Abstract
Colorectal carcinoma (CRC) is a major cause of morbidity and mortality in Western countries. It has so far been molecularly defined mainly by alterations of the Wnt pathway. We show here for the first time that aberrant activities of the signal transducer and activator of transcription STAT3 actively contribute to this malignancy and, thus, are a potential therapeutic target for CRC. Constitutive STAT3 activity was found to be abundant in dedifferentiated cancer cells and infiltrating lymphocytes of CRC samples, but not in non-neoplastic colon epithelium. Cell lines derived from malignant colorectal tumors lost persistent STAT3 activity in culture. However, implantation of colon carcinoma cells into nude mice resulted in restoration of STAT3 activity, suggesting a role of an extracellular stimulus within the tumor microenvironment as a trigger for STAT activation. STAT3 activity in CRC cells triggered through interleukin-6 or through a constitutively active STAT3 mutant promoted cancer cell multiplication, whereas STAT3 inhibition through a dominant-negative variant impaired IL-6-driven proliferation. Blockade of STAT3 activation in CRC-derived xenograft tumors slowed down their development, arguing for a contribution of STAT3 to colorectal tumor growth.
Keywords: Colorectal cancer, JAK/STAT pathway, STAT3, xenograft, cell proliferation
Introduction
Colorectal carcinoma (CRC) is a malignancy with a high and increasing incidence and a high mortality. Almost 200,000 individuals are diagnosed annually with cancer of the colon or the rectum in the United States alone, and nearly 50% of these patients eventually dies of the disease [1]. Surgical resection of tumors, chemotherapy, and radiotherapy constitute current state-of-the-art treatment of CRC and have, in combination, yielded improvements in cure and survival rates.
In recent years, there have been major advances in our understanding of the molecular basis of this tumor and its progression from adenoma to carcinoma. They hold potential for translation into novel strategies for the treatment of CRC. Many investigations focused on the dysregulation of the Wnt pathway, whose components—adenomatous polyposis coli (APC), β-catenin, and axin—are frequently mutated in colorectal tumors. The consequence of these mutations is an accumulation of nonphosphorylated β-catenin and, hence, constitutive transcription of target genes such as cyclin D1, c-myc, c-jun, and tcf1 (reviewed in Ref. [2]). Other mutations frequently observed in colorectal cancer concern k-Ras as a trigger of the growth-promoting mitogen-activated protein (MAP) kinase pathway, tumor-suppressors p53 and Rb, and mediators of transforming growth factor (TGF) β-signaling such as the TGF-β type 2 receptor and transcription factor Smad4 (reviewed in Ref. [3]). In addition, defects in DNA repair genes hmsh2, hmlh1, hpms1, or hpms2 have been implicated in the tumorigenesis of CRC and appear to be associated with microsatellite instability [4,5]. Frequently observed dysregulation concerns telomerase activity, which is present in colon cancer but not in benign lesions [6].
A mechanism explaining the causal connection between oncogenic malfunction of signal mediators and tumorigenesis in self-renewing tissues as the colon is emerging for the Wnt pathway. Constitutive activity of β-catenin signaling due to single or multiple mutations in the APC and β-catenin gene results in dose-dependent differentiation defects in stem cells [7].
Stem cell development and embryonic patterning are controlled by multiple routes of signal transduction, among them the JAK-STAT (Janus kinase-signal transducer and activator of transcription) pathway (reviewed in Ref. [8]). Although the dysregulated activity of STAT factors 3, 1, and 5 has been implicated in various cancers, no studies have addressed it yet in the context of CRC.
STAT3, originally characterized as a mediator of interleukin-6 receptor signaling [9], has extremely widespread functions throughout the organism (reviewed in Ref. [10]) and is the only embryonic lethal knockout within the STAT family [11]. Apart from cytokine and growth factor receptors, several viral or cellular oncogenes such as src, fps, polyoma virus middle T-antigen, and sis are known to activate STAT3 (reviewed in Ref. [12]). Some cancer-derived cell lines depend on constitutively active STAT3 and undergo apoptosis when STAT3 action is blocked [13,14]. Importantly, a constitutively active artificial variant of STAT3 generated by forced dimerization was shown to behave as an oncoprotein, causing tumorigenesis in nude mice [15]. Cell transformation by aberrant STAT3 activity probably involves upregulation of genes promoting cell cycle progression (cyclin D1 and c-myc) and/or preventing apoptosis (bcl-xL, mcl-1, and survivin) [15–18].
STAT3 is dysregulated in many cancers of the hematopoietic system (reviewed in Ref. [19]) as well as in various solid tumors. Constitutive activity of STAT3 has been reported in malignant tumors of the breast [14], head and neck [20], skin [21], brain [22], and prostate [23]. It has also been shown in some studies that STAT3 activation is significantly associated with poor prognosis [20,24] and metastasis [25]. The as-yet poorly characterized mechanistic contributions of STAT3 to the respective tumor phenotype are apparently not uniform and depend on the particular tissue context. In head and neck tumors, for instance, STAT3 acts on the expression of cyclin D1 and is, thus, directly involved in triggering the cell cycle [20]. In contrast, STAT3 activity in melanoma cells was found to downregulate cell proliferation [26]. STAT3 may also promote the transformed state of tumor cells by antiapoptotic signaling (i.e., through upregulation of genes that counteract active cell death). In cell lines from brain, skin, or breast tumors, overexpression of antiapoptotic genes such as bcl-2, bcl-xL, and mcl-1 was found to be associated with constitutive STAT3 activity, and its experimental blockage could induce apoptosis [21,27,28].
Inhibition of signal mediators acting upstream of STAT3, or the use of dominant-negative variants of STAT3 reduced proliferation or enhanced apoptosis in various cell types [24,20,21]. The most promising results with regard to STAT3 as a potential target for tumor treatment came from a study on melanoma xenografts in nude mice. Introduction of a dominant-negative STAT3 variant into preexisting tumors by gene therapy inhibited tumor growth and led to tumor regression [29].
Apart from STAT3, two other members of the STAT family have also been associated with cancer. STAT5, originally termed mammary gland factor, is not only frequently activated in blood cell tumors, but can also play a relevant role in cellular growth control in solid cancers (e.g., of head and neck or prostate) [30,31]. STAT1 is constitutively active along with STAT3 in leukemias (e.g., Ref. [32]) and in solid tumors. It appears to have a negative effect on tumor progression. In breast cancer, activation of STAT1 is a favorable prognostic marker [33].
In this work, we have analyzed the status of STAT activation in biopsies from colorectal cancer tissue, tumor-derived cell lines, and xenograft tumors originating from human colon carcinoma cells, and we present evidence for an active role of STAT3 in oncogenesis.
Materials and Methods
Tumor Biopsies
Tumor biopsies were obtained from colorectal cancer patients after approval of the study by the local ethical committee. Patients were not treated with chemotherapy prior to resection. Biopsies were snap-frozen in liquid nitrogen immediately after resection. Frozen blocks were disintegrated mechanically and whole cell extracts were prepared by suspending the cells in a buffer containing 20 mM N-2 hydroxyethylpiperazine-N′-2-ethanesulfonic acid (Hepes; pH 7.9), 400 mM NaCl, 1 mM EDTA, 20% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 5 µg/ml leupeptin, 0.2 U/ml aprotinin, 5 mM sodium orthovanadate, 10 mM NaF, and 5 M β-glycerophosphate, followed by three to four freeze-thaw cycles in liquid nitrogen and on ice. The extracts were cleared by centrifugation at 20,000g for 30 minutes at 4°C. Supernatants were subjected to protein determinations followed by electrophoretic mobility shift assays (EMSAs) and/or Western blot analyses.
