Abstract
Cleavage of the β-aryl ether linkage is the most important process in lignin degradation. Here we characterize the three tandemly located glutathione S-transferase (GST) genes, ligF, ligE, and ligG, from low-molecular-weight lignin-degrading Sphingomonas paucimobilis SYK-6, and we describe the actual roles of these genes in the β-aryl ether cleavage. Based on the identification of the reaction product by electrospray ionization-mass spectrometry, a model compound of β-aryl ether, α-(2-methoxyphenoxy)-β-hydroxypropiovanillone (MPHPV), was transformed by LigF or LigE to guaiacol and α-glutathionyl-β-hydroxypropiovanillone (GS-HPV). This result suggested that LigF and LigE catalyze the nucleophilic attack of glutathione on the carbon atom at the β position of MPHPV. High-pressure liquid chromatography-circular dichroism analysis indicated that LigF and LigE each attacked a different enantiomer of the racemic MPHPV preparation. The ligG gene product specifically catalyzed the elimination of glutathione from GS-HPV generated by the action of LigF. This reaction then produces an achiral compound, β-hydroxypropiovanillone, which is further degraded by this strain. Disruption of the ligF, ligE, and ligG genes in SYK-6 showed that ligF is essential to the degradation of one of the MPHPV enantiomers, and the alternative activities which metabolize the substrates of LigE and LigG are present in this strain.
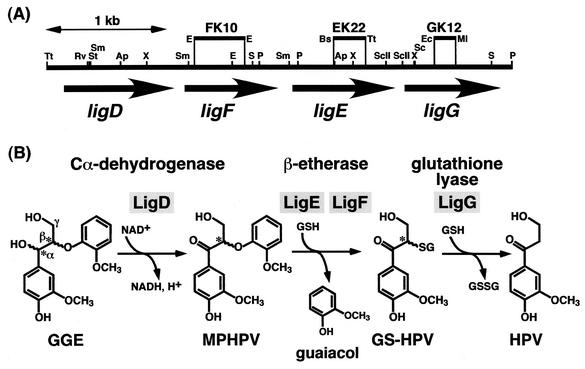
Lignin is a complex phenolic polymer with a variety of intermolecular linkages produced from hydroxycinnamyl alcohols by radical coupling reactions. Lignins are the most abundant aromatic material in nature, and their degradation by microorganisms is therefore an essential step for the carbon cycle on Earth. It is known that degradation of native lignin is initiated by the attack by lignin peroxidase, manganese peroxidase, and laccase secreted by white rot fungi (7). The resulting low-molecular-weight lignin is further degraded and mineralized by bacteria (27, 30). Lignin degradation is therefore accomplished by the cooperative actions of fungal and bacterial enzyme systems. Bacterial lignin degradation systems consist of a wide variety of reaction steps catalyzed by unique and specific enzymes able to cope with the complex structure of lignin. Sphingomonas paucimobilis SYK-6 is able to grow on a wide variety of dimeric lignin compounds with intermolecular linkages present in native lignin, including β-aryl ether, biphenyl, diarylpropane, and pinoresinol (9, 11-15, 20, 21). These compounds were degraded to vanillate or syringate, and they were further degraded via the protocatechuate 4,5-cleavage pathway (8, 13, 16, 17, 19). In the process of lignin degradation, the cleavage of β-aryl ether is the indispensable step, because this intermolecular linkage is the most abundant in lignin (50 to 70%) (1). We have already isolated the ligDFEG gene cluster of S. paucimobilis SYK-6, which is involved in the degradation of β-aryl ether (Fig. 1) (11-15). The ligD gene encoded the Cα-dehydrogenase (LigD), which oxidized a typical model compound of β-aryl ether, guaiacylglycerol-β-guaiacyl ether, to α-(2-methoxyphenoxy)-β-hydroxypropiovanillone (MPHPV). It has been thought that the ether linkage of MPHPV was reductively cleaved by the two kinds of β-etherases, the gene products of ligE (LigE) and ligF (LigF), to produce β-hydroxypropiovanillone (HPV) and guaiacol in the presence of glutathione (11, 12). HPV seemed to be degraded through side chain cleavage and the vanillate degradation pathway. The deduced amino acid sequences of LigE and LigF possessed up to 27% identity with eukaryotic glutathione S-transferases (GSTs), and the identity between LigE and LigF was only 18%. In addition to these genes, another GST gene, ligG, the deduced amino acid sequence of which had approximately 20% identity with those of both LigE and LigF, was found to be located just downstream of ligE (13). However, the ligG gene product (LigG) showed no β-etherase activity, and its role remained unknown. GSTs are well known in eukaryotes as the enzymes that catalyze the formation of the glutathione conjugates of a wide range of compounds. Based on studies of sequence similarity, GSTs have been classified into 11 categories (28). Information regarding bacterial GSTs is limited, although many proteobacteria contain large sets of GST genes with widely divergent sequences (28). Here, we clarify the concrete roles of the tandemly located GST genes of S. paucimobilis SYK-6 in β-aryl ether cleavage.
FIG. 1.
