Abstract
We have used an Escherichia coli K-12 whole-genome array based on the DNA sequence of strain MG1655 as a tool to identify deletions in another E. coli K-12 strain, MC4100, by probing the array with labeled chromosomal DNA. Despite the continued widespread use of MC4100 as an experimental system, the specific genetic relationship of this strain to the sequenced K-12 derivative MG1655 has not been resolved. MC4100 was found to contain four deletions, ranging from 1 to 97 kb in size. The exact nature of three of the deletions was previously unresolved, and the fourth deletion was altogether unknown.
Whole-genome arrays have provided a powerful tool in biology. While work with gene arrays has provided an important tool in determining genome-wide RNA expression levels, arrays have also found other varied uses. Arrays have been used to trace the progression of DNA replication forks (11), to track the genetic adaptation of Escherichia coli to high temperature (18), as an assay for comparative genomic studies (9), and as a tool in conjunction with transposon mutagenesis for determining conditionally essential genes in bacteria (4, 20). We have explored the use of a commercially available genome array based on the E. coli K-12 MG1655 genome as a tool for mapping the deletion endpoints in a commonly used laboratory E. coli K-12 strain, MC4100. Our rationale was that this procedure could provide a set of putative deletions that could then be confirmed with PCR analysis to reconstruct the chromosome by comparison with the MG1655 genome sequence.
The original E. coli K-12 strain was isolated in 1922 (3). A K-12 derivative called MG1655, which was cured of an endogenous lambda phage by UV induction and cured of its conjugal plasmid by growth in the presence of acridine orange, was sequenced and is the basis of a commercially available whole-genome array (7). The MG1655 derivative MC4100 was constructed over 25 years ago when it was used to isolate gene and protein fusions to the lacZ gene product, β-galactosidase, through the use of bacteriophage derivatives (8). MC4100 provided an important host in early gene expression work (reviewed in reference 5). We chose to use one of the newest tools designed for genome-wide gene expression analysis to analyze this strain used in some of the earliest expression analysis experiments. Well beyond providing a proof-in-principle for mapping deletion endpoints, the accurate description of MC4100 is important for the modern understanding of E. coli. For largely historical reasons, MC4100 has been the strain background of choice for many genetic experiments. The genetic nature of MC4100 will continue to be important in relating work with this strain to the sequenced MG1655 K-12 strain. Additionally, the genetic nature of MC4100 will help investigators to decide if MC4100 is an appropriate strain for a given experiment.
The original K-12 strain went through numerous alterations, including X-ray, UV, and chemical mutagenesis, as well as being a genetic recipient in multiple crosses with various E. coli K-12 and E. coli B derivatives to generate the strain MC4100. While the physical analysis of the chromosome by pulsed-field gel electrophoresis identified three deletions, the actual extent of these deletions has remained unclear (10, 14-16).
We used the E. coli whole-genome array to identify the deletion endpoints in a related strain derivative.
The Sigma/Genosys Panorama E. coli array consists of PCR-amplified DNA products corresponding to the 4,290 open reading frames of strain MG1655 applied as duplicate 10-ng spots on a nylon membrane. Our strategy was to identify deletions in a non-MG1655 E. coli strain by isolating and radioactively labeling chromosomal DNA from MC4100 and probing the E. coli MG1655 gene array. As described below, most genes were found to be present based on a ratio of the intensities of 1 (MC4100 intensity/MG1655 intensity). Low ratios indicate putative deletions that could be confirmed by PCR-mediated sequence analysis of the region.
To study MC4100 [F− araD139Δ(argF-lac)U169 rspL150 relA1 flbB5301 fruA25 deoC1 ptsF25], we used a valine-resistant derivative called NLC28 (12). Given the near-total identity of the strains, the derivative is referred to as MC4100 throughout the text. The MC4100 strain was subsequently obtained from the E. coli genetic stock center, and PCR analysis indicated that the deletions were the same as those in NLC28.
