Abstract
Spores of Bacillus anthracis, the causative agent of anthrax, are enclosed by a prominent loose-fitting, balloon-like layer called the exosporium. Although the exosporium serves as the source of surface antigens and a primary permeability barrier of the spore, its molecular structure and function are not well characterized. In this study, we identified five major proteins in purified B. anthracis (Sterne strain) exosporia. One protein was the recently identified collagen-like glycoprotein BclA, which appears to be a structural component of the exosporium hair-like nap. Using a large panel of unique antispore monoclonal antibodies, we demonstrated that BclA is the immunodominant antigen on the B. anthracis spore surface. We also showed that the BclA protein and not a carbohydrate constituent directs the dominant immune response. In addition, the length of the central (GXX)n repeat region of BclA appears to be strain specific. Two other unique proteins, BxpA and BxpB, were identified. BxpA is unusually rich in Gln and Pro residues and contains several different tandem repeats, which also exhibit strain-specific variation. In addition, BxpA was found to be cleaved approximately in half. BxpB appears to be glycosylated or associated with glycosylated material and is encoded by a gene that (along with bclA) may be part of an exosporium genomic island. The other two proteins identified were alanine racemase and superoxide dismutase, both of which were reported to be associated with the surface of other Bacillus spores. Possible functions of the newly identified proteins are discussed.
The genus Bacillus includes a diverse collection of gram-positive, rod-shaped, aerobic bacteria that form an endospore (or spore) upon deprivation of an essential nutrient (10, 30). In this process, an asymmetric septation of the starved vegetative cell produces a large and a small genome-containing compartment called the mother cell and forespore, respectively. The mother cell then engulfs the forespore, thereby surrounding it with two opposing cell membranes. A thick layer of modified peptidoglycan called the cortex is synthesized between the two membranes, and proteins synthesized in the mother cell form multiple layers of a spore coat that covers the cortex. While the coat forms the outermost detectable layer for spores of some species (e.g., Bacillus subtilis), in others (e.g., Bacillus anthracis), the spore is enclosed by an additional layer called the exosporium, a prominent, loose-fitting, balloon-like layer also synthesized by the mother cell (12, 17). After a final stage of maturation, the mother cell lyses to release the mature spore, which is dormant and capable of persisting in the soil for many years until it encounters a germination signal.
Most Bacillus species are not pathogenic to humans. The most notable exception is B. anthracis, the causative agent of anthrax (25). In light of the recent use of B. anthracis spores as a terrorist weapon in the United States and the development of these spores as a weapon of mass destruction by a number of countries, there is a pressing need to learn more about spore components that can be used for rapid detection and targeted for treatment and prevention of anthrax (18). One such component is the exosporium, which serves as a primary permeability barrier for the spore and as the source of spore surface antigens (12). As the outermost surface of the spore, the exosporium will interact with the soil environment, with collection and detection devices, with spore-binding target cells in a mammalian host, and with host defenses. Presently, detailed information about exosporium structure and function is limited.
Previous studies of the exosporium focused on spores of B. anthracis and the opportunistic human pathogen Bacillus cereus. These species along with Bacillus thuringiensis, an insect pathogen, and Bacillus mycoides make up the closely related B. cereus group. All members of this group produce spores with a structurally similar exosporium composed of a basal layer and an external hair-like nap extending up to 600 Å in length (12-14). The basal layer contains four paracrystalline sublayers, each exhibiting a hexagonal, perforate lattice structure (3, 13). The hair-like nap varies in length from species to species (13) and even from strain to strain (22). The exosporium reacts with lectins (6), which appear to bind polysaccharides located in the hair-like nap (23). The exosporium constitutes about 2% of the mass of the spore and contains approximately 50% protein, 20% lipid, 20% neutral polysaccharides, and 10% other components (24). The exosporium contains multiple proteins that are synthesized concurrently with the cortex and coat (3, 7, 28).
More recent studies have attempted to identify individual exosporium proteins. An apparent 72-kDa glycoprotein was identified as a component of either the exosporium or the coat of B. thuringiensis spores (11). Three other proteins (GroEL and homologues of InA and RocA) were found to be associated with, although probably not structural components of, the B. cereus exosporium (5). During the preparation of the manuscript, the discovery of a collagen-like B. anthracis spore surface glycoprotein called BclA (for “Bacillus collagen-like protein of anthracis”) was reported (32). BclA contains multiple collagen-like GXX repeats in its central region and appears to be a structural component of the hair-like nap of the exosporium. BclA also produced the major band on an immunoblot of exosporium proteins probed with mouse antispore polyclonal antibodies.
For several years, our laboratory has also been involved in the characterization of exosporium proteins to identify the binding sites of monoclonal antibodies (MAbs) and short peptides that react specifically with B. anthracis spores. These ligands are being developed for rapid spore detection. In this report, we describe the identification of five proteins, including BclA, present in highly purified B. anthracis exosporium preparations. We provide further characterization of BclA, particularly with respect to the immune response to this protein. We show that BclA is the immunodominant antigen in the B. anthracis exosporium, meaning that most antibodies raised against spores react with BclA. These antibodies bind to the BclA protein and not to its associated carbohydrate. In addition, we find that repeat regions in BclA and one other putative exosporium protein vary in length between B. anthracis strains and thus may contribute to observed strain-specific exosporium structures and possibly strain-specific functions.