Organization of the ligDFEG gene cluster (A) and deduced functions of the gene products in the β-aryl ether cleavage (B). (A) The ligD, ligF, ligE, and ligG genes are indicated by the thick arrows. Vertical bars above the restriction map indicate the positions of the Kmr gene insertions of ligF (FK10), ligE (EK22), and ligG (GK12) mutants. Abbreviations for restriction enzymes: Ap, ApaI; Bs, BstXI; E, EcoRI; Ec, Eco47III; Ml, MluI; P, PstI; RV, EcoRV; S, SalI; Sc, SacI; ScII, SacII; St, StuI; Sm, SmaI; Tt, Tth111I; and X, XhoI. (B) GGE, guaiacylglycerol-β-guaiacyl ether; GSH, glutathione; GSSG, glutathione disulfide. Asterisks indicate the asymmetric carbons.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. S. paucimobilis SYK-6 was grown on Luria-Bertani (LB) medium (Bacto Tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter) or W minimal salt medium (20) containing 10 mM vanillate at 30°C. The SYK-6 ligF, ligE, or ligG insertion mutants were grown on LB medium containing 50 mg of kanamycin per liter.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| S. paucimobilis | ||
| SYK-6 | Wild type; Nalr Smr | 9 |
| FK10 | Mutant derivative of SYK-6; Kmr gene insertion mutant of ligF; Nalr Smr Kmr | This study |
| EK22 | Mutant derivative of SYK-6; Kmr gene insertion mutant of ligE; Nalr Smr Kmr | This study |
| GK12 | Mutant derivative of SYK-6; Kmr gene insertion mutant of ligG; Nalr Smr Kmr | This study |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thiΔ(lac-proAB) F′ [traD36 proAB+ lacIqlacZΔM15] | 29 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3); T7 RNA polymerase gene under the control of the lacUV5 promoter | 25 |
| Plasmids | ||
| pUC19 | Cloning vector; Apr | 29 |
| pBluescript II KS(+) | Cloning vector; Apr | 24 |
| pET21(+) | Expression vector; Apr T7 promoter | Novagen |
| pUC4K | Apr Kmr | 26 |
| pK19mobsacB | oriT sacB Kmr | 23 |
| pBE10 | pVK100 with an approximately 24-kb fragment carrying ligDFEG | 11 |
| pUBX77 | pUC19 with a 1.2-kb fragment carrying ligF | 12 |
| pETF48 | pET21(+) carrying the insert of pUBX77 | This study |
| pUBE13 | pUC19 with a 2.0-kb SalI fragment carrying ligE | 11 |
| pETE10 | pET21(+) with a 1.0-kb SmaI-SacI fragment of pUBE13 | This study |
| pUCP18 | pUC19 with a 1.8-kb PstI fragment carrying ligG | This study |
| pETG9 | pET21(+) with a 0.9-kb SacII-PstI fragment of pUCP18 | This study |
| pBSEM3 | KS(+) with a 3.0-kb EcoRV-MluI fragment carrying ligFE | This study |
| pEMFK04 | pBSEM3 with an insertion of the Kmr gene of pUC4K replacing 0.3- and 0.1-kb EcoRI fragments | This study |
| pMSF37 | pK19mobsacB with a 3.7-kb StuI-Eco47III fragment of pEMFK04 | This study |
| pEMEK03 | pBSEM3 with an insertion of the Kmr gene of pUC4K replacing a 0.3-kb BstXI-Tth111I fragment | This study |
| pMSE38 | pK19mobsacB with a 3.8-kb StuI-Eco47III fragment of pEMEK03 | This study |
| pPGK02 | pUCP18 with an insertion of the Kmr gene of pUC4K replacing a 0.2-kb Eco47III-MluI fragment | This study |
| pMSG28 | pK19mobsacB with a 2.8-kb PstI fragment of pPGK02 | This study |
Abbreviations: Nalr, Smr, Kmr, and Apr, resistance to nalidixic acid, streptomycin, kanamycin, and ampicillin, respectively.
Preparation of substrates.
MPHPV was synthesized chemically according to the method of Adler and Eriksoo (2). HPV and α-O-(β-methylumbelliferyl)acetovanillone (MUAV) were prepared in a previous study (14). The purity of these substrates was estimated to be more than 99% by gas chromatography-mass spectrometry (MS) analysis. Guaiacol was purchased from Tokyo Kasei Kogyo Co. (Tokyo, Japan).
Enzyme assay.
The etherase activities of LigF and LigE toward the fluorescent substrate MUAV were fluorometrically assayed. The 2-ml reaction mixture contained 30 μM MUAV, 1 mM glutathione, and appropriate concentrations of the enzyme in 20 mM Tris-HCl (pH 7.5) (buffer A), and the reaction was carried out at 25°C. Formation of 4-methylumbelliferone (4MU) in the LigF reaction mixture was monitored in a 10-s continuous assay by using an RF-1500 fluorometric analyzer (Shimadzu, Kyoto, Japan). In the case of the LigE reaction, 100 μl of the reaction mixture was taken at 5 min and added to 1.9 ml of 100 mM glycine-NaOH buffer (pH 10.0), and formation of 4MU was determined with the RF-1500 analyzer. One unit of the enzyme was defined as the amount that released 1 μmol of 4MU/min from the substrate. Specific activity was expressed as units per milligram of protein.