Chromosomal DNA was isolated from strains grown in Luria broth, treated with RNase, subjected to phenol-chloroform treatment, and suspended in Tris-EDTA, pH 8.0 (2, 19). DNA was sheared to 1-kb fragments by sonication and radioactively labeled with [α-32P]dCTP by random DNA labeling (Roche), and unincorporated nucleotides were removed with Sephadex G-50 nick spin columns (Pharmacia). Panorama E. coli gene arrays (Sigma/Genosys) were probed overnight with 33 ng of DNA (∼25 million cpm) at 65°C. The array blots were washed and probed according to the manufacturer's recommendations in a hybridization volume of 15 ml. The arrays were visualized by phosphorimaging with the Molecular Dynamics Storm system, and the data were assembled with ArrayVision 6.0 software (Imaging Research Inc.).
Multiple open reading frame deletions can be identified in the K-12 derivative MC4100.
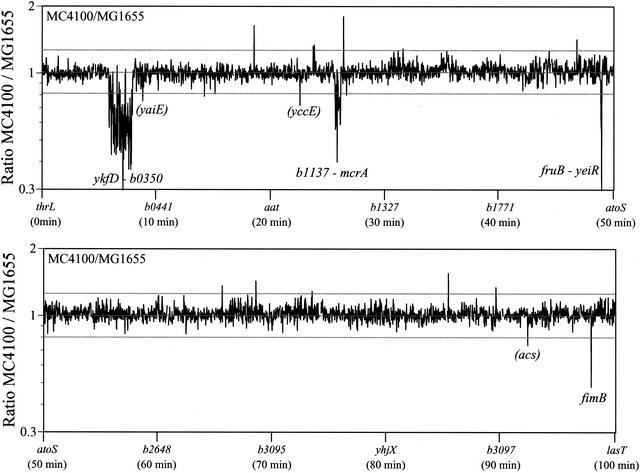
Deletions were identified in MC4100 by calculating a ratio of the normalized spot intensity from MC4100 to MG1655, i.e., MC4100 normalized intensity/MG1655 normalized intensity. Spot intensity was normalized by dividing the background corrected intensity by the overall average intensity for the blot. If a gene is found in both MC4100 and MG1655, the value should equal 1. When the MC4100/MG1655 ratios were plotted in gene order, we found that most values were about 1 (Fig. 1). However, we found over 100 MG1655 open reading frames that appeared to be missing in MC4100 based on low intensity ratios. Examination of the ratios plotted in gene order pinpointed the three multiple open reading frame deletions already known in MC4100 and showed their extent, ykfD-b0350, b1137-mcrA, and fruB-yeiR (Fig. 1; Table 1). The array data also suggested a previously unknown single open reading frame deletion in the fim genes that was confirmed by PCR (see below).
FIG. 1.
Probing the MG1655 genome array with labeled chromosomal DNA from MC4100 provides a sensitive mechanism for detecting deletions. Deletions were identified in MC4100 by calculating a ratio of the change in spot intensity from results found with MG1655, i.e., MC4100 intensity/MG1655 intensity. Array data were exported from ArrayVision into MS Excel for analysis. Duplicate open reading frame values were averaged to give 4,290 data points. Data were normalized within each blot by dividing the intensity of each open reading frame by the average spot intensity for all 4,290 of the individual open reading frames on the blot. For analysis, intensity ratios were calculated and graphed; the intensity of each normalized spot from MC4100 was determined as a ratio to MG1655. Each strain was blotted twice from DNA from separate labeling reactions. Four separate ratios were calculated from the two trials of each strain, and these values were averaged and used to assign putative deletions. The MG1655 sequence version M52 and a list of all of the MG1655 open reading frames were downloaded from the E. coli Genome Project at the University of Wisconsin—Madison. The y axis shows the ratio of the intensity of the 4,290 MG1655 open reading frames listed in gene order for the 100-min chromosome on the x axis. If a gene is found in both MC4100 and MG1655, the value should equal 1. The reference lines indicate 1.5 standard deviations below the mean and the inverse of this value above the mean. PCR amplification and sequencing confirmed the ykfD-b0350, b1137-mcrA, fruB-yeiR, and fimB deletions. PCR amplification indicates that yaiE, yccE, and acs (shown in parentheses) are still present in MC4100 compared to the MG1655 genome sequence.