Cell Lines and Cell Culture
Colon carcinoma cell lines HT-29, SW-480, and CaCo2 were purchased from the ATCC (Manassas, VA). The establishment and characterization of the low passage number cell lines COGA 1, COGA 2, and COGA 3 have been described previously [34]. All cell lines were cultured in RPMI 1640 medium containing 10% fetal calf serum, 200 mM l-glutamine, 100 mM sodium pyruvate, and 1 mg/ml gentamycin. Cells were cultured in coated plasticware (Greiner Labortechnik, Frickenhausen, Germany). For cytokine stimulation, cells were incubated with 10 ng/ml IL-6 medium for 30 minutes. Cells were harvested at 80% confluency after three PBS washes at 4°C. Whole cell extracts were prepared from cell pellets as described above.
DNA Constructs and Retroviral Infection of Cells
A retroviral construct expressing wild-type mouse STAT3 was constructed by excision of the STAT3 coding sequence from pME185m3 (obtained from Fabrice Gouilleux, Paris, France) with EcoRI/NotI, and its ligation in the corresponding sites of pMX-IRES-GFP (obtained from Toshio Kitamura, Tokyo, Japan). The dominant-negative mutation in STAT3 (“STAT3 d.n.”; Y705F) was introduced using a site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The mutated sequence was confirmed by sequencing and subcloned into the retroviral vector as described above. The retroviral construct encoding the constitutively active STAT3 mutant (“STAT3 c.a.”) was obtained by transferring the STAT3 sequence form the pRC/CMV2m3C-A (obtained from Jacqueline Bromberg, New York, NY) into the pXM-IRES-GFP vector.
For retrovirus production, 293GPG packaging cells stably expressing gag-pol and the VSV-G envelope protein [35] were transiently transfected with retroviral vectors encoding the different STAT3 constructs by the standard calcium phosphate precipitation method. Four days after transfection, the growth medium containing virus particles was filtered and used for infection. HT-29 cells were plated at 1 x 106 cells per well in six-well culture dishes 24 hours before infection and incubated three times with a virus-containing supernatant supplemented with 4 µg/ml polybrene (Sigma, St. Louis, MO) for 24 hours. Infection efficiency was analysed by measuring the GFP expression of the HT-29 cells by standard flow cytometric analysis.
EMSA
Whole cell extracts were prepared as described above and subjected to Bradford protein determination. Protein concentrations were adapted to equal levels. For analysis of STAT1/STAT3 complexes, the SIEm67 STAT binding site from the human c-fos promoter was used as a probe. Double-stranded blunt-ended oligonucleotides were annealed (5′-CATTTCCCGTAAATC-3′) and end-labelled using 32P-γ-ATP and T4 polynucleotide kinase to a specific activity of 10,000 cpm/fmol as described [15,36]. For STAT5 analysis, the proximal STAT binding element (5′-AGATTTCTAGGAATTCAAATC-3′) from the bovine β-casein promoter was employed in analogous ways [37]. Binding reactions were performed by incubating 10,000 cpm of radiolabelled probe with 20 µg of cell lysate for 30 minutes at room temperature. For supershift reactions of STAT containing complexes, 2 µg of antibodies to STAT1 (M22 recognizing the C-terminus; Santa Cruz Biotechnology, Santa Cruz, CA), STAT3 (C-20 or H-190, recognizing the N-terminus or C-terminus; Santa Cruz Biotechnology), or Stat5 (C-17; Santa Cruz Biotechnology) was added to the binding reactions before EMSA was performed. Samples were separated by electrophoresis through 6% native polyacrylamide gels and analyzed by autoradiography using BioMax-sensitive films (Kodak, Rochester, New York).
Western Blot Analysis
Western blot analysis was performed as described previously [36,37]. Briefly, 20 µg of whole cell extract was solubilized in a gel loading buffer (62.5 mM Tris/HCl, pH 6.8, 2% SDS, 25% glycerol, 1% phenol blue, and 5% β-mercaptoethanol), boiled for 10 minutes, and separated on a 7.5% or 10% acrylamide SDS gel. After protein transfer, nitrocellulose membranes were blocked in NET-G buffer for 1 hour. STAT3 P-Tyr705 (Cell Signaling Technology, Beverly, MA) and STAT3-α (c-20; Santa Cruz Biotechnology) were incubated for 36 hours in a 1:1000 dilution. Detection was performed with peroxidase-conjugated anti-rabbit IgG (Roth, Karlsruhe, Germany) at a dilution of 1:10,000 for 1 hour. Visualization was performed using an Enhanced Chemiluminescence detection kit (Amersham, Piscataway, NJ).
Immunohistochemistry and Laser Microscopy
Formalin-fixed, paraffin-embedded sections (5 µm) were deparaffinized by subjecting them to two 5-minute changes in 100% xylene and rehydrated by serial incubations in 100%, 96%, 80%, 70%, and 50% xylene, followed by Tris-buffered saline (TBS). For antigen retrieval, sections were autoclaved in 10 mM sodium citrate (pH 6.0) at 121°C for 20 minutes, cooled down to room temperature, and kept for 30 minutes in TBS. Furthermore, the sections were treated with 3.0% H2O2 for 20 minutes to reduce endogenous peroxidase activity. Unspecific binding sites were blocked 5% goat normal serum (Jackson Immunoresearch, West Grove, PA) for 30 minutes. Sections were incubated overnight with a 1:15 dilution for 2 hours (protocol i) or with a 1:50 dilution overnight (protocol ii) of the STAT3 P-Tyr705 antibody (Cell Signaling Technology). Signal detection was achieved by two alternative procedures:
In Figures 2 and 6, the streptavidin-enzyme conjugate-based 4plus Universal Immunoperoxidase Detection System (Biocarta, San Diego, CA) using Jenchrom blue as chromogen (Mobitech, Göttingen, Germany) was applied according to the manufacturer's recommendations. Color development time was in the range of 8 to 12 minutes. Single-photon laser scanning microscopy was performed with an LSM 310 (Carl Zeiss, Jena, Germany) in transmission mode using an argon ion laser at 488 nm. The Zeiss objective Plan-NEO FLUAR 63x/1.30 oil was employed at a scanning time of 30 seconds. The nonprocessed grey values of typical transmission images were converted to inverted values and then further processed to obtain red-green-blue images out of 3 x 28 color values. The chosen false color table was stored as a look-up table and finally saved as a TIFF file [38].
In Figure 7, detection was performed using the Super Sensitive Detection System by BioGenex (San Ramon, CA) and AEC as chromogen. Microscopic pictures were taken by using an Axioskop microscope and Axiovision 4.2 software (Carl Zeiss).
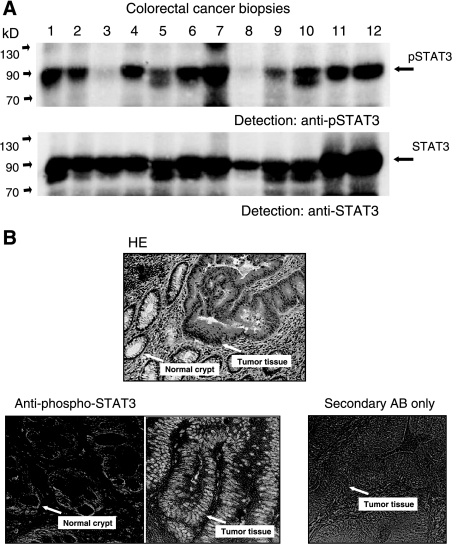
Figure 2.