The etherase activities of LigF and LigE toward MPHPV were determined by measuring the decrease in substrate by high-pressure liquid chromatography (HPLC) analysis. The 1-ml assay mixture contained buffer A, 50 μM MPHPV, 1 mM glutathione, and the cell extract of E. coli BL21(DE3) harboring pETF48 (100 μg of protein/ml) or pETE10 (1 mg of protein/ml). Reactions were carried out at 25°C and stopped by the addition of methanol (final concentration, 25%) at 15 s for the LigF reaction and at 5 min for the LigE reaction. Precipitated protein was removed by centrifugation (15,000 × g for 10 min), and the supernatant was analyzed with an Alliance 2690 Separations Module HPLC system (Waters, Milford, Mass.) equipped with a TSKgel ODS-80TM column (6 by 150 mm; Tosoh, Tokyo, Japan). The mobile phase was a mixture of water (49.5%), acetonitrile (49.5%), and phosphate (1.0%), and the flow rate was 1.0 ml/min. Compounds were detected at 280 or 310 nm. One unit of the enzyme was defined as the amount that degrades 1 μmol of MPHPV/min. Specific activity was expressed as units per milligram of protein.
GST activities of LigF toward 1-chloro-2,4-dinitrobenzene (1 mM), p-nitrobenzyl chloride (1 mM), and 1,2-epoxy-3-p-nitrophenoxypropane (0.5 mM) were assayed by incubating LigF (10 μg of protein/ml) with the substrates in a 2-ml reaction mixture containing 20 mM potassium phosphate buffer (pH 6.5) and 1 mM glutathione at 25°C. Each activity was monitored by the increase in absorbance at 340, 310, and 360 nm derived from the production of glutathione conjugates of 1-chloro-2,4-dinitrobenzene, p-nitrobenzyl chloride, and 1,2-epoxy-3-p-nitrophenoxypropane, respectively, with a DU-7500 spectrophotometer (Beckman, Fullerton, Calif.).
Preparation of cell extract and LigF purification.
Cells of E. coli BL21(DE3) transformants harboring pETF48, pETE10, and pETG9, carrying ligF (DDBJ accession no. D11473), ligE (DDBJ accession no. D11473), and ligG (DDBJ accession no. AB026292), respectively, were grown in 200 ml of LB medium containing 100 mg of ampicillin/liter at 37°C. The expression of the genes was induced for 4 to 5 h by adding isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) when the turbidity of the culture at 660 nm reached 0.5. Cells were harvested by centrifugation and sonicated in buffer A. The cell lysate was centrifuged at 15,000 × g for 15 min. The supernatant was then used as a crude enzyme. For purification of LigF, streptomycin (final concentration, 1%) was added to the supernatant, and it was centrifuged at 100,000 × g for 60 min. Enzyme purification was performed according to the method described below by using a BioCAD700E apparatus (PerSeptive Biosystems, Framingham, Mass.). The crude LigF enzyme was applied to a POROS HQ (quaternized polyethyleneimine) column (4.6 by 100 mm) (PerSeptive Biosystems) previously equilibrated with buffer A containing 100 mM NaCl. The enzyme was eluted with the same buffer, and fractions containing etherase activity toward MUAV were pooled. After these fractions were concentrated and desalted, ammonium sulfate was added to the enzyme solution to a final concentration of 2 M. The enzyme solution was centrifuged at 3,000 × g for 10 min, and the supernatant was applied to a POROS PE (phenyl ether) column (4.6 by 100 mm) (PerSeptive Biosystems) equilibrated with buffer A containing 2 M ammonium sulfate. The enzyme was eluted with 25 ml of a linear gradient of 2.0 to 0 M ammonium sulfate. The fractions containing etherase activity that eluted at 0.93 M were collected. Glycerol was added to a final concentration of 20%, and the purified enzyme was stored at −80°C until used.
Analysis of the reaction product.
The 10-ml assay mixture contained buffer A, 50 μM MPHPV, 100 μM glutathione, and the purified LigF (10 μg of protein/ml) or the cell extract of E. coli BL21(DE3) harboring pETE10 (100 μg of protein/ml). Reactions were carried out at 25°C and were stopped by the addition of methanol (final concentration, 25%) at 10 min for the LigF reaction and at 60 min for the LigE reaction. Precipitated protein was removed by centrifugation (15,000 × g for 10 min), and the supernatant was analyzed with the HPLC system by the method described above and by electrospray ionization (ESI)-MS (HP1100 series LC-MSD; Hewlett-Packard Co., Palo Alto, Calif.). In the analysis by ESI-MS, mass spectra were obtained by negative-mode ESI, with a needle voltage of −3.5 kV and a source temperature of 330°C. The mobile phase was a mixture of water (90%) and methanol (10%), and the flow rate was 0.25 ml/min.