MATERIALS AND METHODS
Bacterial strains, culture methods, and spore preparation.
We used the Sterne veterinary vaccine strain of B. anthracis, which was obtained from John Ezzell, U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Md. The Sterne strain is not a human pathogen, because it lacks a plasmid necessary to produce the capsule of the vegetative cell. Spores were prepared from liquid cultures grown for approximately 48 h at 37°C with shaking in Difco sporulation medium (27). Spores were extensively washed in cold (4°C) distilled H2O, sedimented through cold 50% Renografin (Bracco Diagnostics) to remove vegetative cells and debris, washed three times in cold distilled H2O, and stored in distilled H2O at 4°C and protected from light. Spores were quantitated microscopically with a Petroff-Hausser counting chamber.
Exosporium preparation and deglycosylation.
Following a previously described exosporium purification procedure (13), approximately 1012 spores were washed five times in cold (4°C) distilled H2O, collected by centrifugation for 20 min at 3,000 × g and 4°C, and resuspended in 50 ml of cold TEP buffer containing 50 mM Tris-HCl (pH 7.2), 0.5 mM EDTA, and 2 mM phenylmethylsulfonyl fluoride. The spores were passed once through a chilled (4°C) French press at 30,000 lb/in2. We confirmed by electron microscopic examination that this procedure removed large exosporium fragments without detectable damage to the remainder of the spore. The sample extruded from the French press was centrifuged for 45 min at 1,200 × g and 4°C, and the supernatant was saved. The pellet was resuspended in 50 ml of cold TEP buffer, the sample was centrifuged as described above, the supernatant was saved, and the latter three steps were repeated. The three supernatant samples were pooled, and the pooled sample was centrifuged as described above. The resulting supernatant was centrifuged for 90 min at 40,000 × g and 4°C. The pellet was resuspended in 7 ml of cold distilled H2O and stored at 4°C. Microscopic examination of this exosporium sample confirmed the virtual absence of spores and verified that essentially all particulate matter reacted with a fluorescently labeled exosporium-specific peptide ligand. This peptide was selected from a phage display 7-mer peptide library with B. anthracis spores as the binding target (33). To deglycosylate exosporium glycoproteins, a dried sample of purified exosporium (5 to 10 mg) was treated with trifluoromethanesulfonic acid (TFMS; Aldrich) as previously described (9).
Exosporium solubilization, protein electrophoresis, and immunoblotting.
Exosporium and TFMS-treated exosporium samples (∼15 μg) were solubilized by boiling for 8 min in 30 μl of 125 mM Tris-HCl (pH 6.8) 4% sodium dodecyl sulfate (SDS), 10% (vol/vol) 2-mercaptoethanol, 1 mM dithiothreitol, 0.05% bromophenol blue, and 10% (vol/vol) glycerol (17). Solubilized proteins (∼15 μg per lane) were separated by SDS-polyacrylamide gel electrophoresis (PAGE) with 12.5% polyacrylamide gels (27). For immunoblotting, proteins were electrophoretically transferred from an SDS-polyacrylamide gel to a nitrocellulose membrane and treated as described in the manual for the Bio-Rad Immun-Blot assay kit. Briefly, each blot was blocked with gelatin, probed with the primary MAb at 5 μg/ml for 2 h, and washed. The blot was then probed with a 1:3,000 dilution of goat anti-mouse immunoglobulin G (IgG) (H+L) horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h, washed, and developed with the HRP developer solution.
Amino-terminal protein sequencing, electron microscopy, and mass spectrometry.
After transfer to a sheet of polyvinylidene difluoride, protein bands were detected by staining with Ponceau S, excised, and subjected to 10 cycles of automated Edman degradation with a Beckman model PI 2090E sequencer. Electron microscopy was performed with a Hitachi 7000 instrument, and sample preparation was done essentially as previously described (16). Matrix-assisted laser desorption-ionization mass spectrometry was performed with a Voyager Elite mass spectrometer.
Preparation of MAbs.
MAbs were made by immunizing BALB/c mice subcutaneously in the hindquarters either with 108 B. anthracis (Sterne) spores killed by gamma irradiation (40 Gy) or with approximately 50 μg of purified Sterne exosporium. The first injections were emulsified in complete Freund's adjuvant. At 3-day intervals, four subsequent injections were done with the same amount of either killed spores or exosporium, with both samples in saline. One day after the last injections, the popliteal, inguinal, and para-aortic lymph nodes were surgically removed from the mice. B lymphocytes from the lymph nodes were fused with plasmacytoma P3X63Ag8.653 cells (21) to produce hybridomas, which were cloned and screened by flow cytometry for antispore MAbs. After subcloning and rescreening of selected hybridomas, MAbs were purified from culture supernatants by affinity chromatography on protein G-Sepharose (Pharmacia). Each hybridoma or MAb was isotyped by a modified enzyme-linked immunosorbent assay with goat anti-isotype antibodies (Southern Biotechnology Associates) (20). The hybridoma genes encoding the VH and VL regions of the antispore MAbs were sequenced as described previously (4, 36). Each MAb used in this study displayed unique VH and VL sequences.
