Abstract
Five genes involved in the two initial steps of the tetralin biodegradation pathway of Sphingomonas macrogolitabida strain TFA have been characterized. ThnA1A2 and ThnA3A4, components of the ring-hydroxylating dioxygenase, were encoded in divergently transcribed operons. ThnA1, ThnA2, and ThnA3 were essential for tetralin ring-hydroxylating dioxygenase activity. ThnB was identified as a dehydrogenase required for tetralin biodegradation.
The organic solvent tetralin (1,2,3,4-tetrahydronaphthalene) is widely used as a degreasing agent and solvent for fats, resins, and waxes and as a substitute for turpentine in paints, lacquers, and shoe polishes; tetralin is also used in the petrochemical industry in connection with coal liquefaction (7). Tetralin is a bicyclic molecule composed of an aromatic moiety and an alicyclic moiety, which share two carbon atoms.
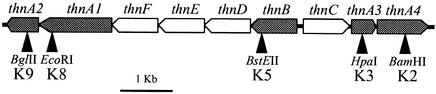
A few bacterial strains able to aerobically grow on tetralin as the only carbon and energy source have been described previously (22). Several reports suggest that some bacteria initially hydroxylate and further oxidize the alicyclic ring, while others initially dioxygenate the aromatic ring, thus indicating that tetralin may be aerobically metabolized in different ways (21, 23). Sphingomonas macrogolitabida strain TFA (formerly Sphingopyxis macrogoltabida) (9, 26, 27) is able to grow on tetralin as the only source of carbon and energy (10). Previous work with this strain characterized a meta-cleavage and subsequent reactions, which cleave both the aromatic and the alicyclic rings of tetralin, thus yielding pyruvate and pimelic acid semialdehyde (2, 8, 9). Four genes (thnC, thnD, thnE, and thnF) coding for these enzymes have been identified and have been shown to be located in two closely linked operons, which are divergently transcribed (Fig. 1). These data suggested that biodegradation of tetralin by strain TFA involves initial oxidation of the aromatic ring to yield 1,2-dihydroxytetralin (1,2-DHT), through reactions presumably catalyzed by a ring-hydroxylating dioxygenase followed by a dehydrogenase. However, these enzymes or the genes coding for them had not yet been identified. In this work, we describe the structural and functional characterization of the genes encoding the enzyme components involved in the first two steps of tetralin biodegradation.
FIG. 1.
Representation of the genomic region of strain TFA involved in tetralin biodegradation. Genes identified in this work are shaded. The locations of the restriction sites where the cassette KIXX was inserted in the mutants are also shown.
In previous work, a collection of mutants unable to grow on tetralin as the only carbon and energy source had been constructed (10). Some of them were located in genomic regions that were very close but outside of the previously sequenced DNA fragments. In an attempt to fully characterize the tetralin biodegradation pathway of strain TFA, three DNA regions surrounding the previously identified genes were sequenced (Fig. 1). Putative gene products encoded by the new sequences were initially compared to those in the databases with the Gapped BLAST program (1). For each open reading frame, sequences showing high similarity to that of strain TFA were aligned by using the CLUSTAL X program (25) with default parameters. A phylogenetic tree was then constructed by the neighbor-joining method (20).
Two potential genes, designated thnA1 and thnA2, whose products showed high similarity to α and β subunits, respectively, of terminal oxygenases of ring-hydroxylating dioxygenases, were identified in the region downstream of thnF. ThnA1 had the highest similarities to an uncharacterized α subunit coded in fragment C of Rhodococcus sp. strain RHA1 (12) (62% identity over a 440-residue overlap) and to the gene product of bphA1b in plasmid pNL1 of Sphingomonas aromaticivorans (19) (62% identity over a 450-residue overlap). Phylogenetic analysis indicated that these proteins clustered within group III, as defined by Nam et al. (17), represented by polycyclic aromatic hydrocarbon dioxygenases (data not shown). However, the three proteins generate a distinctive branch in the dendrogram, not previously detected, which is highly divergent from the other known α subunits (identity <40%). ThnA1 showed strict conservation of the universally conserved Cys and His residues, which have been shown to bind the Rieske-type iron-sulfur center, and the catalytic residues coordinating mononuclear iron identified in the crystallized naphthalene dioxygenase (11). Phylogenetic analysis of ThnA2 revealed a dendrogram very similar to that obtained for ThnA1 (data not shown), although the levels of divergence among β subunits were higher than those among α subunits. These data suggest that, in general, genes coding for the partner α and β subunits have the same origin and evolve together, although the β subunits tolerate more changes in the amino acid sequence.