To determine the enantioselectivities of LigF and LigE toward MPHPV enantiomers, 50 μM MPHPV was incubated with the purified LigF (10 μg of protein/ml) in a 10-ml reaction mixture containing buffer A and 1 mM glutathione at 25°C. A portion of the reaction mixture (1 ml) was taken for HPLC analysis, and conversion of 50% of MPHPV to α-glutathionyl-HPV (GS-HPV) was confirmed by HPLC analysis. After the LigF reaction, crude LigE enzyme (100 μg of protein/ml) was added to this reaction mixture (total volume, 9 ml) and incubated for 60 min at 25°C. The reaction mixture was then analyzed by HPLC.
To analyze which enantiomer of MPHPV was degraded by LigF, the MPHPV preparation remaining after the reaction with LigF was analyzed with an HPLC system (Jasco Corp., Tokyo, Japan) equipped with a CHIRAL OD column (4.6 by 250 mm; Daicel Chemical Industries, Tokyo, Japan). The 5-μg MPHPV preparation was dissolved in tetrahydrofuran and separated on this HPLC system. The mobile phase was a mixture of hexane (74.5%), ethanol (24.5%), and acetic acid (1.0%), and the flow rate was 0.5 ml/min. MPHPV enantiomers were detected at 300 nm by a CD-2095 detector (Jasco). The MPHPV preparation (100 μM) was incubated with the cell extract of E. coli BL21(DE3) harboring pETF48 carrying ligF (100 μg of protein/ml) in a 100-ml reaction mixture containing buffer A and 1 mM glutathione at 25°C. The reaction mixture was acidified with hydrochloric acid, extracted with ethyl acetate, and separated by thin-layer chromatography with chloroform-ethyl acetate-formic acid (10:8:1), and then the spot of MPHPV was cut out and extracted with ethyl acetate. The sample (approximately 3 μg) was finally dissolved in tetrahydrofuran and separated on a chiral HPLC-circular dichroism (CD) system.
For detection of LigG activity, GS-HPV I and II were prepared in a 10-ml reaction mixture containing buffer A, 50 μM MPHPV, 1 mM glutathione, and the purified LigF (10 μg of protein/ml) or the cell extract of E. coli BL21(DE3) harboring pETE10 (100 μg of protein/ml), respectively, at 25°C. A portion of the reaction mixture (1 ml) was taken for HPLC analysis to confirm that half of the MPHPV preparation was transformed to GS-HPV. The mixtures containing GS-HPV I or II were incubated with the cell extract of E. coli BL21(DE3) harboring pETG9 (10 μg of protein/ml) for 10 min or with the same extract (500 μg protein/ml) for 60 min, respectively, in a 9-ml reaction mixture at 25°C. These mixtures were analyzed by HPLC as described above. HPV generated in these reactions was identified by comparing the retention time and the UV-visible spectrum with those of the authentic HPV.
Analytical methods.
The protein concentration was determined by the method of Bradford (6). The expression of the genes was examined by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) (10). The molecular mass of the native LigF was determined by Superdex 200 HR10/30 (Pharmacia Biotech, Milwaukee, Wis.) gel filtration column chromatography with BioCAD700E. Elution was performed with 50 mM potassium phosphate buffer (pH 7.5) containing 0.15 M NaCl at a flow rate of 0.8 ml/min. The molecular weight was estimated on the basis of the calibration curve of reference proteins. The isoelectric point of LigF was determined by isoelectric focusing with an Ampholine PAG plate (pH 3.5 to 9.5) (Pharmacia Biotech), using a model Mutiphor II electrophoresis system (Pharmacia Biotech). Gas chromatography-MS analysis was performed as described previously (17).
Construction of insertion mutants of S. paucimobilis SYK-6.
DNA manipulations were carried out essentially as described previously (4, 22). Construction of each gene-disrupted plasmid was carried out as follows. The 0.3- and 0.1-kb EcoRI fragments were replaced with the 1.3-kb kanamycin resistance (Kmr) gene of pUC4K in the 3.0-kb EcoRV-MluI fragment carrying ligFE in order to disrupt ligF in plasmid pBSEM3. The 3.7-kb ligF-inactivated fragment of the resulting plasmid was cloned into pK19mobsacB (23) to construct pMSF37. The 0.3-kb BstXI-Tth111I fragment was replaced with the Kmr gene in pBSEM3 to disrupt ligE. The 3.8-kb ligE-inactivated fragment was cloned in pK19mobsacB to construct pMSE38. The 0.2-kb Eco47III-MluI fragment was replaced with the Kmr gene to disrupt ligG in the 1.8-kb PstI fragment in pUCP18. The 2.8-kb ligG-inactivated fragment of the resulting plasmid was cloned in pK19mobsacB to construct pMSG28. These plasmids were introduced into SYK-6 by electroporation. The selection of each gene-disrupted mutant was made as described previously (17). To examine the disruption of each gene, Southern hybridization analysis was performed. The total DNAs of candidates for the ligF and ligE mutants and for the ligG mutant were digested with EcoRV and MluI and with SalI, respectively. The 1.3-kb SalI fragment carrying the Kmr gene, the 1.7-kb XhoI fragment carrying ligF and part of ligE, and the 0.6-kb SacI-SalI fragment carrying part of ligG were labeled with the digoxigenin system (Roche Molecular Biochemicals, Mannheim, Germany) and used as probes.