Analysis of STAT3 tyrosine phosphorylation in colorectal cancer tissue. (A) Examination of lysates from tumor biopsies for activated STAT3 by Western blot analysis. Samples were separated by PAGE, transferred to nitrocellulose, and probed with an antibody specifically recognizing STAT3 phosphorylated on tyrosine 705 (top). Comparable loading of the lanes and identity of STAT3 was confirmed by reprobing the blot with anti-STAT3 (bottom). (B) Histologic examination of a colorectal tumor sample for STAT3 activity. Sections from a representative tumor biopsy were stained with hematoxylin/eosin (top) or immunoreacted with antibody to phospho-STAT3 (bottom). Sections showing differentiated normal cells in crypt structures or dedifferentiated tumor tissues, respectively (arrows), were treated with an antibody specific for STAT3 phosphorylated at tyrosine 705. As a control for antibody and detection specificity, a typical section was stained with peroxidase-coupled secondary antibody without prior treatment with anti-pSTAT3 Tyr 705 (bottom right).
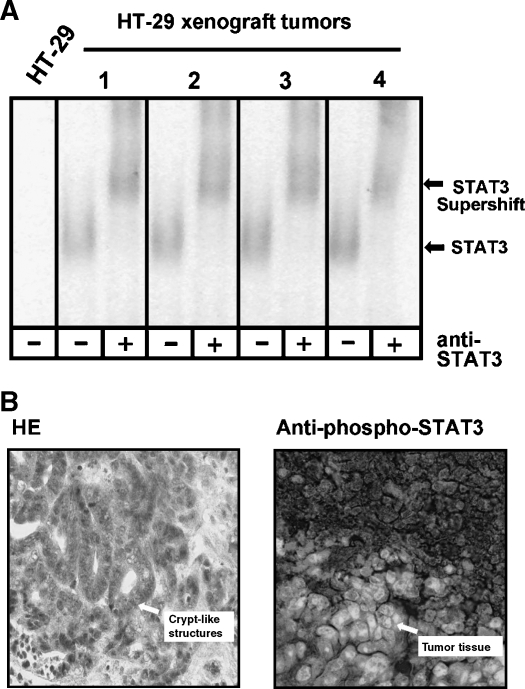
Figure 6.
Determination of STAT3 acticity in HT-29 xenograft tumors. (A) Left: Test by EMSA for the formation of specific complexes on the SIE m67 STAT recognition element. Extracts of HT-29-derived tumors from nude mice (1–4) were incubated with the radiolabelled probe and analyzed by native PAGE and autoradiography. Right: Identification of STAT3 in the DNA-protein complex from tumor “1” by incubation with a specific anti-STAT3 antibody as indicated. Positions of STAT3-containing complexes and the antibody-dependent supershifted complex are marked by arrows. (B) Histologic examination of an HT-29-derived xenograft tumor for STAT3 activity. Sections from a representative tumor induced by injection of HT-29 cells into nude mice were stained with hematoxilin/eosin (top) or immunoreacted with an antibody specific for STAT3 phosphorylated at tyrosine 705 (bottom). Typical sites of crypt-like structures and tumor tissues with substantial nuclear concentration of activated STAT3 are indicated by arrows.
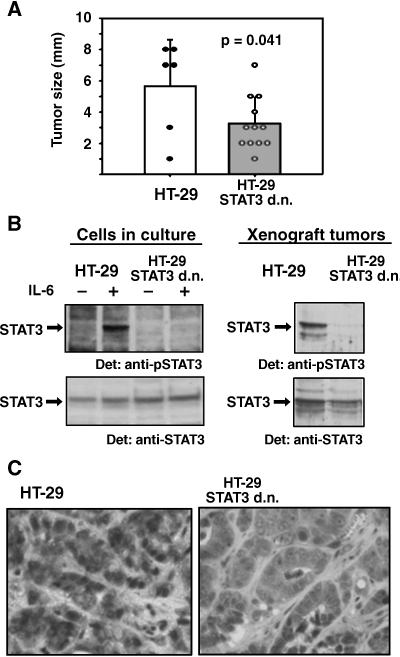
Figure 7.
Influence of a dominant-negative STAT3 variant on the growth of HT-29 xenograft tumors. (A) Size of explanted tumors from nude mice injected with HT-29 cells (white bar) and HT-29 cells stably expressing dominant-negative STAT3 (grey bar). Closed and open circles represent the measured sizes of the individual HT-29 and HT-29 STAT3 d.n. tumors, respectively. Bars with standard errors represent average tumor sizes; P was determined by Student's t test. (B) (Inducible) STAT3 tyrosine phosphorylation in cells before implantation into nude mice (left) and on development of xenograft tumors after 21 days (right). Cells were optionally incubated with 10 ng/ml IL-6 for 30 minutes as indicated. After cell or tumor tissue lysis, Western blot analysis was performed with antibody to phosphorylated STAT3 (top) or STAT3 (bottom), respectively. (C) Immunohistologic detection of tyrosine-phosphorylated STAT3 in sections from HT-29 xenograft tumors (left) and HT-29 STAT3 d.n. tumors (right).
Long-Term Cell Proliferation Assay
Aliquots of 105 cells were suspended in 2 ml of RPMI 1640 medium supplemented as described above and allowed to adhere in individual wells of six-well cell culture plates. They were grown in the absence or presence of 20 ng/ml IL-6 for up to 9 days. Medium with or without cytokine was changed every 2 days. At intervals, cells from individual wells were detached from the plastic surface in 0.5 ml of 0.05% trypsin/0.02% EDTA (Biochrom, Berlin, Germany) for 2 minutes at 37°C before 1.5 ml of medium was added to restore the original volume. Aliquots of cell suspensions were counted using a Neubauer chamber and numbers of cells were extrapolated to the total volume of the culture.
Xenograft Experiments
Aliquots of 105 HT-29 cells were suspended in a volume of 100 µl of PBS and injected subcutaneously into the neck and (optionally) right and left flank (n = 2–4 per animal) of male 5- to 8-week-old athymic Hsd:NMRI-nu/nu mice (Harlan, Indianapolis, IN). Animals were kept under PF isolation conditions and standard diet. The experimental endpoint was 21 days after tumor cell application and the animals were sacrificed. If applicable, tumor sizes were measured with the aid of a ruler and were calculated within a group as arithmetic means with standard error of the mean (SEM). For statistical analysis, the SigmaStat software (Systat, Erkrath, Germany) was used. Differences were identified with Student's t test and considered significant at P < .05. All operations on live animals were performed under isoflurane anesthesia (CuraMed Pharma, Karlsruhe, Germany).
For biochemical and immunohistologic analyses, tumor specimens were further treated analogous to the human biopsies.
Results
STAT3, STAT1, and STAT5 Are Constitutively Active in Colorectal Cancer Tissue
Thirty-two colorectal tumor biopsies were obtained in the course of tumor resections and subjected to the analysis of STAT activity. The patients were staged according to operation and pathologic findings with UICC TNM classification: 11 in stage T2, 16 in stage T3, and 5 in stage T4. Regarding the differentiation grade, 29 tumors were classified G2, two as G3, and one as G4. The surgical specimens were investigated for their status of STAT activity by EMSA. Figure 1A shows a representative selection of tumor tissue samples tested for the formation of protein complexes on the high-affinity DNA binding element for STAT3 and STAT1, the SIEm67 site originating from the human c-fos promoter.