In an operon divergently transcribed and downstream of thnC, two additional genes potentially coding for a ferredoxin and a ferredoxin reductase were designated thnA3 and thnA4, respectively. The thnA3 gene apparently codes for a Rieske-type ferredoxin of 104 residues, which showed strict conservation of the Cys and His residues coordinating the irons of [2Fe-2S]. It had the highest similarities to DbtAb of Burkholderia sp. strain DBT1 (accession no. AF404408) (51% identity); BphA3 on plasmid pNL1 of Sphingomonas aromaticivorans (19) (50% identity); PhnR of Sphingomonas chungbukensis strain DJ77 (accession no. AF061802) (48.5% identity), which presumably is involved in phenanthrene degradation; and NsaA3, involved in degradation of naphthalene sulfonate in Sphingomonas sp. strain BN6 (accession no. U65001.3) (50% identity), with which it constituted a distinctive branch of ferredoxins. Interestingly, the gene arrangements of the most similar ferredoxins are the same: they are located just downstream of genes coding for extradiol dioxygenases, very similar to ThnC, the extradiol dioxygenase of the tetralin biodegradation pathway (2), which suggests that these ferredoxins and extradiol dioxygenases have common origins and evolved together.
Sequence analysis of ThnA4 showed high similarity to ferredoxin reductases bearing a binding domain for a [2Fe-2S] center coordinated by four cysteines in addition to the NAD+ and the FAD binding domains. These domains were also conserved in ThnA4, thus suggesting that the ring-hydroxylating dioxygenase involved in tetralin biodegradation is a class III dioxygenase (3). However, ThnA4 had the highest similarity to reductases of multicomponent toluene or phenol monooxygenases (up to 45% identity). Taken together, these data suggest that components of the electron transfer system of different origins can be recruited to functionally supply electrons to terminal oxygenases, which, in turn, may have a different evolutionary origin.
In the intervening region between thnC and thnD, a fifth open reading frame translated in the same direction as thnD was identified and designated thnB, since its putative gene product showed homology to known cis-dihydrodiol dehydrogenases of aromatic compound catabolic pathways. The similarity was highest (65% identity) to PhnB of Burkholderia sp. strain RP007, which is involved in phenanthrene degradation (14).
Since no mutant of the previously obtained collection had an insertion located in thnA2, a new insertion mutant was constructed. To this end, the plasmid pIZ673, with a 5.1-kb fragment comprising thn′DEFA1A2 and sequences downstream plus a KIXX cassette (from pUC4-KIXX; Pharmacia) (10) located in the 69th codon of thnA2, was used to electrotransform strain TFA in a BTX Electro cell manipulator 600 (Biotechnologies & Experimental Research, Inc., San Diego, Calif.) at 2.5 kV and 246 Ω. Transformants that had acquired the insertion by recombination with homologous sequences in the plasmid (5 × 105 μg of plasmid−1) were isolated as previously described (10). The locations of the KIXX insertions in these transformants were confirmed by Southern hybridization, and one transformant, designated K9, was selected for further characterization. Out of the five insertion mutants (Fig. 1), K3, K5, K8, and K9 were unable to grow with tetralin as the only carbon and energy source, thus suggesting that thnA1, thnA2, thnA3, and thnB were strictly required for tetralin utilization. On the other hand, growth of mutant K2 on tetralin was reduced to 31% of that of wild-type TFA. The growth phenotype suggested that thnA4 was also involved in tetralin utilization, although it was not strictly required.