Gene cloning and expression and protein purification.
Sequence data for the B. anthracis (Ames strain) genome were obtained from The Institute for Genome Research (TIGR) web site at http://www.tigr.org. Based on this sequence, DNA primers were designed to separately PCR amplify the entire bclA, bxpA, and bxpB open reading frames (ORFs) of the Sterne strain (see text for gene descriptions). Vegetative cells from a fresh colony of the Sterne strain were added to each PCR mixture as a source of template DNA. PCR amplifications were performed with Pfu DNA polymerase (Stratagene), using the reaction mixture recommended by the supplier. The PCR conditions were 95°C for 5 min; 95°C for 30 s, 55°C for 30 s, and 68°C for 2.5 min for 30 to 35 cycles; and finally 68°C for 5 min. The PCR products (after trimming with appropriate restriction enzymes) were individually inserted into the cloning site of the expression vector pET15b (Novagen). The inserted DNA regions in the resulting recombinant plasmids were sequenced, with three independent isolates per plasmid checked to confirm the sequences. A bclA-containing plasmid was transformed into Escherichia coli BL21(DE3) to express the cloned gene according to the pET system manual (Novagen). The expressed BclA protein, with a six-His tag, was purified by Ni-nitrilotriacetic acid agarose (Qiagen) affinity chromatography according to the manufacturer's instructions, with the protein eluted with a combination of 8 M urea and 100 mM EDTA.
RESULTS
Identification of exosporium proteins and encoding genes.
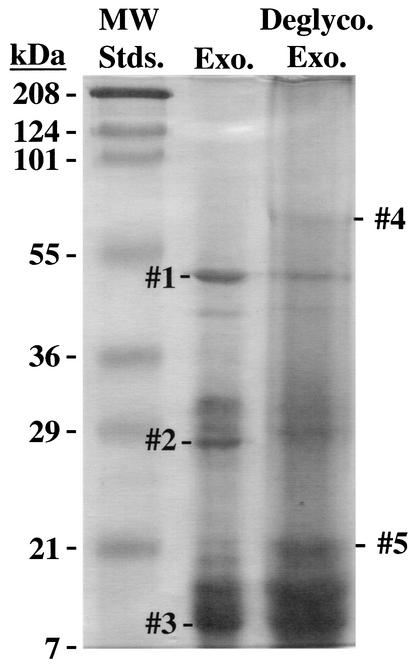
We used a highly purified sample of B. anthracis exosporia to identify protein components. Preliminary experiments indicated that some exosporium proteins and/or glycoproteins were entrapped in high-molecular-mass complexes, which could be disrupted by deglycosylation with TFMS. Thus, we examined proteins in both untreated and TFMS-treated exosporium samples. Each sample was solubilized, loaded in duplicate (along with a lane of molecular mass standards) on each half of an SDS-polyacrylamide gel, and proteins were separated by electrophoresis. One-half of the gel was stained with Coomassie brilliant blue to visualize proteins (Fig. 1). The other half of the gel was used to transfer proteins to a sheet of polyvinylidene difluoride, which was subsequently stained to locate protein bands. Five major protein bands were excised from the polyvinylidene difluoride sheet, with bands 1 to 3 and bands 4 and 5 obtained by using untreated and TFMS-treated exosporium samples, respectively (Fig. 1). Amino-terminal sequences were determined for each protein band, and these sequences were used to find matches with proteins encoded by the genome sequence of the Ames strain of B. anthracis (provided by TIGR).
FIG. 1.
Separation of B. anthracis (Sterne strain) exosporium and deglycosylated exosporium proteins by SDS-PAGE. Proteins were stained with Coomassie brilliant blue. The bands used for protein identification are numbered 1 to 5. MW Stds., molecular mass standards; Exo., exosporium; Deglyco. Exo., deglycosylated exosporium.
Band 1.
The amino-terminal sequence for band 1 [MEEAPFY(CYIR)DT] matched the sequence MEEAPFYRDT found at the start of a 389-codon ORF that was recently predicted to encode alanine racemase (31). The gel mobility of band 1 is consistent with the calculated molecular mass of 43,662 Da for this enzyme. The sequence of a likely intrinsic transcriptional terminator (35) was identified 76 bp upstream of the translational initiation codon. No clear promoter sequences, derived from B. subtilis (15), could be identified between the putative terminator and start codon.
Band 2.
The amino-terminal sequence for band 2 (AVPIGGXTLP) matched the sequence AVPIGGHTLP found at codons 97 to 106 in a 304-codon ORF that was predicted to encode iron/manganese superoxide dismutase (31). Apparently this enzyme was proteolytically cleaved (after an Arg residue) to produce a 208-amino acid protein, which retains the entire catalytic region of the enzyme. Except for Bacillus species, most superoxide dismutases for which sequence data are available do not contain a region similar to the first approximately 100 amino acids of the B. anthracis enzyme. The gel mobility of band 2 is consistent with the calculated molecular mass of 24,600 Da for the truncated protein (residues 97 to 304). No clear promoter sequences were found between the start codon and a likely intrinsic terminator located 64 bp upstream.