Cell extracts of SYK-6 and its mutants were prepared from cells grown on LB medium and on the same medium containing 50 mg of kanamycin per liter, respectively, by the method described above. Each cell extract (1 mg of protein/ml) was incubated with 50 μM MPHPV at 25°C in a 10-ml reaction mixture containing buffer A and 2 mM glutathione and was analyzed by HPLC.
RESULTS AND DISCUSSION
Reaction of the β-aryl ether cleavage catalyzed by LigF and LigE.
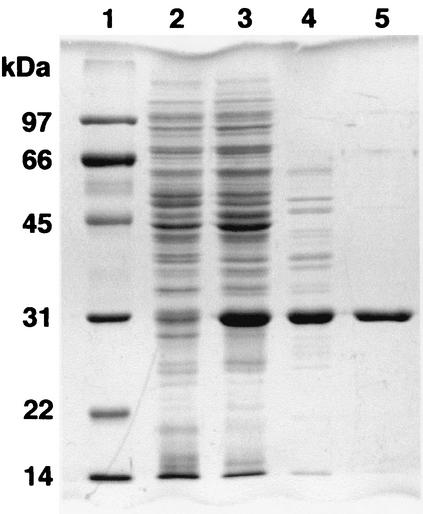
The ligF gene was expressed in E. coli BL21(DE3) harboring pETF48. A 30-kDa protein was observed by SDS-PAGE, and this protein was purified to near homogeneity and approximately 11-fold with a recovery of 57%, using a series of quarternized polyethyleneimine and phenyl ether column chromatography procedures (Table 2 and Fig. 2). LigF was estimated to be a homotetramer (128 kDa) by gel filtration column chromatography. When the fluorescent β-etherase assay substrate MUAV was used, the optimum pH and temperature were estimated to be 7.5 and 15°C, respectively. The isoelectric point of LigF was 5.2. LigF showed no GST activities toward 1-chloro-2,4-dinitrobenzene, p-nitrobenzyl chloride, or 1,2-epoxy-3-p-nitrophenoxypropane, which are used as the general substrates for GSTs.
TABLE 2.
Purification of LigF from E. coli BL21(DE3) harboring pETF48
| Fraction | Total protein (mg) | Total activity (mU) | Sp act (mU/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 50.0 | 1,700 | 33.9 | 100 | 1.0 |
| HQ | 6.14 | 1,380 | 225 | 81 | 6.6 |
| PE | 2.66 | 966 | 363 | 57 | 11 |
FIG. 2.
SDS-PAGE analysis of protein fractions. Proteins were separated on an SDS-12% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: 1, molecular mass markers; 2, crude extract of E. coli BL21(DE3) harboring pET21(+) (10 μg of protein); 3, cell extract of E. coli BL21(DE3) harboring pETF48 (10 μg of protein); 4, HQ fraction (3 μg of protein); 5, PE fraction (1.5 μg of protein). Molecular masses are given on the left.
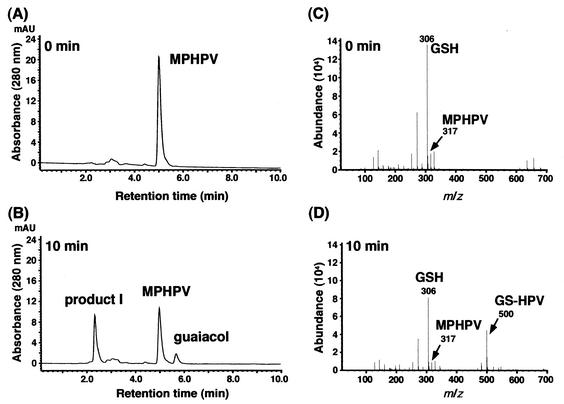
To address the reaction mechanism of β-aryl ether cleavage by LigF, the standard model β-aryl ether compound MPHPV was used for the following experiments. MPHPV was incubated with the purified LigF, and the reaction products were analyzed by HPLC and ESI-MS. HPLC analysis showed the accumulation of guaiacol with a retention time of 5.70 min and of an unknown product with a retention time of 2.35 min from MPHPV (Fig. 3B). This result was observed only in the presence of glutathione. ESI-MS indicated the generation of a fragment at m/z 500, which corresponds to the mass of the deprotonated molecular ion of the glutathione conjugate of HPV, GS-HPV, as illustrated in Fig. 1. Thus, this result strongly suggested that LigF catalyzed the nucleophilic attack of glutathione to the carbon atom at the β position (Cβ) of MPHPV. Interestingly, only 50% of the MPHPV was strictly transformed by LigF, even if an excess amount of LigF was incubated with MPHPV for 6 h. As the Cβ of MPHPV is the asymmetric carbon, the MPHPV preparation used in this study was estimated to contain two enantiomers. Therefore, one of the enantiomers of MPHPV appeared to be degraded by LigF. On the other hand, ligE was expressed in E. coli BL21(DE3) harboring pETE10, and a 31-kDa protein was observed by SDS-PAGE. The reaction product of MPHPV incubated with the crude LigE enzyme was analyzed by HPLC and ESI-MS. The generation of deprotonated ion at m/z 500 was observed, and one-half of MPHPV was transformed in the presence of glutathione (data not shown). These results suggested that LigE is also a GST that catalyzes the nucleophilic attack of glutathione at the Cβ of one of the MPHPV enantiomers. The specific activity of the crude LigF enzyme toward MPHPV (1 U/mg) was approximately 170 times higher than that of the crude LigE enzyme (6 mU/mg). On the other hand, the specific activity of LigF for MUAV was also more than 1,000 times higher than that of LigE. Significantly higher etherase activity of LigF was suggested, but the expression of ligE was lower than that of ligF. Purification of LigE is necessary to compare the actual specific activities of LigF and LigE.