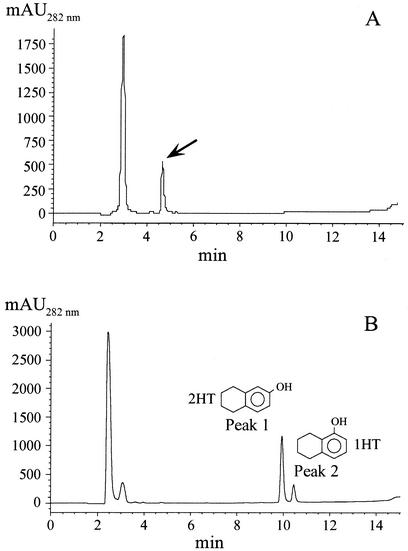
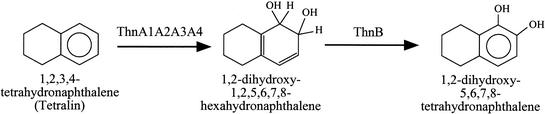
Mutant K5 was grown in minimal medium containing 8 mM β-hydroxybutyrate and tetralin provided in the gas phase, a condition that allows growth of Thn− mutants and expression of thn genes (2, 8). In an attempt to detect any metabolite accumulated during growth, the supernatant was analyzed by high-performance liquid chromatography (HPLC) with an HP 1100 series chromatograph (Hewlett Packard, Waldbronn, Germany) equipped with a diode array detector and using a Hewlett Packard reversed-phase column (ODS Hypersil; 5 μm, 250 by 2 mm). The separation method involved a gradient from 40 to 100% acetonitrile in water for 15 min at a flow rate of 0.2 ml min−1. A compound that eluted at min 4.7, which was not detected in cultures of wild-type TFA, was very evident in cultures of mutant K5 (Fig. 2A). The supernatant was acidified and further analyzed by HPLC. Under similar separation conditions, the peak eluting at min 4.7 was no longer detected, and two new peaks appeared at 9.9 and 10.4 min, which were not detected in supernatants of wild-type TFA treated in the same way (Fig. 2B). The remaining peaks were also detected in supernatants of wild-type TFA and are presumably due to medium composition or to an unrelated metabolite excreted by the cells (data not shown). With commercially available 1-hydroxy-5,6,7,8-tetrahydronaphthalene (1HT) and 2-hydroxy-5,6,7,8-tetrahydronaphthalene (2HT) as standards (Sigma Chemical Co., St. Louis, Mo.), 2HT eluted exactly at the same time and had the same absorption spectrum as compound 1, while characteristics of 1HT fully matched those of compound 2 (data not shown). Since dihydrodiols are known to be unstable under acidic conditions and to be transformed in a mixture of the rearomatized monohydroxylated derivatives (5, 23), accumulation of 1HT and 2HT by acidification suggested that the metabolite that accumulated in the ThnB mutant was the dihydrodiol derivative of tetralin 1,2-dihydroxy-1,2,5,6,7,8-hexahydronaphthalene and, therefore, that ThnB catalyzes the dehydrogenation reaction, leading to production of 1,2-dihydroxytetralin (see Fig. 5).
FIG. 2.
(A) HPLC chromatogram of the culture medium after growth of the KIXX::thnB insertion mutant on 8 mM β-hydroxybutyrate and tetralin as carbon sources. The arrow points out the peak detected after growth of the mutant. (B) HPLC chromatogram of the same culture medium after acidification up to pH 2.2.
FIG. 5.
Initial reactions of the tetralin biodegradation pathway of Sphingomonas macrogolitabida strain TFA, which lead to formation of a catecholic intermediate.
After growth in minimal medium containing 8 mM β-hydroxybutyrate and tetralin, dioxygenase activity was tested in the wild type and thnA mutants by monitoring the capacity to transform indole to ultimately yield indigo (18) in a resting cell assay. Wild-type TFA accumulated indigo at a rate of 603 nM min−1. The thnA1, thnA2, and thnA3 insertion mutants did not accumulate indigo at all, thus suggesting that these mutants lacked any detectable dioxygenase activity. Interestingly, the mutant with mutation in thnA4 did accumulate indigo at a rate 55% of that of wild-type TFA. This result suggested that ThnA4 is not strictly required, although it contributes to maximal dioxygenase activity. The slight reduction in dioxygenase activity of the thnA4 mutant was sufficient to result in an evident reduction in growth rate with tetralin as the only carbon and energy source, thus suggesting that the first reaction may be a rate-limiting step in the tetralin biodegradation pathway.
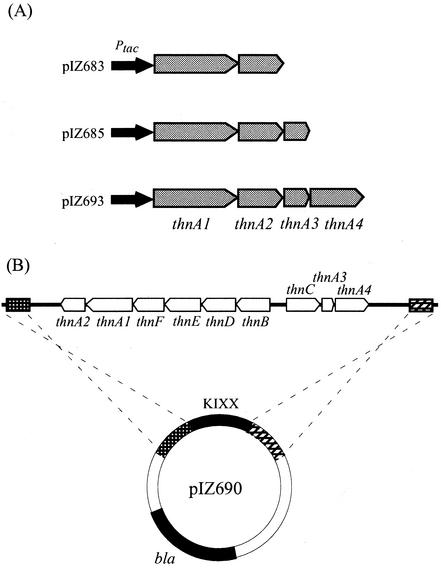
To directly confirm the role of ThnA gene products in the initial dioxygenation of tetralin, the function of these products was tested in a genetic background lacking all other known thn genes by monitoring production of 1HT and 2HT from tetralin. To this end, different combinations of thnA genes were assembled in the broad-host-range vector pIZ1016, a derivative of pBBR1MCS-5 (13) constructed by cloning an NcoI-Sall fragment bearing the tac promoter and lacIq from pMM40 (16), which drives transcription of downstream genes in response to isopropyl-β-d-thiogalactopyranoside (IPTG) (Fig. 3A). A mutant derivative bearing a deletion of all known thn genes was also constructed. To this end, plasmid pIZ690, bearing a KIXX cassette flanked by DNA fragments from TFA that are outside of the thn cluster, was constructed (Fig. 3B). Transformation of strain TFA with pIZ690 resulted in isolation of the kanamycin-resistant strain T-690, which had a 12.2-kb deletion covering all identified thn genes, as confirmed by Southern hybridization (data not shown). Each pIZ1016 derivative bearing different thnA genes was transferred to the mutant T-690 by triparental matings with plasmid pRK2013 (6) as a donor of transfer functions.