TABLE 1.
Deletions in MC4100
| Putative missing open reading frame(s) | Deletion coordinates (size) | PCR primers |
|---|---|---|
| ykfD-b0350 | 274,723-371,962 (97,239 bp) | NLC1034, AGGGATGAATTACTCTCAGG |
| NLC1035, CCACGTCAACGATCATATCC | ||
| b1137-mcrA | 1,195,433-1,210,636 (15,203 bp) | NLC1036, GCACAAGGCAAGAAGATCAC |
| NLC1037, AGTGGATAGTAATTGACACG | ||
| fruB-yeiR | 2,260,297-2,266,975 (6,678 bp) | NLC1032, GATTCACGCATCAGCAAGCC |
| NLC1033, ATTAATCGATCACTTAATGG | ||
| fimBa | 4,538,595-4,539,613 (1,018 bp) | NLC1060, GTGCCGTTGCTGCTAAAATG |
| NLC1061, CCTGATAATGCAGATCAAGCAG | ||
| yaiE | None | NLC1048, CTTCACACTTGCCCGGTAAT |
| NLC1064, CCATGGACGTTGTGTCATTG | ||
| yccE | None | NLC1062, ACATTCAGCGTTTTCGGAAT |
| NLC1063, AAACGCATCTACCAGCGAGT | ||
| acs | None | NLC1058, ATTGGAATACCGCGTGTGAC |
| NLC1059, ACCATCTTCAGCGTTTTTCG |
The actual deletion compared to MG1655 was 1,018 bp, but the deletion was associated with the insertion of an IS1 element.
The (argF-lac)U169 deletion of MC4100 is larger than previously thought, encompassing the ykfD-b0350 genes.
A large number of the low ratio values (i.e., putative missing open reading frames) grouped in the region of the large MC4100 (argF-lac)U169 deletion encompassing the lactose genes (ykfD-b0350 in Fig. 1 and Table 1). PCR amplification with primers specific to genes outside the putative deletion (ykfC and mhpE) along with DNA sequencing defined the endpoint of the large (argF-lac)U169 deletion in MC4100. This deletion was found to be much larger than expected from previous work with pulsed-field gel electrophoresis that had suggested that the deletion was approximately 86 kb in size (15). DNA sequencing indicated that 97,239 bp were deleted via homologous recombination between 10-bp direct repeats (GTC TGG CTG G), leaving only one of the repeats.
Interdigitated within the open reading frames that were shown to be missing by PCR were some open reading frames with ratios that were close to the value expected for genes that are present, e.g., within 1 standard deviation of the mean. These “false-positive” values are most easily explained by technical reasons such as cross-hybridization with similar genes in the genome, although we cannot rule out the unlikely possibility that certain genes moved to new positions in the chromosome. Homologous sequences could come from a variety of sources and allow cross-hybridization sufficient for the false positives identified within the deletions. The six IS1 elements found in MG1655 are all on the array, making the missing IS1 elements within the MC4100 deletions appear to be present. Paralogs could also allow cross-hybridization to give false-positive values within deletions: for example, the putative permease YagG (b0270), which appeared to be present within one of the deletions, could cross-hybridize with seven other known and putative permeases found on the MG1655 array, ranging from 25 to 45% identity across the genes. Because cross-hybridization could also stem from contributions from many different open reading frames found throughout the chromosome, a one-for-one documentation comparing each false positive with another open reading frame is not possible.
A deletion of the b1137-mcrA genes indicates that MC4100 lacks the e14 element found in MG1655.
e14 is a genetic element that can move into and out of the chromosome and is found in a specific location in the E. coli K-12 genome. Multiple putative missing open reading frame values grouped in the region of the e14 element found in MG1655 (6) (b1137-mcrA in Fig. 1 and Table 1). Amplification and sequencing with PCR primers specific to genes flanking the region (icdA and b1160) confirmed that 15,203 bp including the e14 element were missing in MC4100 (Table 1). The loss of e14 is further supported by the observation that MC4100 does not possess the restriction and modification system normally ascribed to e14 (17; M. Sibley and L. Raleigh, personal communication).