FIG. 3.
Identification of the reaction products from MPHPV catalyzed by LigF. Purified LigF was incubated with the MPHPV preparation in the presence of glutathione. (A and B) HPLC chromatograms at 0 and 10 min of incubation, respectively. Compounds were detected at 280 nm. (C and D) ESI-MS spectra of the reaction mixtures at 0 and 10 min of incubation, respectively. GSH, glutathione; AU, absorbance units.
Enantioselectivity of LigF and LigE.
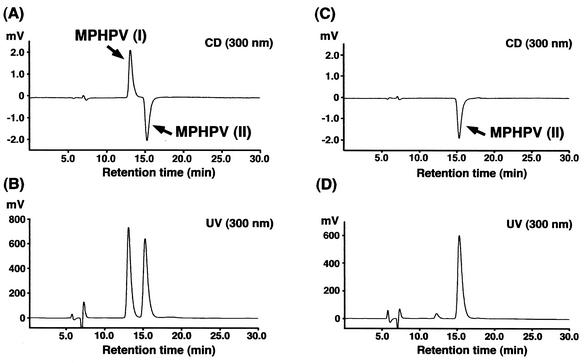
It has been assumed that native lignin is optically inactive, and the presence of racemic forms of arylglycerol-β-aryl ether in lignin was recently indicated (3). In order to investigate the enantioselectivity of LigF and LigE, after the incubation of MPHPV with purified LigF in the presence of glutathione, crude LigE enzyme was added to a reaction mixture. In these reactions, HPLC analysis indicated that all of the MPHPV preparation was completely converted to GS-HPV, and it thus seemed that LigF and LigE attacked different enantiomers of MPHPV. A chiral HPLC-CD system was employed to confirm this hypothesis. The MPHPV preparation was found to contain equimolar amounts of two enantiomers, based on the peak area calculations (Fig. 4A and B). We identified these enantiomers, which were eluted at 13.1 and 15.2 min, as MPHPV I and II, respectively. After incubation of the MPHPV preparation with LigF, the reaction mixture was extracted with ethyl acetate and separated by thin-layer chromatography. The spot of remaining MPHPV was collected, extracted, and analyzed with the chiral HPLC-CD system. The elution profile showed that only the peak of MPHPV II remained, indicating that LigF specifically attacked MPHPV I (Fig. 4C and D). Thus, we concluded that LigF and LigE are GSTs that specifically attack MPHPV I and II, respectively.
FIG. 4.
Chiral HPLC-CD analysis of LigF enantioselectivity. (A and B) The MPHPV preparation was separated on a chiral column by HPLC and was detected with CD (A) and UV (B) detectors. (C and D) After incubation of the MPHPV preparation with LigF, the remaining MPHPV was analyzed by HPLC and detected with CD (C) and UV (D) detectors.
Disruption of ligF, ligE, and ligG in S. paucimobilis SYK-6.
In order to determine the actual roles of ligE and ligF and to obtain insight into the role of ligG in the degradation of β-aryl ether compounds, each of these genes in SYK-6 was disrupted. Gene inactivation was carried out by using the ligE, ligF, and ligG disruption plasmids pMSE38, pMSF37, and pMSG28, which were constructed by replacing the internal segment of each gene in pK19mobsacB by a Kmr gene (Fig. 1). The ligE, ligF, and ligG mutations were confirmed by Southern hybridization analysis with the probes described in Materials and Methods (Fig. 5A). While almost all of the MPHPV preparation was degraded by the cell extract of the wild-type strain in the presence of glutathione, the cell extract of the ligF insertion mutant was unable to degrade half of the MPHPV preparation (Fig. 5B). All of the remaining MPHPV was degraded by the purified LigF, indicating that ligF is essential to the degradation of MPHPV I. On the other hand, the inactivation of ligE had only a slight effect on the MPHPV degradation (Fig. 5B). This result indicated that ligE is not essential to MPHPV II degradation and that at least one alternative GST seems to be involved in MPHPV II degradation, since degradation of MPHPV by SYK-6 depended on the presence of glutathione. Disruption of ligG did not affect the rate of MPHPV transformation. However, a trace amount of GS-HPV was detected (data not shown), suggesting the involvement of LigG in the transformation of GS-HPV.