Band 3.
The amino-terminal sequence for band 3 [VSPPK(IP)(AP)TFD] matched the sequence VSPPKPPTFD, which was found at codons 134 to 143 in a 248-codon ORF that would encode a 29,174-Da protein with no significant similarity to any protein in the current Entrez Nucleotide database. (A 250-amino acid homologue is encoded by the unfinished TIGR sequence of the B. cereus ATCC 10987 genome.) The 248-codon ORF begins with a Met residue and is preceded by a consensus Shine-Dalgarno sequence that is appropriately positioned for efficient translational initiation (34) (Fig. 2A). This ORF is located 52 bp downstream from a consensus sequence for the late mother-cell-specific sigma factor, σK (15). The ORF ends 30 bp upstream from a G+C-rich hyphenated dyad symmetry followed by a T-rich sequence, which likely functions as an intrinsic transcriptional terminator for a monocistronic operon (Fig. 2B) (35). The protein product of this operon is apparently proteolytically cleaved before residue 134 (after an Arg residue) to create the VSPPKPPTFD amino terminus. The molecular mass of a protein encoded by residues 134 to 248 is 12,842 Da, which is consistent with the gel mobility of band 3.
FIG. 2.
Relevant features of the bxpA operon of B. anthracis (Ames strain). (A) Sequence of the genomic DNA upstream of the bxpA ORF. The elements of the consensus σK promoter are overlined, the Shine-Dalgarno (SD) sequence is underlined, and the initiation codon is boxed. (B) Sequence of the genomic DNA downstream of the bxpA ORF. Arrows indicate the dyad symmetry elements of a putative intrinsic transcriptional terminator, and the stop codon is boxed. (C) Amino acid sequence of BxpA. Arrows indicate three pairs of identical or nearly identical tandem repeats, with dots indicating mismatches. Brackets enclose the 14 amino acids deleted in the Sterne version of BxpA. A vertical arrowhead indicates the site of proteolytic cleavage that produced the amino-terminal sequence VSPPKPPTFD detected in this study.
In addition to the internal cleavage, there are two other striking features about the 248-amino-acid protein. First, it has a highly unusual amino acid composition, with 54 Gln and 38 Pro residues. The Gln residues are mostly in the amino-terminal half of the protein, while the Pro residues are fairly evenly distributed (Fig. 2C). Second, the protein is composed of three pairs of identical (5 of 5) or nearly identical (33 of 40 and 13 of 14) tandem direct repeats (Fig. 2C). Only the 13/14-amino-acid repeat elements are located in the carboxy-terminal half of the protein, just downstream of the VSPPKPPTFD sequence. To determine if this highly repetitive structure, deduced from the genome sequence of the Ames strain of B. anthracis, was maintained in the Sterne strain, we cloned and sequenced the latter strain's corresponding ORF. This sequence revealed that the ORFs were identical except for a 14-codon deletion in the Sterne strain, which precisely removes the last 13/14-amino-acid repeat element (Fig. 2C). We have called the 248/234-amino-acid protein BxpA, for Bacillus exosporium protein A, and its encoding gene is bxpA.
Band 4.
The amino-terminal sequence for band 4 (AFDPNLVGPT) matched residues 20 to 29 of the recently described 382-amino-acid exosporium glycoprotein BclA (32). The amino-terminal 19 amino acids of BclA are presumably removed by proteolytic processing to produce a mature form of the protein. Band 4 (BclA) was detected only after deglycosylation of the exosporium. Without deglycosylation, BclA was contained in >250-kDa material that only poorly entered the SDS-polyacrylamide gel (see below). Deglycosylated BclA (residues 20 to 382) is predicted to have a molecular mass of 33,309 Da; however, the gel mobility of band 4 indicated a molecular mass of ∼70 kDa. This discrepancy may be explained by reduced gel mobility caused by the low isoelectric point (pI = 3.46) of the protein, an unusual protein structure, or some other unrecognized factor. It does not appear that residual glycosylation after TFMS-treatment is a significant factor in reducing gel mobility (see below).
The most striking feature of BclA is a collagen-like region between residues 41 and 250, which contains 70 repeats of GXX, including 54 repeats of GPT (32). Between residues 41 and 232, a 42-amino-acid motif [(GPT)5GDTGTT]2 is repeated three times, with a varying number of GPT triplets (from 2 to 8) separating each motif (Fig. 3). To determine if the repetitive collagen-like region found in the Ames genomic sequence was maintained in the Sterne strain, we cloned and sequenced the bclA gene of Sterne. This sequence showed that the Sterne bclA gene encoded a 400-amino-acid protein, in which an additional six GPT repeats (i.e., 18 amino acids) were present in the collagen-like region. Three additional GPT repeats were inserted before each of the first and second [(GPT)5GDTGTT]2 motifs (Fig. 3). The calculated molecular mass of the mature Sterne protein (residues 20 to 400) is 34,840 Da, which is still much less than expected from its gel mobility.