FIG. 3.
(A) Schematic representation of thnA genes cloned in pIZ1016. (B) Scheme of plasmid pIZ690 showing recombination with homologous genomic regions of strain TFA to produce the 12.2-kb deletion covering the thn genes, which resulted in strain T-690.
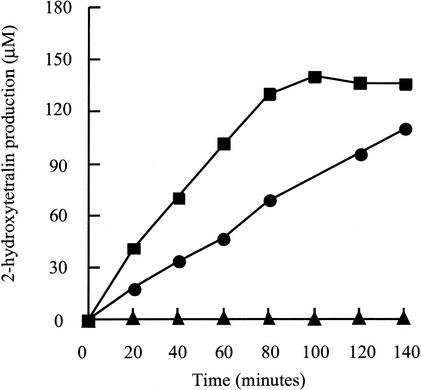
Production of 2-HT from tetralin by the resulting transconjugants was tested after induction with IPTG in a resting cell assay. To this end, induced transconjugants were concentrated to an optical density at 600 nm of 4 in 10 ml of minimal medium. After addition of 300 μM tetralin, samples of the supernatant at different time points were acidified, and the concentration of 2-HT was determined by HPLC from the area of the corresponding peak, by using a conversion coefficient at λ282 of 8.4 milli-absorbance units (mAU) · s μM−1, previously calculated with commercially available 2-HT. As shown in Fig. 4, strain T-690 producing only the components of the terminal oxygenase from pIZ683 did not produce 2-HT. When the ferredoxin ThnA3 was also present in T-690(pIZ685), significant production of 2-HT was observed. However, maximal 2-HT production was observed only when the four ThnA products were synthesized from pIZ693. These data confirm the strict requirement of ThnA1A2A3 for tetralin dioxygenase activity and, together with the characteristics of the ThnA4 mutant, show direct involvement of this reductase in the activity. The fact the ThnA4 is partially dispensable may be explained by functional complementation with other reductases of the host cell. Similar results have been reported for different multicomponent ring-hydroxylating dioxygenases (4, 14, 15, 24), which further supports the view that functional electron transport systems to the terminal oxidase can be reconstituted with components from different origins.
FIG. 4.
Tetralin dioxygenase activity of Sphingomonas macrogolitabida T-690 bearing the plasmid pIZ683 (thnA1A2; ▴), pIZ685 (thnA1A2A3; •), or pIZ693 (thnA1A2A3A4; ▪). Gene expression was induced by addition of 1 mM IPTG.
The results presented in this report allowed structural and functional characterization of the genes involved in the first two steps of tetralin biodegradation by Sphingomonas macrogolitabida strain TFA, leading to formation of an intermediate with a catecholic structure, 1,2-dihydroxytetralin (Fig. 5), whose metabolization through extradiol cleavage was reported previously (2, 8, 9), thus extending the characterization of the degradation pathway of this organic solvent.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to the DDBJ, EMBL, and GenBank nucleotide sequence databases and annotated as updates of the sequence under accession no. AF157565.
Acknowledgments
This work was supported by the Spanish Comisión Interministerial de Ciencia y Tecnología (grant BIO96-0908), by a fellowship of Fundación Cámara to E.M.-R., and by fellowships of the Spanish Ministerio de Educación to M.J.H. and O.M.-P.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andújar, E., M. J. Hernáez, S. R. Kaschabek, W. Reineke, and E. Santero. 2000. Identification of an extradiol dioxygenase involved in tetralin biodegradation: gene sequence analysis and purification and characterization of the gene product. J. Bacteriol. 182:789-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batie, C. J., D. P. Ballou, and C. C. Correll. 1991. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases, p. 546-566. In F. Müller (ed.), Chemistry and biochemistry of flavoenzymes, vol. 3. CRC Press, Boca Raton, Fla.