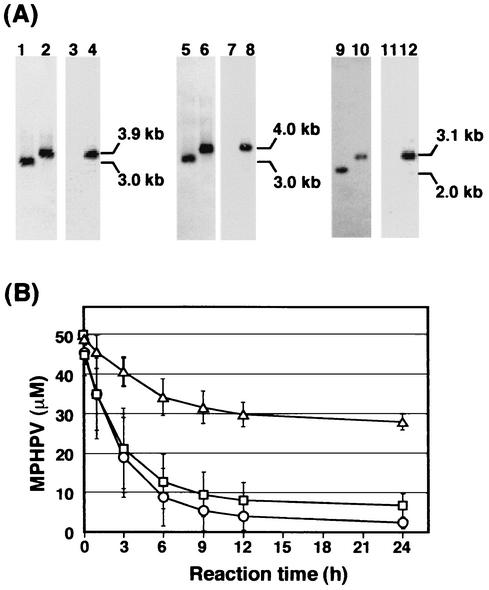
FIG. 5.
Disruption of ligF, ligE, and ligG in S. paucimobilis SYK-6. (A) Southern hybridization analysis of the insertion mutants. Lanes: 1, 3, 5, and 7, total DNA of SYK-6 digested with EcoRV and MluI; 2 and 4, total DNA of the ligF insertion mutant (FK10) digested with EcoRV and MluI; 6 and 8, total DNA of the ligE insertion mutant (EK22) digested with EcoRV and MluI; 9 and 11, total DNA of SYK-6 digested with SalI; and 10 and 12, total DNA of the ligG insertion mutant (GK12) digested with SalI. The 1.3-kb SalI fragment carrying the Kmr gene (lanes 3, 4, 7, 8, 11, and 12), the 1.7-kb XhoI fragment carrying ligF and part of ligE (lanes 1, 2, 5, and 6), and the 0.6-kb SacI-SalI fragment carrying part of ligG (lanes 9 and 10) were used as probes. (B) Degradation of MPHPV by cell extracts of SYK-6 (circles), FK10 (triangles), and EK22 (squares) in the presence of glutathione. The rate of transformation of MPHPV by the cell extract of GK12 was at the same level as that for SYK-6. Error bars indicate standard deviations.
ligG encodes glutathione lyase.
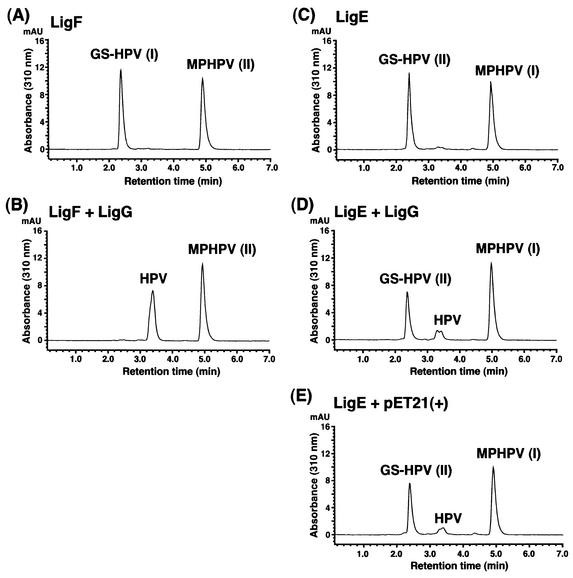
To estimate the function of LigG, the ligG gene was expressed in E. coli BL21(DE3) harboring pETG9. SDS-PAGE analysis indicated the expression of a 32-kDa protein. GS-HPV I and II were prepared from the MPHPV preparation with the purified LigF and the crude LigE enzyme, respectively, in the presence of glutathione (Fig. 6A and C). When the crude LigG enzyme (10 μg of protein/ml) was incubated with GS-HPV I and II, LigG converted only GS-HPV I into HPV (Fig. 6B). When 500 μg of protein of crude LigG enzyme per ml was used, a small amount of HPV was generated from GS-HPV II (Fig. 6D). However, the cell extract of E. coli BL21(DE3) harboring pET21(+) (500 μg of protein per ml) also produced a similar amount of HPV from GS-HPV II as did LigG (Fig. 6E). These results suggested that ligG encodes a glutathione lyase for GS-HPV I. The deduced amino acid sequence of ligG revealed 20% identity with the omega class GSTs. GSTO 1-1 from humans in this class has glutathione-dependent thiol transferase and dehydroascorbate reductase activities that are not associated with other human GSTs (5). An active-site Cys-32 of GSTO 1-1 can form a disulfide bond with glutathione, and Pro-33 is thought to promote the positioning of Cys-32 thiol for the stabilization of the thiolate form. The presence of the Cys-15-Pro-16 sequence at the N-terminal end of LigG suggested a functional relationship between LigG and omega class GSTs.
FIG. 6.
Detection of the glutathione lyase activity of LigG. (A and C) HPLC chromatograms of the substrates, GS-HPV I and GS-HPV II, generated from the MPHPV preparation by the actions of LigF and LigE enzymes, respectively, in the presence of glutathione. (B) HPLC chromatogram of the reaction product of GS-HPV I incubated for 10 min with the cell extract of E. coli BL21(DE3) harboring pETG9 (10 μg of protein/ml). (D and E) HPLC chromatograms of the reaction products of GS-HPV II incubated for 60 min with the cell extract of E. coli BL21(DE3) harboring pETG9 (500 μg of protein/ml) and with the cell extract of E. coli BL21(DE3) harboring pET21(+) (500 μg of protein/ml), respectively. AU, absorbance units.