FIG. 3.
Comparison of the BclA collagen-like regions of the Ames and Sterne strains of B. anthracis. Amino acids 41 to 232 and 41 to 250 are depicted for the Ames and Stern strains, respectively.
Band 5.
The amino-terminal sequence for band 5 [(SMD)FSSDCEFT] matched the sequence MFSSDCEFT found at the start of a 167-codon ORF that would encode a 17,331-Da protein with no significant similarity to any protein in the current Entrez Nucleotide database. (A nearly identical 167-amino-acid homologue is encoded by the unfinished TIGR sequence of the B. cereus ATCC 10987 genome.) This ORF begins with a Met residue and is preceded by an appropriately positioned Shine-Dalgarno sequence (i.e., AAAGGAG/8-base spacer/ATG). The ORF ends with tandem stop codons, which are immediately followed by the sequence of a likely intrinsic transcriptional terminator (5′ AAAGCGGTATTTCTAATACAGAAATACCGCTTTTTTAT, with dyad symmetry elements underlined). Although a near consensus sequence for a σK promoter was identified 31 bp upstream of the ORF, it is possible that the ORF is the last gene in a polycistronic operon, for which we cannot yet identify a likely promoter. Band 5, with a gel mobility consistent with its predicted molecular mass, was routinely observed at much higher levels in deglycosylated exosporium samples. This result suggests that the protein in band 5 is either glycosylated or associated with glycosylated material. We have called the 167-amino-acid protein BxpB, for Bacillus exosporium protein B, and its encoding gene is bxpB. This protein does not contain the type of repetitive sequences that characterize BclA and BxpA. Furthermore, we have found that the BxpB sequences of the Ames and Sterne strains are identical (data not shown).
Screening antispore MAbs for binding to exosporium proteins.
Antibodies are too large to penetrate the exosporium (8, 12); therefore, antibodies that react with intact spores of B. anthracis presumably recognize exosporium antigens. Such antibodies, particularly antispore MAbs that react with a single antigen, are excellent reagents for the identification of exosporium proteins. To examine the proteins in our exosporium samples for reactivity to antispore MAbs, we prepared a panel of 20 unique MAbs raised against either irradiated (killed) spores (10 MAbs) or purified exosporia (10 MAbs) of the Sterne strain of B. anthracis. Each of the 20 MAbs was shown to bind to multiple (i.e., Sterne and ΔAmes) strains of B. anthracis spores by fluorescence-activated cell sorting and by fluorescence microscopy (data not shown). Nineteen of the MAbs were of the IgG class, with all subclasses represented, and one (which was raised against exosporia) was of the IgM class.
The 20 MAbs were used to immunoblot exosporium and deglycosylated exosporium proteins, which had been separated by SDS-PAGE. Twelve of the 20 MAbs (7 of 10 and 5 of 10 raised against spores and exosporia, respectively) gave essentially identical results. Each reacted with a >250-kDa band in the exosporium sample (Fig. 4, lane 6) and with an ∼70-kDa band, apparently band 4 (BclA), in the deglycosylated sample (Fig. 4, lane 4). Thus, antigenic material (BclA) was released from high-molecular-mass complexes by deglycosylation. With extended color development of the immunoblots, it was observed that each of the 12 MAbs also reacted with a wide band of material that ran immediately above band 4 in the deglycosylated sample (Fig. 4, lane 5). This material was not stained with Coomassie brilliant blue and in fact excluded background staining with this dye (Fig. 4, lane 3). The wide band was shown to contain polysaccharide by staining with the Bio-Rad Immun-Blot kit for glycoprotein detection (data not shown). The nature of the association of the polysaccharide with deglycosylated BclA has not been determined. However, it appears that BclA is included in the wide band, in a manner that prevents staining with Coomassie brilliant blue (described below).
FIG. 4.
Reactivity of antispore MAbs to the dominant exosporium antigen. Twelve of the unique antispore MAbs reacted virtually identically with samples of B. anthracis (Sterne) exosporium (Exo.) and deglycosylated exosporium (Degly. Exo.). Results for one of these MAbs (BD8) are shown. Lanes 1 to 3 (from Fig. 1) show proteins separated by SDS-PAGE and stained with Coomassie brilliant blue. Lanes 4 and 5 show immunoblots of deglycosylated exosporium, with color development for approximately 3 and 30 min, respectively. Lane 6 shows the immunoblot of untreated exosporium with color development for approximately 30 min. Lane 7 shows prestained molecular mass protein standards (MW Stds.) (same as in lane 1), which were included in the immunoblot.
Seven of the remaining MAbs, including the IgM species, did not bind proteins in either the exosporium or deglycosylated exosporium sample. One MAb (which was raised against exosporia) reacted with a presently unidentified protein in both samples (data not shown).
MAb binding to purified BclA.