It remains unclear how the e14 element was lost. The e14 element could have excised and been lost, or the intervening region could have been lost by host-mediated homologous recombination between the 166-bp near-perfect direct repeats that flanked the element in MG1655. DNA sequencing indicates that the near-perfect repeat in mcrA was maintained and not the repeat found in the icdA gene.
A deletion of the fruB-yeiR genes likely accounts for the fruA25 allele of MC4100.
Multiple putative missing open reading frames fell in consecutive genes including a portion of the fruBKA operon encoding functions for fructose transport and catabolism (fruB-yeiR in Fig. 1 and Table 1). A deletion had previously been suggested to be associated with the fruA25 allele of MC4100 (15). PCR primers were designed for the open reading frames flanking the putative deletion, fruK and b2174, and sequencing of the resulting PCR product confirmed a 6,678-bp deletion by comparison to the MG1655 genome sequence. The deletion removes all of fruB and the first of 29 amino acids of the coding region from the 312-amino-acid FruK protein. Therefore, the fruA25 allele actually leaves the fruA gene intact but removes the fruBKA promoter.
The genome array can detect a single open reading frame deletion.
Inspection of the array data indicated a few single open reading frames that gave low ratio values, yaiE, yccE, acs, and fimB (Fig. 1). Using PCR, we confirmed that fimB, the open reading frame which gave the lowest ratio value, did indeed have a deletion: DNA sequencing indicated that a 1,018-bp deletion removed 533 bp of the fimB gene along with 5 bp of the adjacent fimE gene. The fimB-fimE deletion was associated with an IS1 insertion. IS1 insertion has previously been shown to sometimes be associated with the deletion of adjacent DNA sequence (21). Our ability to detect the fimB deletion indicates that using chromosomal DNA to probe whole-genome blots provides a sensitive tool for detecting deletions that are less than a kilobase in size that include a portion of an open reading frame. Of additional significance, we found that by detecting a deletion in fimB we were able to identify the insertion of foreign DNA sequences. This suggests that array techniques may also be utilized for the detection of heterologous sequences such as pathogenicity and fitness islands. Unlike techniques such as restriction fragment length polymorphism, this array technique could identify newly inserted DNA even if there was not a net change in the size of a given region. Because the 1,018-bp deletion of the MG1655 sequence was replaced with 767 bp of heterologous IS1 DNA sequence, a net physical drop of only 251 bp was realized; a 251-bp deletion would likely be missed with most restriction mapping techniques involving rare cutting enzymes.
There were some single open reading frames that gave high ratios of MC4100/MG1655 intensity (Fig. 1). There are multiple possible explanations for high ratio values. Overrepresented values could indicate that the strain in question contains more copies of these open reading frames than does MG1655. While it seems unlikely, we cannot formally rule out the chance that they result from amplification of single open reading frames. Amplification of certain genes can occur when a gene is under strong selection (1, 22). Additionally, it is formally possible that these genes were deleted in the isolate of MG1655 that we obtained from the American Type Culture Collection.
Experiments with MC4100 suggest the limits of the Panorama array for detecting deletions.
PCR with primers flanking yaiE, yccE, and acs resulted in the same sizes of fragments for the strains MG1655 and MC4100, indicating that no deletion of these genes occurred. It is unclear why some open reading frames display low ratio values. Knowing the lowest MC4100/MG1655 ratios where no deletion really exists is important in extending this array technology to other non-MG1655 E. coli strains. Using the array technology to predict putative missing genes in unsequenced E. coli strains would require establishing an appropriate threshold for ratio values. The threshold limit for assigning putative deletions should err on the side of having some false negatives to minimize the chance of missing deletions. Based on results with MC4100, a threshold of 1.5 standard deviations below the mean seems appropriate, because this is just within the minimum value obtained with the three false-negative open reading frames yaiE, yccE, and acs (Fig. 1). This threshold would introduce an extra three open reading frames as potential false negatives.