Conclusions.
We uncovered the actual functions of the three tandemly localized GST genes, ligFEG, in SYK-6. LigF and LigE are the enantioselective GSTs that cleave the β-aryl ether linkage of one of the different enantiomers of β-aryl ether compounds, and LigG is the glutathione lyase for the glutathione conjugate produced by LigF. These results lead to the reaction sequences of the degradation of β-aryl ether compounds by SYK-6 shown in Fig. 1. In the case of the tetrachlorohydroquinone dehalogenase (PcpC) (18), which is a GST of a pentachlorophenol degrader, Sphingobium chlorophenolicum ATCC 39723, this enzyme catalyzes both the glutathione conjugation to generate 2,3,5-trichloro-6-glutathionylhydroquinone from tetrachlorohydroquinone and the elimination of glutathione from 2,3,5-trichloro-6-glutathionylhydroquinone to give trichlrohydroquinone. Because of the deficiency of the glutathione lyase activity in LigF, glutathione lyase encoded by ligG should be required in the gene cluster for the degradation of β-aryl ether compounds.
Among the three GSTs characterized here, only ligF was essential to MPHPV degradation. LigE and LigG were able to attack MPHPV II and GS-HPV I, respectively, but these compounds were metabolized by the alternative activities in SYK-6. It is most likely that an additional set of enantioselective GSTs are involved in these reaction steps. Isolation of the enzyme gene(s) involved in MPHPV II degradation is under way in our laboratory.
Acknowledgments
We thank M. Nakamura for providing the authentic MPHPV and JASCO Corporation for performing the chiral HPLC-CD analysis.
REFERENCES
- 1.Adler, E. 1957. Structural elements of lignin. Ind. Eng. Chem. 49:1377-1383. [Google Scholar]
- 2.Adler, E., and E. Eriksoo. 1955. Guaiacylglycerol and its β-guaiacyl ether. Acta Chem. Scand. 9:341-342. [Google Scholar]
- 3.Akiyama, T., K. Magara, Y. Matsumoto, G. Meshitsuka, A. Ishizu, and K. Lundquist. 2000. Proof of the presence of racemic forms of arylglycerol-β-aryl ether structure in lignin: studies on the stereo structure of lignin by ozonation. J. Wood Sci. 46:414-415. [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Board, P. G., M. Coggan, G. Chelvanayagam, S. Easteal, L. S. Jermiin, G. K. Schulte, D. E. Danley, L. R. Hoth, M. C. Griffor, A. V. Kamath, M. H. Rosner, B. A. Chrunyk, D. E. Perregaux, C. A. Gabel, K. F. Geoghegan, and J. Pandit. 2000. Identification, characterization, and crystal structure of the omega class glutathione transferases. J. Biol. Chem. 275:24798-24806. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Gold, M. H., and M. Alic. 1993. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol. Rev. 57:605-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katayama, Y., S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77-79. [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Masai, E., Y. Katayama, S. Kawai, S. Nishikawa, M. Yamasaki, and N. Morohoshi. 1991. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J. Bacteriol. 173:7950-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masai, E., Y. Katayama, S. Kubota, S. Kawai, M. Yamasaki, and N. Morohoshi. 1993. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 323:135-140. [DOI] [PubMed] [Google Scholar]
- 13.Masai, E., Y. Katayama, S. Nishikawa, and M. Fukuda. 1999. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J. Ind. Microbiol. Biotechnol. 23:364-373. [DOI] [PubMed] [Google Scholar]
- 14.Masai, E., Y. Katayama, S. Nishikawa, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1989. Detection and localization of a new enzyme catalyzing the β-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 249:348-352. [DOI] [PubMed] [Google Scholar]
- 15.Masai, E., S. Kubota. Y. Katayama, S. Kawai, M. Yamasaki, and N. Morohoshi. 1993. Characterization of the Cα-dehydrogenase gene involved in the cleavage of β-aryl ether by Pseudomonas paucimobilis. Biosci. Biotechnol. Biochem. 57:1655-1659. [DOI] [PubMed] [Google Scholar]
- 16.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy, D. L., S. Navarrete, W. S. Willett, P. C. Babbitt, and S. D. Copley. 1996. Exploration of the relationship between tetrachlorohydroquinone dehalogenase and the glutathione S-transferase superfamily. Biochemistry 35:14634-14642. [DOI] [PubMed] [Google Scholar]
- 19.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng, X., E. Masai, Y. Katayama, and M. Fukuda. 1999. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 65:2789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 24.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 26.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicuña, R. 1988. Bacterial degradation of lignin. Enzyme Microbiol. Technol. 10:646-655. [Google Scholar]
- 28.Vuilleumier, S., and M. Pagni. 2002. The elusive roles of bacterial glutathione S-transferases: new lesson from genomes. Appl. Microbiol. Biotechnol. 58:138-146. [DOI] [PubMed] [Google Scholar]
- 29.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann, W. 1990. Degradation of lignin by bacteria. J. Biotechnol. 13:119-130. [Google Scholar]