- 4.Bergeron, J., D. Ahmad, D. Barriault, A. Larose, and M. Sylvestre. 1994. Identification and mapping of the gene translation products invoved in the first steps of the Comamonas testosteroni B-356 biphenyl/chlorobiphenyl biodegradation pathway. Can. J. Microbiol. 40:743-753. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D. R., J. Blacker, B. Byrne, H. Dalton, M. Hand, S. C. Kelly, R. A. More O'Ferrall, S. Nagaraja Rao, N. D. Sharma, and G. N. Sheldrake. 1994. Acid-catalysed aromatization of benzene cis-1,2-dihydrodiols: a carbocation transition state poorly stabilized by resonance. J. Chem. Soc. Chem. Commun. 1994:313-314. [Google Scholar]
- 6.Figursky, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaydos, R. M. 1981. Naphthalene, p. 698-719. In M. Grayson and D. Eckroth (ed.), Kirk-Othmer encyclopedia of chemical technology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 8.Hernáez, M. J., E. Andújar, J. L. Ríos, S. R. Kaschabek, W. Reineke, and E. Santero. 2000. Identification of a serine hydrolase which cleaves the alicyclic ring of tetralin. J. Bacteriol. 182:5448-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernáez, M. J., B. Floriano, J. J. Ríos, and E. Santero. 2002. Identification of a hydratase and a class II aldolase involved in biodegradation of the organic solvent tetralin. Appl. Environ. Microbiol. 68:4841-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernáez, M. J., W. Reineke, and E. Santero. 1999. Genetic analysis of biodegradation of tetralin by a Sphingomonas strain. Appl. Environ. Microbiol. 65:1806-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Rasmaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 15:571-586. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1, demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 13.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 14.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin, V. J. J., and W. W. Mohn. 1999. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J. Bacteriol. 181:2675-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merrick, D., W. Paul, and M. J. Merrick. 1988. Construction of multicopy expression vectors for regulated overproduction of proteins in Klebsiella pneumoniae and other enteric bacteria. J. Gen. Microbiol. 134:1779-1784. [DOI] [PubMed] [Google Scholar]
- 17.Nam, J.-W., H. Nojiri, T. Yoshida, H. Habe, H. Yamane, and T. Omori. 2001. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 65:254-263. [DOI] [PubMed] [Google Scholar]
- 18.O'Connor, K. E., A. D. W. Dobson, and S. Hartmans. 1997. Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl. Environ. Microbiol. 63:4287-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romine, M., L. C. Stillwell, K.-K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 21.Schreiber, A. F., and U. K. Winkler. 1983. Transformation of tetralin by whole cells of Pseudomonas stutzeri AS39. Eur. J. Appl. Microbiol. Biotechnol. 18:6-10. [Google Scholar]
- 22.Sikkema, J., and J. A. M. de Bont. 1991. Isolation and initial characterization of bacteria growing on tetralin. Biodegradation 2:15-23. [Google Scholar]
- 23.Sikkema, J., and J. A. M. de Bont. 1993. Metabolism of tetralin (1,2,3,4-tetrahydronaphthalene) in Corynebacterium sp. strain C125. Appl. Environ. Microbiol. 59:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon, M. J., T. D. Osslund, R. Saunders, B. D. Ensley, S. Suggs, A. Harcourt, W.-C. Suen, D. L. Cruden, D. T. Gibson, and G. J. Zylstra. 1993. Sequencing of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB-9816-4. Gene 127:31-37. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yabuuchi, E. 2002. Correction of the connecting vowel and gender of the specific epithet in the name Sphingomonas macrogoltabidus Takeuchi et al. 1993 to Sphingomonas macrogolitabida. Int. J. Syst. E vol. Microbiol. 52:1039.. [DOI] [PubMed] [Google Scholar]
- 27.Yabuuchi, E., Y. Kosako, N. Fugiwara, T. Naka, I. Matsunaga, H. Ogura, and K. Kobayashi. 2002. Emendation of the genus Sphingomonas Yabuuchi et al. 1990 and junior objective synonymy of the species of three genera, Sphingobium, Novosphingobium and Sphingopyxis, in conjunction with Blastomonas ursincola. Int. J. Syst. E vol. Microbiol. 52:1485-1496. [DOI] [PubMed] [Google Scholar]