Analysis of the deletion endpoint identified by PCR also suggests the portion of a gene that must be missing to register by the array technique (Table 2). Our results suggest that deletions that remove only a portion of a gene can be detected. However, very small deletions leaving 80% of an open reading frame intact will appear to be present. Because the results are calculated as a ratio of signals in comparing a tester strain to MG1655, the percentage of the open reading frame that is missing, and not the actual number of base pairs, is important. Therefore, smaller deletions will be detectable in small open reading frames that might go missing in larger open reading frames.
TABLE 2.
How much of a gene must be missing to register as missinga
| Gene | Total gene size (bp) | Missing bp | % Remaining | Array analysis indication |
|---|---|---|---|---|
| fimE | 594 | 5 | 99 | Present |
| fruK | 936 | 87 | 91 | Present |
| b2174 | 747 | 138 | 82 | Present |
| b0350 | 813 | 630 | 23 | Missing |
| ykfD | 1,425 | 1,217 | 15 | Missing |
| fimB | 600 | 533 | 11 | Missing |
Results with MC4100 were used to estimate how much of an open reading frame must be lost to register as missing by examining the six open reading frames that were split in the four MC4100 deletions (two deletion junctions were between open reading frames). Because the MC4100 data are calculated as ratios, the percent loss of an open reading frame is important and not the actual number of missing base pairs.
Our analysis of the deletions in MC4100 identified 133 open reading frames that are not essential for E. coli under standard laboratory conditions on minimal medium (Table 3). The genes that are missing in MC4100 compared to MG1655 include examples from most of the gene categories established for the MG1655 genome (7).
TABLE 3.
Genes deleted in MC4100 by functional class
| Functional class | No. | Gene name(s) |
|---|---|---|
| Putative regulatory proteins | 10 | yagI, yagL, yagW, ykgA, ykgD, b0294, b0316, b0318, b0330, b0346 |
| Cell structure | 3 | fimB,afimE,ayagD |
| Phage, transposon, or plasmid | 11 | insA2, insA3, insB2, insB3, lit, pin, b0281, b0299, b1140, b1145, b1157 |
| Transport and binding proteins | 5 | betT, codB, cynX, fruB,alacY |
| Putative transport proteins | 3 | yeiO, ykgG, b0263 |
| Energy metabolism | 3 | b0288, b0328, fruKa |
| Transcription, RNA processing and degradation | 1 | mcrAa |
| Translation, posttranslational modification | 1 | b0296 |
| Cell processes (including adaptation, protection) | 4 | betA, betB, betI, ykgC |
| Nucleotide biosynthesis and metabolism | 1 | codA |
| Amino acid biosynthesis and metabolism | 2 | argF, yagF |
| Carbon compound catabolism | 9 | cynR, lacA, lacI, lacZ, mhpB, yagG, yeiQ, b0271, b0349 |
| Central intermediary metabolism | 8 | cynS, cynT, yagC, yagE, yagT, ykfD,ab0331, b0333 |
| Structural proteins | 1 | eaeH |
| Putative enzymes | 4 | b0323, b0324, b0325, b0350a |
| Hypothetical, unclassified, unknown | 57 | yagA, yagB, yagJ, yagK, yagP, yagQ, yagR, yagS, yagU, yagV, yagX, yagY, yagZ, yahA, ycfA, ycfK, yeiP, yeiR,aykgB, ykgE, ykg, ykgH, b0279, b0280, b0295, b0298, b030, b0303, b0309, b0317, b0319, b0320, b0321, b0322, b0326, b0327, b0329, b0332, b0334, b0335, b0347, b1137,ab1138, b1141, b1142, b1143, b1144, b1146, b1147, b1148, b1149, b1150, b1151, b1152, b1153, b1155, b2174a |
Partially deleted gene.
