |
|---|---|---|---|---|---|---|---|---|
| ATCC 824(pSO595del) | 2 | 5.43 ± 0.36 | 7.9 ± 0.9 | 15.4 ± 1.5 | 5.1 ± 0.3 | 4.3 ± 0.4 | 5.6 ± 0.3 | 1.9 ± 0.4 |
| ATCC 824(pCTFA2AS) | 2 | 4.26 ± 0.19 | 0.9 ± 0.01 | 4.8 ± 0.2 | 7.8 ± 0.5 | 8.2 ± 0.5 | 10.4 ± 0.6 | 10.4 ± 0.6 |
| ATCC 824(pCTFB1AS) | 2 | 4.03 ± 0.00 | 0.0 ± 0.0 | 2.7 ± 0.3 | 8.0 ± 0.0 | 7.8 ± 0.1 | 11.9 ± 0.2 | 11.9 ± 0.2 |
| ATCC 824(pCOAT11AS) | 2 | 4.28 ± 0.0 | 0.9 ± 0.5 | 4.6 ± 0.5 | 8.7 ± 0.1 | 5.9 ± 0.2 | 8.6 ± 0.4 | 8.4 ± 0.6 |
Mean ± standard error of the mean.
All concentrations are millimolar.
Ratios of product concentrations over the maximum cell density (A600) were calculated in order to normalize some key data for comparison purposes.
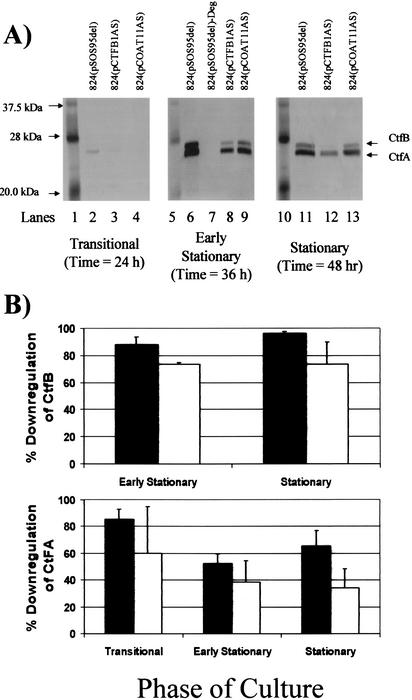
To confirm that the drastic changes in metabolism seen in cultures of ATCC 824(pCTFB1AS) were due to the downregulation of CoAT and to investigate the effectiveness of 824(pCOAT11AS) in downregulating both the CtfA and CtfB subunits from a single asRNA molecule, Western analysis was performed with these strains and the plasmid control strain 824(pSOS95del). An example of the Western blots and a quantitative summary of the results from these Western blots are shown in Fig. 6. In the transitional phase, both ATCC 824(pCTFB1AS) and 824(pCOAT11AS) exhibited downregulation of CtfA (the only subunit seen in the plasmid control lane in this phase) compared to the plasmid control strain [824(pSOS95del)]. ATCC 824(pCTFB1AS) and 824(pCOAT11AS) exhibited downregulation of both CtfA and CtfB in the early stationary phase compared to the plasmid control, with lower levels of CtfB seen in 824(pCTFB1AS). In addition, a protein sample from a degenerate strain (due to pSOL1 loss [4]) of ATCC 824(pSOS95del) was included in the early-stationary-phase blot as a negative control. There was no detection of CoAT in this lane, further confirming that the bands seen in this blot as well as the ones from the transitional and stationary-phase blots are indeed CtfA and CtfB. In the stationary phase, both ATCC 824(pCTFB1AS) and 824(pCOAT11AS) showed significant downregulation of both CtfA and CtfB, with ATCC 824(pCTFB1AS) exhibiting more downregulation of both subunits. The combination of downregulation of CoAT in the Western blot analysis of ATCC 824(pCTFB1AS) and 824(pCOAT11AS) and the severely diminished acetone production in these strains show that CoAT is indeed the limiting enzyme in the acetone formation pathway.