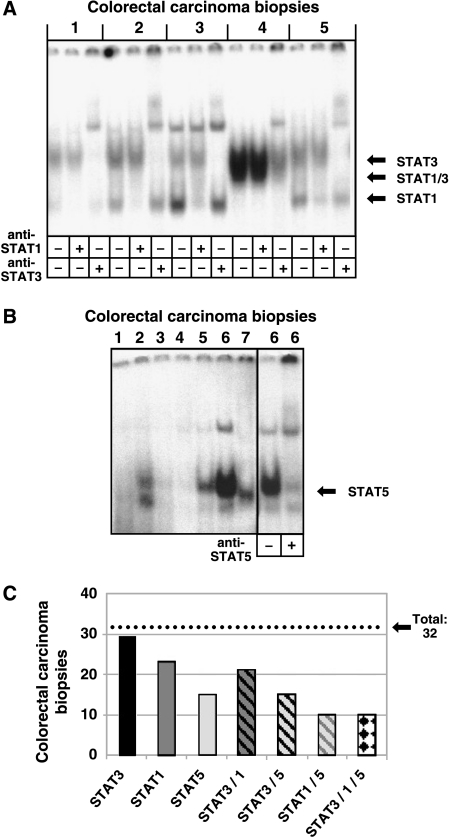
Figure 1.
Analysis of STAT activity by EMSA in extracts of colorectal carcinoma biopsies. (A) Extracts from colorectal tumor tissue samples (1–5) were incubated with the double-stranded 32P-labelled SIE m67 STAT binding site. Complexes were resolved on a 6% native polyacrylamide gel and visualized by autoradiography. The identity of STAT-containing complexes was determined by including specific antibodies in the binding reactions as indicated. According to the results from the supershift experiments, positions of the specific STAT complexes are indicated by arrows. (B) Extracts from colorectal tumor biopsies (1–7) were analyzed for the formation of DNA-protein complexes on the 32P-labelled proximal STAT5 binding element from the bovine β-casein promoter. A representative tumor tissue extract (6) was subjected to a supershift experiment by simultaneous incubation with a STAT5-specific antibody as indicated. (C) Incidence of persistent STAT activity in the colorectal tumor samples investigated in this work. Bars represent the respective numbers of biopsies (from 32 individual colorectal tumors) positive for the indicated STAT activity (activities) as analyzed by EMSA.
In the majority of cases, one, two or three distinct specific DNA-protein complexes were resolved. To define the identity of the individual complexes, all tumor samples were analyzed by supershift analysis employing antibodies to either STAT3 or STAT1. These experiments revealed that the uppermost complex contained STAT3 and the lowermost one contained STAT1, respectively. The intermediate complex contained both STAT3 and STAT1, hence representing a heterodimer of the two factors.
Next, we addressed the status of STAT5 activation. Lysates from all 32 biopsies were tested for STAT binding to the proximal STAT5 cognate element from the bovine β-casein promoter. Figure 1B shows the result for a representative selection of biopsies. To verify the identity of the complex observed in about half the samples analyzed, a DNA binding experiment were performed in the presence or absence of a specific antibody to STAT5. Antibody-induced disappearance of the specific DNA-protein complex and the occurrence of a supershift band confirmed the presence of activated STAT5.
Figure 1C summarizes STAT activity data obtained with the 32 colorectal tumor biopsies under investigation. Notably, 29 of 32 examined tumor samples showed STAT3 DNA binding activity. For STAT1 and STAT5, the respective findings were 23/32 and 15/32. In many tumor samples, simultaneous activation of more than one STAT was observed. All samples with activated STAT5 also contained constitutive STAT3 activity. In 10 of 32 samples, all three STAT factors analyzed were found to be activated. There was no striking correlation between tumor stage and the respective STAT activity pattern. Of the T2 tumors investigated, 10/11 were positive for STAT3, 6/11 for STAT1, and 5/11 for STAT5. For T3 tumors, the figures were STAT3: 16/16, STAT1: 14/16, and STAT5: 6/16. Results for T4 tumors were: STAT3: 2/5, STAT1: 3/5, and STAT5: 1/5.
STAT3 was the STAT protein with the highest incidence of constitutive activity in colorectal cancer biopsies. We therefore concentrated on it and analyzed its phosphorylation state in tumor tissues both by Western blot analysis and immunohistology. Figure 2A shows a Western blot analysis of 12 representative tumor tissue lysates with an antibody specific for STAT3 phosphorylated on Tyr705. In all samples that had been positive for STAT3 DNA binding, signals from phosphorylated STAT3 were observed. The identity of the bands as STAT3 was confirmed by reprobing the blots with anti-STAT3 (Figure 2B).
We addressed by immunohistochemistry, using an antibody to phospho-STAT3, which cell types within the tumor are responsible for biochemically detected signals of activated STAT3 in the tumor specimens. Five representative STAT3-positive tumor samples were subjected to this kind of analysis and yielded similar results. Figure 2B shows sections from one tumor that were stained with hematoxilin and eosin (H&E) (A) or antibody to phospho-STAT3 (B), respectively. STAT3 is predominantly active in the transformed cells of dedifferentiated epithelium and, thus, is massively abundant in their nuclei. Signals are only apparent to a much lesser extent in the epithelium of neighboring non-neoplastic intestinal crypts. This result rules out the possibility that STAT3 activity in the biopsies is mainly the result of lymphocyte function and inflammatory processes. It rather, together with the biochemical data, indicates an involvement of STAT3 in the development or maintenance of the cancerous cell state in the intestinal epithelium.
Once Established in Culture, Colon Cancer Cells Lose Constitutive STAT Activity
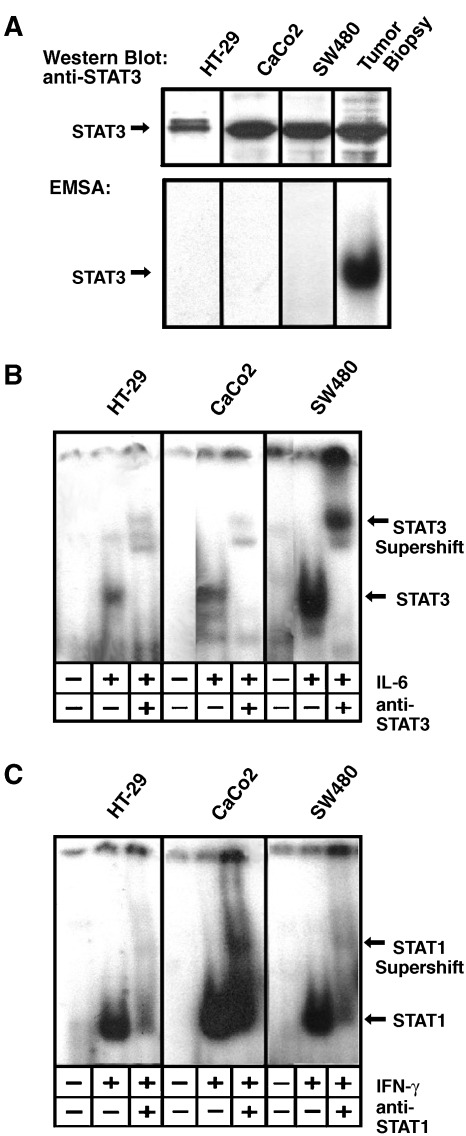
Having observed strong STAT activation in malignantly transformed colon epithelium cells, we employed cell lines to study the origin and consequences of this activity. First, three established human colon carcinoma cell lines were analyzed for STAT3 expression and specific DNA binding activity in comparison with a clinical tumor specimen. All three cell lines investigated showed STAT3 expression to a level comparable with tumor tissues (Figure 3A).