Whole-genome arrays provide an important tool in comparative genomics by providing a tool to extend the genome sequence of a type strain to other isolates.
Arrays have proved useful in assessing the evolution of pathogenic bacteria (reviewed in reference 9). By probing the MG1655 array with E. coli isolates of known genealogical relationship, it was previously deduced that 67 additions and deletions account for the distribution of genes in the MG1655 strain (13). In the work presented here we show that the MG1655 gene array could be used as a tool to map specific deletion endpoints in other K-12 strain derivatives in conjunction with PCR. We chose the K-12 derivative MC4100 because of its widespread use and because of its historical significance in bacterial genetics. The technique was found to be reproducible and accurate at identifying deletions of a wide range of sizes that could be confirmed by PCR amplification and sequencing.
Acknowledgments
This work was supported by NIH training grant 5T32CA09139 (J.E.P.). N.L.C. and T.E.T. are employees of the Howard Hughes Medical Institute.
We thank Mary Berlyn, Lise Raleigh, and Marion Sibley for providing strains and information. We thank the members of the Craig lab for comments on the manuscript.
REFERENCES
- 1.Andersson, D. I., E. S. Slechta, and J. R. Roth. 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282:1133-1135. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. Greene Publishing Associates, Inc., and John Wiley & Sons, Inc., New York, N.Y.
- 3.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 4.Badarinarayana, V., P. W. Estep III, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. [DOI] [PubMed] [Google Scholar]
- 5.Bassford, P., J. Beckwith, M. Berman, E. Brickman, M. Casadaban, L. Guarente, I. Saint-Girons, A. Sarthy, M. Schwartz, H. Shuman, and T. Silhavy. 1980. Genetic fusions of the lac operon: a new approach to the study of biological processes, p. 245-261. In J. H. Miller and W. S. Reznikoff (ed.), The operon. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 6.Berlyn, M. K., K. B. Low, and K. E. Rudd. 1996. Linkage map of Escherichia coli K-12, edition 9, p. 1715-1902. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol 104:541-555. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. R., and J. M. Musser. 2001. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 9:547-553. [DOI] [PubMed] [Google Scholar]
- 10.Heath, J. D., J. D. Perkins, B. Sharma, and G. M. Weinstock. 1992. NotI genomic cleavage map of Escherichia coli K-12 strain MG1655. J. Bacteriol. 174:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khodursky, A. B., B. J. Peter, M. B. Schmid, J. DeRisi, D. Botstein, P. O. Brown, and N. R. Cozzarelli. 2000. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci. USA 97:9419-9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKown, R. L., C. S. Waddell, L. A. Arciszewska, and N. L. Craig. 1987. Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. Proc. Natl. Acad. Sci. USA 84:7807-7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochman, H., and I. B. Jones. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19:6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perkins, J. D., J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1992. SfiI genomic cleavage map of Escherichia coli K-12 strain MG1655. Nucleic Acids Res. 20:1129-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkins, J. D., J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1993. XbaI and BlnI genomic cleavage maps of Escherichia coli K-12 strain MG1655 and comparative analysis of other strains. J. Mol. Biol. 232:419-445. [DOI] [PubMed] [Google Scholar]
- 16.Peters, J. E., and N. L. Craig. 2000. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol. Cell 6:573-582. [DOI] [PubMed] [Google Scholar]
- 17.Raleigh, E. A., N. E. Murray, H. Revel, R. M. Blumenthal, D. Westaway, A. D. Reith, P. W. Rigby, J. Elhai, and D. Hanahan. 1988. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 16:1563-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riehle, M. M., A. F. Bennett, and A. D. Long. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turlan, C., and M. Chandler. 1995. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 14:5410-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whoriskey, S. K., V. H. Nghiem, P. M. Leong, J. M. Masson, and J. H. Miller. 1987. Genetic rearrangements and gene amplification in Escherichia coli: DNA sequences at the junctures of amplified gene fusions. Genes Dev. 1:227-237. [DOI] [PubMed] [Google Scholar]