FIG. 6.
Downregulation of CtfA and CtfB subunits of CoAT by CoAT-asRNA. (A) CoAT Western blots from the transitional, early stationary, and stationary phases of C. acetobutylicum static-flask fermentations. The CtfA and CtfB subunits of CoAT as well as the closest marker bands are indicated for all blots. The culture phase from which the samples on each blot were taken are indicated below each blot. Lanes 1, 5, and 10, biotinylated protein marker from the horseradish peroxidase protein marker detection pack from New England Biolabs; lanes 2, 6, and 11, ATCC 824(pSOS95del); lanes 3, 8, and 12, ATCC 824(pCTFB1AS); lane 7, degenerate ATCC 824(pSOS95del); lanes 4, 9, and 13, ATCC 824(pCOAT11AS). (B) Percent downregulation of CtfA and CtfB subunits of CoAT by CoAT-asRNA. The percent downregulation of CtfA and CtfB in ATCC 824(pCTFB1AS) (▪) and 824(pCOAT11AS) (□) was calculated as the percent decrease of the desired gel band intensity of Western blots in the asRNA-expressing strain compared to that in the plasmid control strain [824(pSOS95del)] at the same culture phase. The standard error of measurement was calculated from two to four different blots.
To further pursue the hypothesis that the component/nucleotide ratio can be used to design effective asRNA constructs in C. acetobutylicum, we calculated the percentage of overall protein downregulation for the asRNA produced by pCTFB1AS (ctfb1-asRNA) on CtfB downregulation and the asRNA generated by pCOAT11AS (coat11-asRNA) separately on CtfA and CtfB downregulation (Fig. 5). Although ctfb1-asRNA and coat11-asRNA contain the same antisense region directed toward CtfB downregulation, they differ in their ability to downregulate this protein. ctfb1-asRNA is clearly the most effective in downregulating CtfB, with a percent overall protein downregulation of 92% ± 4%, while coat11-asRNA was less effective at downregulating CtfB (74% ± 12%). This is most likely due to the fact that coat11-asRNA contains an extra antisense region (i.e., the region encoding for an antisense molecule targeted against CtfA [Fig. 1]), which hinders the ability for the coat11-asRNA's ctfB antisense region to bind to ctfB mRNA as effectively as ctfb1-asRNA's ctfB antisense region does. Nonetheless, both asRNAs downregulate their target proteins effectively compared to other asRNAs in C. acetobutylicum (Fig. 5), and it appears that using the component/nucleotide ratio is a promising design parameter for developing effective asRNA constructs in C. acetobutylicum.
Downregulation of CtfA by coat11-asRNA (44% ± 24%) was not nearly as effective as coat11-asRNA's downregulation of CtfB. In fact, coat11-asRNA's ability to downregulate CtfA was approximately the same as ctfb1-asRNA's ability to downregulate CtfA (68% ± 9%), which does not contain a region that is complementary to ctfA mRNA. This suggests that downregulation of CtfA by coat11-asRNA might be due to the effect of the ctfB antisense region on the rest of the polycistronic message rather than the effect of the ctfA antisense region of coat11-asRNA.
DISCUSSION
An important goal of this study was to develop a method for designing effective asRNA constructs for the downregulation of enzymes in C. acetobutylicum. We chose to vary the length of asRNAs directed toward AADC and analyze each asRNA's ability to downregulate AADC in vivo. Based on the structural data on each adc-asRNA as well as the asRNAs discovered or designed previously, we discovered a correlation between the component/nucleotide ratio and percentage of overall protein downregulation. Based on this correlation, we then designed asRNAs directed towards the downregulation of CoAT. Results from CoAT-asRNA experiments corroborate the usefulness of this parameter as a predictor of asRNA effectiveness. The reason for this ability to predict asRNA effectiveness might derive from the likely relationship between the component/nucleotide ratio and asRNA stability. As the component/nucleotide ratio decreases, the number of free nucleotides decreases, and thus, more nucleotides are intramolecularly bound, making it more difficult for RNases to degrade the asRNA molecule.