Figure 3.
Analysis of the STAT activity status in colon carcinoma cell lines. (A) Western blot analysis (top) and EMSA analysis (bottom) of cell lines HT-29, CaCo2, and SW 480. A STAT3-positive tumor sample (compare Figure 1) served as a positive control. STAT3 expression was tested by subjecting cell extracts to a Western blot probed with anti-STAT3. STAT3 activation was analyzed by assaying for the formation of specific complexes on the SIE m67 DNA binding site. (B) Test for IL-6-inducible activation of STAT3 in cell lines HT-29, CaCo2, and SW 480. Cells were either left untreated or incubated with 10 ng/ml IL-6 for 30 minutes as indicated. Lysates were subjected to an EMSA using the m67 element as a probe. The identity of STAT3-containing complexes was verified by supershift experiments employing an antibody to STAT3 as indicated. Positions of STAT3 and STAT3-DNA complexes are marked with arrows. (C) Test for IFN-γ-inducible activation of STAT3 in cell lines HT-29, CaCo2, and SW 480. Cells were either left untreated or incubated with 10 ng/ml IFN-γ for 30 minutes as indicated. Lysates were analyzed as in (B), using an antibody to STAT1 for supershift experiments as indicated. Positions of STAT1 and STAT1-DNA complexes are marked with arrows.
Interestingly, and in striking contrast to the findings with surgical tumor specimens, neither HT-29 nor CaCo2 or SW480 cells showed any constitutive binding of STAT3 to the SIE m67 STAT binding element (Figure 3B, leftmost lanes of the respective panels). We therefore asked if cytokines are involved in the activation of STAT3. Cells were stimulated with various cytokines known to trigger STAT3 through the shared gp130 cytokine receptor chain. Oncostatin M, leukemia-inhibitory factor, and interleukin-11 did not evoke any detectable STAT3 activity (data not shown). In contrast, IL-6 induced profound STAT3 activity in all three colon carcinoma cell lines tested as indicated by specific supershifting with the STAT3-specific antibody (Figure 3B). Because persistent activities of STAT3 and STAT1 were found together in many tumor samples investigated (compare Figure 1), we also addressed if STAT1 was inducible by cytokine stimulation. As shown in Figure 3C, interferon-γ induced specific DNA binding of STAT1 in all three colon carcinoma cell lines. These data suggest that constitutive STAT activity in cancerous colon epithelial cells is a consequence of receptor activation by extracellular stimuli.
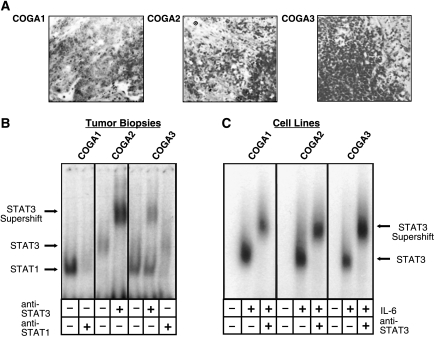
The striking differences with regard to constitutive STAT3 activity between CRC biopsies and colon cancer cell lines prompted us to include in this study low passage cell lines that have been shown to closely mirror many properties of the original tumors (e.g., phenotype, markers, genetic changes) [34]. Three COGA cell lines, derived from colorectal tumors of different grades, were examined for specific DNA-binding of STAT3 by EMSA experiments and compared with their respective parental tumors (Figure 4). First, we studied clinical biopsies histologically. Representatives of serial sections were examined by methylene blue staining. To make sure that the succeeding biochemical analysis reflects the properties of actual tumor tissues, only portions of the biopsies with obvious cancerous characteristics (Figure 4A) were processed further. Regarding their constitutive STAT activity status, biopsies from the tumors that had given rise to cell lines COGA 1, 2, and 3 were found representative for the spectrum of tumor samples previously analyzed (Figure 4B): The COGA 2 and 3 tumors showed STAT3 activity as proven by antibody supershifts; the COGA 1 and 3 tumors revealed a faster migrating complex, which, in analogy to Figure 1, represents STAT1. Figure 4C shows that, despite the preservation of various other properties, all three COGA lines examined did not exhibit constitutive but only IL-6-inducible STAT3 activity. Extending the findings presented in Figure 3, these results demonstrate that tumor-borne constitutive STAT3 activity is consistently lost on cultivation of colon carcinoma cell lines and regained by cytokine stimulation.
Figure 4.
Comparative analysis for STAT3 activity in colon carcinoma tissue and low passage colon cell lines derived from the respective tumors. (A) Histologic representation (methylene blue staining) of sections from tumors that were analyzed for STAT3 activity in (B) and gave rise to the COGA cell lines studied in (C). (B) EMSA of samples from tumors that gave rise to the COGA cell lines studied in (C). The SIE m67 STAT binding element was used as a probe; supershift experiments were performed by employing an antibody to STAT3 as indicated. Arrows mark the positions of the respective complexes. Only portions of the biopsies were processed for sample preparation that had previously been confirmed as cancerous by histologic examination of flanking sections (compare A). (C) EMSA analysis of cell lines originating from the tumors assayed in (A) and (B). Cells were either left quiescent or stimulated with 10 ng/ml IL-6 before lysis and treatment as in (B).
STAT3 Drives Accelerated Proliferation in Colon Carcinoma Cells
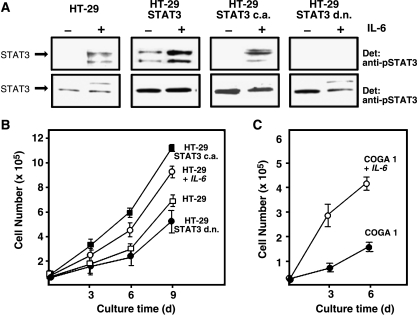
To address the possible functional involvement of STAT3 in the malignant transformation of colon epithelium cells, we introduced a STAT3 cDNA as well as a constitutively active and a dominant-negative derivative into the colon carcinoma cell line HT-29 by retroviral infection. The constitutively active STAT3 construct (“STAT3 c.a.”) had been generated by introduction of a disulfide bridge, which forces the formation of stable dimers [15]. A dominant-negative STAT3 derivative (“STAT3 d.n.”) was constructed by replacing Tyr705 with a Phe residue. Because the retroviral vector encoded GFP, infection efficiencies could be checked by FACS analysis for fluorescence and were routinely above 70% (data not shown). Expression of the heterologous STAT3 proteins as well as tyrosine phosphorylation in response to IL-6 stimulation was analyzed by Western blot analysis (Figure 5A). The introduction of a wild-type STAT3 cDNA into HT-29 cells led to a robust overexpression of the protein in comparison to parental cells and, concomitant with the high abundance of STAT3, to a certain degree of constitutive tyrosine phosphorylation. Tyrosine phosphorylation of STAT3 in response to incubation with IL-6 was enhanced further only to a limited extent, probably due to saturation of the IL-6 receptor system.