To identify the antigen to which the majority of antispore MAbs bound, we prepared purified BclA. We cloned the bclA ORF (residues 1 to 400) from the Sterne strain into an expression vector in Escherichia coli. We overexpressed and affinity purified BclA, which carried an amino-terminal six-His tag plus several residues from the vector. The expected molecular mass of the six-His-tagged BclA protein (39,000 Da) was confirmed (±0.3%) by mass spectrometry, indicating the absence of glycosylation of the protein in E. coli. Upon SDS-PAGE, purified BclA migrated as a 70-kDa species (Fig. 5, lane 3), indistinguishable from band 4. Thus, the anomalously slow gel mobility of band 4 (BclA) was not due to residual glycosylation after TFMS treatment.
FIG. 5.
Reactivity of antispore MAbs to purified BclA. Fifteen of our antispore MAbs reacted with purified BclA, which was synthesized in E. coli and devoid of glycosylation. The results with one of these MAbs (EA4-10-4) are shown. Lanes 1 to 3 show molecular mass protein standards (MW Stds.), deglycosylated exosporium (Degly. Exo.) proteins, and purified BclA (3 μg), respectively, which were analyzed by SDS-PAGE and stained with Coomassie brilliant blue. Lane 4 shows the immunoblot of purified BclA (3 μg). Lane 5 shows prestained molecular mass protein standards (same as in lane 1), which were included in the immunoblot.
Purified BclA was immunoblotted with the same panel of 20 MAbs examined above. The same 12 MAbs that reacted with band 4 and the >250-kDa band in the TFMS-treated exosporium sample (Fig. 4) reacted with purified BclA (Fig. 5, lane 4). In addition, the 3 (of 10) MAbs raised against spores that did not bind a band in the untreated and TFMS-treated exosporium samples did react with purified BclA. However, this reactivity was relatively weak, which may explain the failure to detect their binding to BclA in exosporium samples. In any event, all MAbs that react with band 4 and the >250-kDa band also bind to BclA produced by E. coli, demonstrating that the reactivity is to the protein and not a carbohydrate component. In addition, these results establish that BclA is the immunodominant antigen on the spore surface, because a large majority of antispore antibodies react with this protein.
DISCUSSION
We identified five of the major proteins in highly purified exosporium samples prepared from spores of the Sterne strain of B. anthracis. One of the proteins was the recently identified collagen-like glycoprotein, BclA, which appears to be a structural component of the exosporium hair-like nap (32). It was reported that BclA in exosporium samples reacts strongly with mouse polyclonal antibodies and to a single mouse MAb raised against Sterne spores (32). In this study, we examined the immunoreactivity of BclA, especially as a purified protein, with a panel of 20 unique mouse MAbs that react with spores produced by multiple strains of B. anthracis. These MAbs were raised against either Sterne spores or purified Sterne exosporium. All 10 of the antispore MAbs raised against spores and half of the antispore MAbs raised against exosporium reacted with purified BclA, which had been synthesized in E. coli to preclude glycosylation. These results demonstrate that BclA is the immunodominant antigen on the B. anthracis spore surface and that the protein and not a carbohydrate constituent directs the dominant immune response. Preliminary studies indicate that MAbs raised against spores of either B. thuringiensis or B. cereus do not recognize the dominant epitope(s) on BclA of B. anthracis (data not shown). Thus, immunodominant spore epitopes are likely to be species specific, presumably reflecting unique functions.
BclA contains a long, centrally located region of triplet repeats (GXX), most of which are GPT. The multiple Thr residues in the repeat region are likely sites of O-linked glycosylation. Extensive glycosylation within the repeat region could contribute to the formation of an extended BclA conformation that determines the length of the hair-like nap (19). In this study, we discovered that the repeat region of BclA of the Sterne strain is 18 amino acids (i.e., six GPT repeats) longer than the repeat region predicted for BclA in the Ames strain of B. anthracis. Such a difference could result in a longer hair-like nap on Sterne spores, compared with Ames spores. Furthermore, differences in the length of the BclA repeat region could account for the differences in the length of hair-like nap observed between different strains of B. anthracis and between different species in the B. cereus group (13, 22). It will be interesting to determine if different lengths of hair-like nap affect spore properties, particularly virulence, and if there are selective pressures for a particular nap length.
With regard to the function of BclA and the hair-like nap, a ΔbclA mutation in the Sterne strain was recently constructed (32). This deletion mutation removes residues 50 to 173 in the triplet repeat region of BclA and prevents the formation of a detectable hair-like nap on spores. When analyzed, the ΔbclA mutation did not affect virulence in a mouse subcutaneous model or spore resistance to a limited number of harsh reagents and treatments. These results suggested that BclA does not contribute to the resistance properties of spores or participate in virulence under the conditions tested (32). However, these results should be interpreted with caution for several reasons. Although ΔbclA spores appear to be devoid of a hair-like nap, a shorter (difficult to detect) version of BclA could still be present on the remaining exosporium basal layer of mutant spores and contribute to resistance and virulence. Furthermore, because the spore's resistance to the chemical and physical challenges employed in the analysis above is attributed to spore features other than the hair-like nap of the exosporium, it is not surprising that altering the nap was without effect (26). It seems likely that different and more physiologically relevant assays will be required to detect the roles that BclA and the hair-like nap play in spore survival and infectivity. Finally, it seems improbable that a structure as elaborate as the hair-like nap, which functions as a large boundary area and as a major antigenic determinant of the spore, will not affect important interactions with the spore's natural environment and/or with cells of the host immune system during infection.