Although the downregulation of AADC was effective, acetone concentrations in the adc-asRNA-producing strains were not as drastically altered compared to those in the plasmid control as originally hypothesized. However, cultures of strains containing CoAT-asRNA plasmids showed substantially lower levels of acetone and CoAT subunit amounts than the plasmid control. These results suggest that CoAT is the rate-limiting enzyme in the acetone formation pathway of C. acetobutylicum.
Interestingly, butanol levels in CoAT-asRNA-expressing strains were also significantly decreased compared to the plasmid control, which suggests that aad expression might also be altered by CoAT-asRNA. This is not surprising in view of the fact that aad resides on the same polycistronic message as ctfA and ctfB. In fact, recent pH-controlled fermentation experiments with a C. acetobutylicum strain, which overexpresses ctfb1-asRNA and aad, exhibited butanol formation comparable to that with 824(pSOS95del) (data not shown). These results suggest that the mechanism of antisense action in these strains is the RNase degradation of the polycistronic message initiated by asRNA binding rather than the blocking of translation by the asRNA binding of target ribosome binding sites.
This is the first study to our knowledge to examine the ability of expressing a single asRNA directed against two different proteins. coat11-asRNA was able to downregulate CtfB, but its effects on CtfA appear to be relatively small. A potential reason for this is that the ctfA mRNA might be embedded in the secondary structure of the polycistronic message that is hindering the ability of coat11-asRNA to bind its target region in the ctfA mRNA. Another potential reason might derive from the fact that the portion of coat11-asRNA targeted for CtfA downregulation is transcribed after the ctfB antisense portion. Since the ctfB antisense portion is transcribed first, it may bind its target and fix coat11-asRNA's position on the polycistronic message and not allow the ctfA antisense portion either the time or the flexibility to bind the second intended target mRNA.
Acknowledgments
This work was supported by a National Science Foundation grant (BES-9905669).
We thank Abbot Laboratories for the donation of clarithromycin, G. N. Bennett for the donation of AADC and CoAT primary antibodies, and Payam Roshandel for technical assistance.
REFERENCES
- 1.Berghammer, H., and B. Auer. 1993. “Easypreps”: fast and easy plasmid minipreparation for analysis of recombinant clones in E. coli. BioTechniques 14:524-528. [PubMed] [Google Scholar]
- 2.Buday, Z., J. C. Linden, and M. N. Karim. 1990. Improved acetone-butanol fermentation analysis using subambient HPLC column temperature. Enzyme Microb. Technol. 12:24-27. [Google Scholar]
- 3.Cary, J. W., D. J. Petersen, E. T. Papoutsakis, and G. N. Bennett. 1990. Cloning and expression of Clostridium acetobutylicum ATCC 824 acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase in Escherichia coli. Appl. Environ. Microbiol. 56:1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornillot, E., R. V. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, R. P., and E. T. Papoutsakis. 1999. Antisense RNA strategies for the metabolic engineering of Clostridium acetobutylicum. Appl. Environ. Microbiol. 65:936-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engdahl, H. M., T. A. Hjalt, and E. G. Wagner. 1997. A two unit antisense RNA cassette test system for silencing of target genes. Nucleic Acids Res. 25:3218-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fierro-Monti, I. P., S. J. Reid, and D. R. Woods. 1992. Differential expression of a Clostridium acetobutylicum antisense RNA: implications for regulation of glutamine synthetase. J. Bacteriol. 174:7642-7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer, R. J., J. Helms, and P. Durre. 1993. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J. Bacteriol. 175:6959-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridovich, I. 1972. Acetoacetate decarboxylase, p. 255-270. In P. D. Boyer (ed.), Carboxylation and decarboxylation (nonoxidative) isomerization, vol. 6. Academic Press, Inc., New York, N.Y.