Figure 5.
Expression and activation of heterologous STAT3 variants in HT-29 cells and effects of STAT3 activity on cell proliferation. (A) Western blot analysis of tyrosine phosphorylation (top) and expression (bottom) of retrovirally introduced STAT3 variants in HT-29 cells. Cells were either left untreated or incubated with 10 ng/ml IL-6 for 30 minutes as indicated, lysed, and probed with antibody to phosphorylated STAT3 (top) or STAT3 (bottom), respectively. (B) Proliferation of HT-29 derivatives expressing constitutively active or dominant-negative STAT3 variants. Samples of 5 x 104 cells of each cell line were seeded into individual wells of six-well cell culture plates in a volume of 2 ml. Medium was changed every 2 days. Where indicated, IL-6 was present in the medium at a concentration of 20 ng/ml throughout the entire cultivation period. After 3, 6, and 9 days, aliquots were counted using a Neubauer chamber and cell numbers were extrapolated to the total culture volume. Results are from four equivalent experiments. (C) Proliferation of COGA 1 cells in dependence of IL-6 stimulation. The experiment was performed as described under (A) with the following variations: individual cultures were started at a cell number of 2 x 104; cell numbers were determined after cultivation for 3 and 6 days. Results are from four independent experiments.
STAT3 c.a. and STAT3 d.n. were also expressed to higher levels as the endogenous STAT3 on retroviral gene transfer. STAT3 c.a. was phosphorylated in response to IL-6 stimulation to a lesser degree than wild-type STAT3. This effect may be due to a somewhat poorer association of STAT3 c.a. with the phosphorylated gp130 receptor chain and/or impaired accessibility of the phosphorylation site for the JAK kinase. Alternatively, negative feedback regulation in stably transfected HT-29 cells might limit further enhancement of STAT3 activity in the presence of the already active artificial dimer. STAT3 d.n. entirely blocked IL-6-dependent phosphorylation of endogenous STAT3.
We then analyzed the effects of the heterologous STAT3 constructs on the proliferative behavior of the HT-29 derivatives. The development in cell number was monitored over a cultivation period of 5 days (Figure 5B). The presence of IL-6 in the medium resulted in faster cell proliferation, suggesting an involvement of gp130-triggered JAK/STAT signaling in growth control. Confirming this notion, expression of STAT3 c.a. led to a profound acceleration of cell proliferation, whereas expression of STAT3 d.n. resulted in a significant decrease in the proliferation rate. Infection of HT-29 cells with the empty retroviral GFP vector had no significant effect on cell proliferation (data not shown). These results show that STAT3 promotes cell division and proliferation of the colon carcinoma cell line, HT-29. To exclude the possibility that these findings are the consequence of specific properties of the HT-29 cell line, we studied the influence of IL-6 on the proliferative behavior of COGA 1 cells. We observed a strong stimulatory effect of IL-6 on cell division (Figure 5C), which, as recent transfection experiments with COGA cell lines show, is mediated by STAT3 (S.A.T., unpublished data).
Constitutive STAT3 Activity Is Restored in Tumors Derived from a Colon Carcinoma Cell Line
A xenograft experiment was performed to further analyze the differences between colon carcinoma cells in culture and in the tumor context with regard to STAT3 activity. A total of 105 HT-29 cells were injected into immunodeficient nude mice subcutaneously. All eight injected animals developed tumors that were subsequently analyzed for STAT3 activity. The EMSA experiment shown in Figure 6A indicates that the HT-29-derived tumors had regained constitutive DNA binding activity in comparison to the cells used for the injection. Specific supershifting of the band appearing in all tumors with an antibody to STAT3 confirmed its identity as a STAT3-containing complex. Histologic examination of the tumors revealed the formation of crypt-like structures, which, on staining with an antibody to tyrosine-phosphorylated STAT3, displayed abundance and nuclear localization of activated STAT3 (Figure 6B). These results show that the constitutively active state of STAT3 in colon carcinoma cells is not lost inevitably by cultivation, but can be restored by injecting tumor cells into mice (Figure 6B). Similar observations were made with xenografts derived from the implantation of COGA 1 cells (data not shown).
Suppression of STAT3 Activity in Colon Carcinoma Xenograft Tumors Reduces Tumor Growth
To address directly in how far STAT3 activation influences the behavior of CRC cells in vivo, we tested the effect of suppressing STAT3 function in xenografts. Both HT-29 cells and HT-29 cells stably expressing STAT3 d.n. were implanted into mice to induce tumors. Aliquots of 105 cells were injected subcutaneously into the necks and/or flanks of nude mice. The numbers of injection sites were n = 7 for HT-29 cells and n = 15 for HT-29 STAT3 d.n. cells. Within 3 weeks, tumors developed at 6 of 7 (HT-29), or 12 of 15 injection sites (HT-29 STAT3 d.n.), respectively. Twenty-one days postinjection, animals were sacrificed, tumors were explanted, and their respective sizes were determined. As shown in Figure 7A, the average size of HT-29 STAT3 d.n.-derived tumors (3 mm) was significantly smaller compared to that of tumors originating from HT-29 parental cells (5.5 mm).
To confirm that this effect correlated with differences in STAT3 activation, the level of STAT3 tyrosine phosphorylation in representative HT-29 and HT-29 STAT3 d.n. tumors was analyzed. As shown in Figure 7B (right-hand side), STAT3 activation in HT-29 STAT3 d.n. tumors was very low compared to tumors originating from the parental cell line. Obviously, the tumors had retained the original properties of the cell lines, HT-29 and HT-29 STAT3 d.n., with regard to inducible STAT3 activation (Figure 7B, left hand side).
Finally, we compared STAT3 activation in tumors from HT-29 and HT-29 STAT3 d.n. cells by immunohistochemistry, employing an antibody to tyrosine-phosphorylated STAT3 (Figure 7C). Whereas tumors derived from HT-29 cells were highly positive for phospho-STAT3 and showed a strong signal accumulation in cell nuclei, tumors of HT-29 STAT3 d.n. cells showed only minor indications of STAT3 activation and no stained nuclei.
These results are highly suggestive for a prominent role of STAT3 in the growth of xenograft colorectal tumors.
Discussion
STAT3 appears to be an important player in the pathogenesis of various human cancers. Like in a number of other solid tumors, it is constitutively active in the large majority of human colorectal cancer tissues.
Obviously, STAT3 may contribute to oncogenesis in the colon epithelium by exerting a promoting effect on the cell cycle. For colon carcinoma, this notion is supported by the finding that heterologous overexpression of STAT3 or cytokine-mediated activation of endogenous STAT3 in colon carcinoma cell lines accelerates cell proliferation. The effect is specific for colon cancer cells and certain other transformed cell types such as, for example, glioblastoma cells [27] and squamous epithelial cells [39], but is not universal because similar experiments with tumor cell lines derived from the mammary gland did not result in comparable enhancement of cell division (E.P., unpublished work). Antiapoptotic signaling may also contribute to the effects of inappropriate STAT3 function in colon carcinoma cells. Preliminary experiments indicate an elevated abundance level of Cyclin D1 and Bcl-2 protein in our tumor biopsies compared to adjacent tissue (data not shown). In situ techniques, however, are required to verify whether such changes coincide spatially with STAT3 activity within the heterogeneous tumor tissue. STAT3 activity may also be associated with enhanced cell motility and invasiveness. It is known that STAT3 deficiency in keratinocytes leads to impaired motility and defects in wound healing [40], and we have observed that overexpression of STAT3 in HT-29 cells increases cell invasiveness in in vitro tests (S.A.T., unpublished results).