BclA and two other proteins identified in our exosporium preparations, BxpA and BxpB, did not exhibit significant similarity to any proteins encoded by ORFs in the current Entrez Nucleotide database (although homologues were encoded in the unfinished TIGR sequence of the B. cereus ATCC 10987 genome). The lack of similarity to proteins in B. subtilis, a species phylogenetically similar to B. anthracis (30) and for which the genome has been sequenced, is significant. It indicates that BclA, BxpA, and BxpB are necessary to form a B. anthracis spore component, namely the exosporium, that is absent in spores of B. subtilis. Consistent with being exosporium components, the genes encoding BclA and BxpA (apparently in single-gene operons) are each preceded by the consensus sequence for a promoter recognized by the late-mother-cell-specific sigma factor, σK. The gene encoding BxpB is also preceded by a possible σK promoter, which deviates by a single base from the σK consensus sequence. However, the assignment of a potential promoter for bxpB is complicated by the likelihood that bxpB is the last gene of a presently undefined polycistronic operon.
BxpA shares with BclA the atypical feature of repeat elements. However, the repeats in BxpA are markedly different from those in BclA. There are fewer repeats, the repeats are distinct, the repeat elements are longer (e.g., 5, 40, and 14 amino acids), and the three longest repeats are in tandem. The origin and function of the repeat elements are still unknown, but their unusually rich Gln and Pro content suggests a distinctive role. Interestingly, the VrrA protein of B. anthracis also contains Gln+Pro-rich repeat regions. This protein of unknown function is encoded by the vrrA gene, which was discovered as an atypical region of genetic variability among B. anthracis strains and related species (1). Based on similarity to a sheath protein of filarial parasites, it was suggested that the Gln+Pro-rich repeats of VrrA are involved in covalent cross-linking of protein subunits (1). BxpA also shares another feature with BclA—the proteins in the Sterne and Ames strains are different lengths. BxpA from Sterne contains a 14-amino-acid deletion that precisely removes the downstream element of the 14-amino-acid repeat region predicted for the Ames protein. If the repeat elements indeed play distinctive roles, then BxpA of Sterne may function differently from its counterpart in Ames. A unique feature of BxpA is that it is proteolytically cleaved approximately in half, which produces an amino-terminal domain with the 5- and 40-amino-acid repeats and a carboxy-terminal domain with the 14-amino-acid repeat (at least in Ames). The cleavage of BxpA is unlikely to be an in vitro artifact, because exosporia were purified in the presence of an inhibitor of trypsin-like proteases, the type that would presumably cleave BxpA after an Arg residue. In this study, we identified the carboxy-terminal domain of BxpA in our exosporium samples. The location of the amino-terminal domain remains to be determined.
Compared to BclA and BxpA, BxpB is rather unremarkable. It does not contain repeats, it is the same length in the Sterne and Ames strains (167 amino acids), and it is not proteolytically processed. However, BxpB appears to be glycosylated or associated with glycosylated material, because its recovery as a 17.3-kDa protein is greatly enhanced by deglycosylation. It is possible that BxpB is associated with the high-molecular-mass exosporium complexes containing glycosylated BclA, which are disrupted by deglycosylation. An indirect but still intriguing connection between BxpB and BclA is that they are encoded by nearby genes in the B. anthracis genome. The bxpB and bclA genes, which are transcribed in the same direction, are separated by only 10 kb. However, they are not in the same operon. Approximately a dozen genes, most transcribed in the opposite direction, separate bxpB and bclA. These intervening genes include the rmlACBD operon, which encodes the l-rhamnose biosynthetic enzymes, and genes that encode activities for polysaccharide biosynthesis (e.g., a glycosyl transferase). The significance of this finding is that the B. anthracis exosporium contains polysaccharides, perhaps components of glycoproteins such as BclA and possibly BxpB, that are rich in l-rhamnose (data not shown). Thus, the BclA-BxpB region of the genome may constitute an exosporium island, a cluster of genes dedicated to the synthesis of the exosporium.
The remaining two proteins identified in our exosporium preparations are alanine racemase and iron/manganese superoxide dismutase. The latter enzyme is proteolytically processed to remove an amino-terminal domain (residues 1 to 96 of 304) that is unique to Bacillus superoxide dismutases. The significance of this processing is unknown, but catalytic activity is retained by the large fragment of the enzyme, which was the protein detected in our experiments. The cleavage of superoxide dismutase after an Arg residue, like that of BclA, is also unlikely to be an in vitro artifact because of the inclusion of a trypsin-like protease inhibitor during exosporium purification. The discovery of both alanine racemase and superoxide dismutase in the outermost layer of the spore is not particularly surprising. Alanine racemase has been reported to be associated with the surface of other Bacillus spores (27). Its presence may be either fortuitous and related to the production of d-alanine needed for cortex synthesis (2, 29) or by design and needed for the interconversion of the germinant l-alanine and the germination inhibitor d-alanine (27). In the case of superoxide dismutase, it has been proposed that this enzyme associates with the outer surface of B. subtilis spores and participates in the oxidative cross-linking of outer coat proteins (16, 17). The enzyme could have been captured by the exosporium after spore coat maturation or similar maturation of the exosporium. Once captured by the exosporium, for whatever reason, both alanine racemase and superoxide dismutase could function as structural elements.