- 10.Gerischer, U., and P. Durre. 1992. mRNA analysis of the adc gene region of Clostridium acetobutylicum during the shift to solventogenesis. J. Bacteriol. 174:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris, L. M., R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2000. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol. Bioeng. 67:1-11. [PubMed] [Google Scholar]
- 12.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hjalt, T., and E. G. Wagner. 1992. The effect of loop size in antisense and target RNAs on the efficiency of antisense RNA control. Nucleic Acids Res. 20:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hjalt, T. A., and E. G. Wagner. 1995. Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res. 23:580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes, J. B., C. Y. Wang, R. Bhadra, A. Richardson, G. N. Bennett, and F. B. Rudolph. 1998. Reduction of 2,4,6-trinitrotoluene by Clostridium acetobutylicum through hydroxylamino-nitrotoluene intermediates. Environ. Toxicol. Chem. 17:343-348. [Google Scholar]
- 16.Lee, S. Y., and S. Rasheed. 1990. A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques 9:676-679. [PubMed] [Google Scholar]
- 17.Mermelstein, L. D., and E. T. Papoutsakis. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mermelstein, L. D., E. T. Papoutsakis, D. J. Petersen, and G. N. Bennett. 1993. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for increased solvent production by the enhancement of acetone formation enzyme activities using a synthetic acetone operon. Biotechnol. Bioeng. 42:1053-1060. [DOI] [PubMed] [Google Scholar]
- 19.Nair, R. V., G. N. Bennett, and E. T. Papoutsakis. 1994. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 176:871-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patzel, V., and G. Sczakiel. 2000. In vitro selection supports the view of a kinetic control of antisense RNA-mediated inhibition of gene expression in mammalian cells. Nucleic Acids Res. 28:2462-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patzel, V., and G. Sczakiel. 1998. Theoretical design of antisense RNA structures substantially improves annealing kinetics and efficacy in human cells. Nat. Biotechnol. 16:64-68. [DOI] [PubMed] [Google Scholar]
- 23.Persson, C., E. G. Wagner, and K. Nordstrom. 1988. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 7:3279-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen, D. J., and G. N. Bennett. 1990. Purification of acetoacetate decarboxylase from Clostridium acetobutylicum ATCC 824 and cloning of the acetoacetate decarboxylase gene in Escherichia coli. Appl. Environ. Microbiol. 56:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen, D. J., J. W. Cary, J. Vanderleyden, and G. N. Bennett. 1993. Sequence and arrangement of genes encoding enzymes of the acetone-production pathway of Clostridium acetobutylicum ATCC 824. Gene 123:93-97. [DOI] [PubMed] [Google Scholar]
- 26.Rittner, K., C. Burmester, and G. Sczakiel. 1993. In vitro selection of fast-hybridizing and effective antisense RNAs directed against the human immunodeficiency virus type 1. Nucleic Acids Res. 21:1381-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Theys, J., W. Landuyt, S. Nuyts, L. Van Mellaert, E. Bosmans, A. Rijnders, W. Van Den Bogaert, A. van Oosterom, J. Anne, and P. Lambin. 2001. Improvement of Clostridium tumour targeting vectors evaluated in rat rhabdomyosarcomas. FEMS Immunol. Med. Microbiol. 30:37-41. [DOI] [PubMed] [Google Scholar]
- 29.Tummala, S. B., C. Tomas, L. M. Harris, N. E. Welker, F. B. Rudolph, G. N. Bennett, and E. T. Papoutsakis. 2001. Genetic tools for solventogenic Clostridia, p. 105-123. In H. Bahl and P. Duerre (ed.), Clostridia: Biotechnology and medical applications. John Wiley & Sons, Inc., New York, N.Y.
- 30.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westheimer, F. H. 1969. Acetoacetate decarboxylase from Clostridium acetobutylicum, p. 231-241. In J. M. Lowenstein (ed.), Lipids, vol. 14. Academic Press, Inc., New York, N.Y.
- 32.Wiesenborn, D. P., F. B. Rudolph, and E. T. Papoutsakis. 1989. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl. Environ. Microbiol. 55:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]