Although aberrant STAT3 activity is now considered as functionally important for the occurrence and/or maintenance of various malignant tumors, it is poorly understood by which mechanisms excessive STAT3 activity in cancer is triggered. Unlike for other signaling molecules, no naturally occurring genetic mutations or amplifications of STAT3 associated with oncogenesis have been identified so far [41], indicating that persistent STAT3 activity is most probably due to dysregulated events upstream of STAT3 within the signaling pathways (e.g., activation or mutation of physiological STAT activators such as the Janus kinases). Alternatively, STAT3 activation in cancer could result from overexpression of growth factors and their receptors, or inhibition of negative regulators [13]. Various recent findings, including our own, point to an important role of cytokines and growth factors secreted by tumor cells.
In contrast to tumor cell lines originating from other tumors, all colon carcinoma cell lines employed in our study were negative for STAT3 activity in the absence of cytokine stimulation. Interestingly, they showed profound activity on implantation and xenograft tumor formation in immunodeficient mice. We suggest that diffusible factors are responsible for this phenomenon. Recent reports have identified autocrine loops involving factors that trigger JAK/STAT pathways as crucial for malignant cell behavior in several tumor models. In breast carcinoma cells, the mechanism of autocrine-mediated STAT3 activation, which correlates with cell proliferation, has been identified [42]. In brain tumors, STAT3 is considered to play a central role in autocrine activation of the vascular endothelial growth factor (VEGF) system [22]. Different examples for IL-6-triggered autocrine stimulation of tumor cells have been published recently such as acute myeloid leukemia cells [43], as well as carcinoma cells of the kidney [44] and the prostate [45]. It is up to future works to identify the factors responsible for the activation of STAT3 in CRC, and to exploit their potential suitability as diagnostic markers or therapeutic targets.
In addition to the tumor biopsies, we also analyzed several samples of non-neoplastic adjacent tissues obtained from the same patients and observed significant STAT3 activity in more than half of the cases (data not shown). These findings suggest the possibility that STAT3 activation may occur before histologic alterations of the intestinal tissue become detectable, and are reminiscent of similar results reported for prostate cancer [23].
CRC is a particularly well studied malignancy with regard to genetic alterations involved in the process of oncogenesis. Dysregulation of the Wnt signaling pathway is of major importance for the development of the malignant cellular phenotype: Mutations of APC or β-catenin cause permanent stimulation of transcription factors, Tcf-4 and Lef-4 (reviewed in Ref. [46]). It is interesting to note that STAT3 appears to exert crosstalk with the Wnt pathway: Mutant APC [47] as well as Tcf-4 and Lef-4 [48] are able to associate or cooperate with STAT3, raising the possibility that Tcf-4/Lef-4-driven transcriptional promotion of the cell cycle through c-myc or D-type cyclins [49] is modulated or enhanced by STAT3. Cell lines HT-29 and COGA used in this study show activated Wnt signaling due to mutations in APC and other mediators [34,50], and are thus interesting model systems to investigate the relationships between pathogenic STAT and Wnt signal transduction in the context of CRC.
STAT3 has recently emerged as an attractive target for tumor treatment. In cultured tumor cells, blockage of STAT3 was found to downmodulate proliferation and to induce programmed cell death. Transfection of a dominant-negative form of STAT3 led to growth inhibition and apoptosis of breast, brain, and prostate cancer cells [14,27,41,51]. Most interestingly, growth of experimental melanomas could be suppressed efficiently in mice by introduction of a functionally deficient STAT3 variant [29]. Interruption of the probable IL-6 autocrine loop in prostate cancer xenografts by administration of a neutralizing anti-IL-6 antibody induced tumor regression [52]. Our findings suggest that targeting STAT3 may also be an interesting option to fight CRC and other cancers in which STAT3 activation is linked with tumor progression.
Acknowledgements
We thank Ursula Möller for help with immunohistochemistry, Veronika Sexl for support with animal work and critical reading of the manuscript, and Stephan Patt, Gerd Raabe, Martin Schmidt, and Bernd Wiederanders for valuable discussions.
Abbreviations
- CRC
colorectal carcinoma
- EMSA
electrophoretic mobility shift assay
- IL
interleukin
- JAK
Janus kinase
- STAT
signal transducer and activator of transcription
Footnotes
Part of this work was funded by the “Interdisziplinäres Zentrum für Klinische Forschung—IZKF Jena, Project B 307-01035.” We acknowledge traveling grants by the Boehringer Ingelheim Fund to F.C. and C.O.
References
- 1.Declan Fleming RY. Colorectal cancer screening and follow-up. Surg Oncol. 1998;7:125–137. doi: 10.1016/s0960-7404(99)00034-1. [DOI] [PubMed] [Google Scholar]
- 2.Oving IM, Clevers HC. Molecular causes of colon cancer. Eur J Clin Invest. 2002;32:448–457. doi: 10.1046/j.1365-2362.2002.01004.x. [DOI] [PubMed] [Google Scholar]
- 3.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 4.Ma AH, Xia L, Littman SJ, Swinler S, Lader G, Polinkovsky A, Olechnowicz J, Kasturi L, Lutterbaugh J, Modrich P, et al. Somatic mutation of hPMS2 as a possible cause of sporadic human colon cancer with microsatellite instability. Oncogene. 2000;19:2249–2256. doi: 10.1038/sj.onc.1203568. [DOI] [PubMed] [Google Scholar]
- 5.Duval A, Hamelin R. Genetic instability in human mismatch repair deficient cancers. Ann Genet. 2002;45:71–75. doi: 10.1016/s0003-3995(02)01115-2. [DOI] [PubMed] [Google Scholar]
- 6.Boldrini L, Faviana P, Gisfredi S, Zucconi Y, Di Quirico D, Donati V, Berti P, Spisni R, Galleri D, Materazzi G, et al. Evaluation of telomerase in the development and progression of colon cancer. Int J Mol Med. 2002;10:589–592. [PubMed] [Google Scholar]
- 7.Kielman MF, Rindapaa M, Gaspar C, van Poppel N, Breukel C, van Leeuwen S, Taketo MM, Roberts S, Smits R, Fodde R. APC modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- 8.O'Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the JAK/STAT pathway. Cell. 2002;109(Supplement):S121–S131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 9.Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 10.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 13.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 15.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 16.Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, Li Y, Wang JM, Yang-Yen HF, Karras J, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epling-Burnette PK, Zhong B, Bai F, Jiang K, Bailey RD, Garcia R, Jove R, Djeu JY, Loughran T, Jr, Wei S. Cooperative regulation of Mcl-1 by Janus kinase/Stat and phosphatidylinositol 3-kinase contribute to granulocyte-macrophage colony-stimulating factor-delayed apoptosis in human neutrophils. J Immunol. 2001;166:7486–7495. doi: 10.4049/jimmunol.166.12.7486. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Devgan G, Darnell JE, Jr, Bromberg JF. Constitutively activated Stat3 protects fibroblasts from serum withdrawal and UV-induced apoptosis and antagonizes the proapoptotic effects of activated Stat1. Proc Natl Acad Sci USA. 2001;98:1543–1548. doi: 10.1073/pnas.041588198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravandi F, Talpaz M, Kantarjian H, Estrov Z. Cellular signalling pathways: new targets in leukaemia therapy. Br J Haematol. 2002;116:57–77. doi: 10.1046/j.1365-2141.2002.03236.x. [DOI] [PubMed] [Google Scholar]
- 20.Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, Tomita K, Komiyama S, Weinstein IB. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
- 21.Niu G, Bowman T, Huang M, Shivers S, Reintgen D, Daud A, Chang A, Kraker A, Jove R, Yu H. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene. 2002;21:7001–7010. doi: 10.1038/sj.onc.1205859. [DOI] [PubMed] [Google Scholar]
- 22.Schaefer LK, Ren Z, Fuller GN, Schaefer TS. Constitutive activation of Stat3alpha in brain tumors: localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2) Oncogene. 2002;21:2058–2065. doi: 10.1038/sj.onc.1205263. [DOI] [PubMed] [Google Scholar]
- 23.Dhir R, Ni Z, Lou W, DeMiguel F, Grandis JR, Gao AC. Stat3 activation in prostatic carcinomas. Prostate. 2002;51:241–246. doi: 10.1002/pros.10079. [DOI] [PubMed] [Google Scholar]
- 24.Benekli M, Xia Z, Donohue KA, Ford LA, Pixley LA, Baer MR, Baumann H, Wetzler M. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood. 2002;99:252–257. doi: 10.1182/blood.v99.1.252. [DOI] [PubMed] [Google Scholar]
- 25.Horiguchi A, Oya M, Shimada T, Uchida A, Marumo K, Murai M. Activation of signal transducer and activator of transcription 3 in renal cell carcinoma: a study of incidence and its association with pathological features and clinical outcome. J Urol. 2002;168:762–765. [PubMed] [Google Scholar]
- 26.Kortylewski M, Heinrich PC, Mackiewicz A, Schniertshauer U, Klingmüller U, Nakajima K, Hirano T, Horn F, Behrmann I. Interleukin-6 and oncostatin M- induced growth inhibition of human A375 melanoma cells is STAT-dependent and involves upregulation of the cyclin-dependent kinase inhibitor p27/Kip1. Oncogene. 1999;18:3742–3753. doi: 10.1038/sj.onc.1202708. [DOI] [PubMed] [Google Scholar]
- 27.Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 28.Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 29.Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, Jove R, Yu H. Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res. 1999;59:5059–5063. [PubMed] [Google Scholar]
- 30.Leong PL, Xi S, Drenning SD, Dyer KF, Wentzel AL, Lerner EC, Smithgall TE, Grandis JR. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene. 2002;25:2846–2853. doi: 10.1038/sj.onc.1205385. [DOI] [PubMed] [Google Scholar]
- 31.Ahonen TJ, Xie J, LeBaron MJ, Zhu J, Nurmi M, Alanen K, Rui H, Nevalainen MT. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem. 2003;278:27287–27292. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- 32.Kirito K, Nagashima T, Ozawa K, Komatsu N. Constitutive activation of Stat1 and Stat3 in primary erythroleukemia cells. Int J Hematol. 2002;75:51–54. doi: 10.1007/BF02981979. [DOI] [PubMed] [Google Scholar]
- 33.Widschwendter A, Tonko-Geymayer S, Welte T, Daxenbichler G, Marth C, Doppler W. Prognostic significance of signal transducer and activator of transcription 1 activation in breast cancer. Clin Cancer Res. 2002;8:3065–3074. [PubMed] [Google Scholar]
- 34.Vécsey-Semjen B, Becker KF, Sinski A, Blennow E, Vietor I, Zatloukal K, Beug H, Wagner E, Huber LA. Novel colon cancer cell lines leading to better understanding of the diversity of respective primary cancers. Oncogene. 2002;21:4646–4662. doi: 10.1038/sj.onc.1205577. [DOI] [PubMed] [Google Scholar]
- 35.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kammer W, Lischke A, Moriggl R, Groner B, Ziemiecki A, Gurniak CB, Berg LJ, Friedrich K. Homodimerization of interleukin-4 receptor a chain can induce intracellular signaling. J Biol Chem. 1996;271:23634–23637. doi: 10.1074/jbc.271.39.23634. [DOI] [PubMed] [Google Scholar]
- 37.Lischke A, Moriggl R, Brändlein S, Berchtold S, Kammer W, Sebald W, Groner B, Liu X, Hennighausen L, Friedrich K. The interleukin-4 receptor activates STAT5 by a mechanism that relies upon the function of common γ chain. J Biol Chem. 1998;273:31222–31229. doi: 10.1074/jbc.273.47.31222. [DOI] [PubMed] [Google Scholar]
- 38.Halbhuber KJ, König K. Modern laser scanning microscopy in biology, biotechnology and medicine. Ann Anat. 2003;185:1–20. doi: 10.1016/S0940-9602(03)80002-X. [DOI] [PubMed] [Google Scholar]
- 39.Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, Tweardy DJ. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, Yoshikawa K, Akira S, Takeda J. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke WM, Zin X, Lin HJ, Huang M, Liu R, Reynolds RK, Lin J. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene. 2001;20:7925–7934. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Shaw PE. Autocrine-mediated activation of STAT3 correlates with cell proliferation in breast carcinoma lines. J Biol Chem. 2002;277:17397–17405. doi: 10.1074/jbc.M109962200. [DOI] [PubMed] [Google Scholar]
- 43.Schuringa JJ, Wierenga AT, Kruijer W, Vellenga E. Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood. 2000;95:3765–3770. [PubMed] [Google Scholar]
- 44.Angelo L, Talpaz M, Kurzrock R. Autocrine interleukin-6 production in renal cell carcinoma: evidence for the involvement of p53. Cancer Res. 2002;62:932–940. [PubMed] [Google Scholar]
- 45.Giri D, Ozen M, Ittmann M. Interleukin-6 is an autocrine growth factor in human prostate cancer. Am J Pathol. 2001;159:2159–2165. doi: 10.1016/S0002-9440(10)63067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morin PJ. Beta-catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 47.Norris AL, Clissold PM, Askham JM, Morrison EE, Moncur P, McCall SH, Coletta PL, Meredith DM, Markham AF. Truncated adenomatous polyposis coli (APC) tumour suppressor protein can undergo tyrosine phosphorylation. Eur J Cancer. 2000;36:525–532. doi: 10.1016/s0959-8049(99)00305-6. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita S, Miyagi C, Carmany-Rampey A, Shimizu T, Fuji R, Schier AF, Hirano T. Stat3 controls movements during zebrafish gastrulation. Dev Cell. 2002;2:363–375. doi: 10.1016/s1534-5807(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 49.Kolligs FT, Bommer G, Goke B. Wnt/beta-catenin/Tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion. 2002;66:131–144. doi: 10.1159/000066755. [DOI] [PubMed] [Google Scholar]
- 50.Hsi LC, Angerman-Stewart J, Eling TE. Introduction of full-length APC modulates cyclooxygenase-2 expression in HT-29 human colorectal carcinoma cells at the translational level. Carcinogenesis. 1999;20:2045–2049. doi: 10.1093/carcin/20.11.2045. [DOI] [PubMed] [Google Scholar]
- 51.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–6666. [PubMed] [Google Scholar]
- 52.Smith PC, Keller ET. Anti-interleukin-6 monoclonal antibody induces regression of human prostate cancer xenografts in nude mice. Prostate. 2001;48:47–53. doi: 10.1002/pros.1080. [DOI] [PubMed] [Google Scholar]