The studies presented here represent a part of the early phase of identifying the structural components of the B. anthracis exosporium and the genes necessary to produce them. Within the next few years, it is likely that all of the major exosporium components will be biochemically and genetically defined, which will permit the construction of mutations that alter these components. We predict that the analysis of these mutations will reveal many important functions for the exosporium, particularly a key role in virulence.
Acknowledgments
We thank Kelly Morrison and Lori Coward in the UAB Cancer Center Shared Facilities for Protein Analysis and Mass Spectrometry and Leigh Millican in the UAB High Resolution Imaging Facility for valuable assistance. We also acknowledge important contributions by Dakin Williams, David Pritchard, James Daubenspeck, and Melissa Swiecki.
Sequencing of the B. anthracis genome by TIGR was accomplished with support from ONR, DOE, NIAID, and DERA. This work was supported by NIH grant AI50566, DARPA grant MDA972-01-1-0030, and ARO grant DAAD19-00-1-0032.
REFERENCES
- 1.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atrih, A., and S. J. Foster. 2001. Analysis of the role of bacterial endospore cortex structure in resistance properties and demonstration of its conservation amongst species. J. Appl. Microbiol. 91:364-372. [DOI] [PubMed] [Google Scholar]
- 3.Beaman, T. C., H. S. Pankratz, and P. Gerhardt. 1971. Paracrystalline sheets reaggregated from solubilized exosporium of Bacillus cereus. J. Bacteriol. 107:320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedict, C. L., and J. F. Kearney. 1999. Increased junctional diversity in fetal B cells results in a loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity 10:607-617. [DOI] [PubMed] [Google Scholar]
- 5.Charlton, S., A. J. G. Moir, L. Baillie, and A. Moir. 1999. Characterization of the exosporium of Bacillus cereus. J. Appl. Microbiol. 87:241-245. [DOI] [PubMed] [Google Scholar]
- 6.Cole, H. B., J. W. Ezzell, Jr., K. F. Keller, and R. J. Doyle. 1984. Differentiation of Bacillus anthracis and other Bacillus species by lectins. J. Clin. Microbiol. 19:48-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desrosier, J. P., and J. C. Lara. 1984. Synthesis of the exosporium during sporulation of Bacillus cereus. J. Gen. Microbiol. 130:935-940. [Google Scholar]
- 8.Du, C., and K. W. Nickerson. 1996. Bacillus thuringiensis HD-73 spores have surface-localized Cry1Ac toxin: physiological and pathogenic consequences. Appl. Environ. Microbiol. 62:3722-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edge, A. S. B., C. R. Faltynek, L. Hof, L. E. Reichert, Jr., and P. Weber. 1981. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal. Biochem. 118:131-137. [DOI] [PubMed] [Google Scholar]
- 10.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Patrone, M., and J. S. Tandecarz. 1995. A glycoprotein multimer from Bacillus thuringiensis sporangia: dissociation into subunits and sugar composition. Mol. Cell. Biochem. 145:29-37. [DOI] [PubMed] [Google Scholar]
- 12.Gerhardt, P. 1967. Cytology of Bacillus anthracis. Fed. Proc. 26:1504-1517. [PubMed] [Google Scholar]
- 13.Gerhardt, P., and E. Ribi. 1964. Ultrastructure of the exosporium enveloping spores of Bacillus cereus. J. Bacteriol. 88:1774-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hachisuka, Y., K. Kojima, and T. Sato. 1966. Fine filaments on the outside of the exosporium of Bacillus anthracis spores. J. Bacteriol. 91:2382-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 16.Henriques, A. O., L. R. Melsen, and C. P. Moran, Jr. 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 18.Inglesby, T. V., T. O'Toole, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Gerberding, J. Hauer, J. Hughes, J. McDade, M. T. Osterholm, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2002. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA 287:2236-2252. [DOI] [PubMed] [Google Scholar]
- 19.Jentoft, N. 1990. Why are proteins O-glycosylated? Trends Biochem. Sci. 15:291-294. [DOI] [PubMed] [Google Scholar]
- 20.Kearney, J. F., R. Barletta, Z. S. Quan, and J. Quintans. 1981. Monoclonal vs. heterogeneous anti-H-8 antibodies in the analysis of the anti-phosphorylcholine response in BALB/c mice. Eur. J. Immunol. 11:877-883. [DOI] [PubMed] [Google Scholar]
- 21.Kearney, J. F., A. Radbruch, B. Liesegang, and K. Rajewsky. 1979. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J. Immunol. 123:1548-1550. [PubMed] [Google Scholar]
- 22.Kramer, M. J., and I. L. Roth. 1968. Ultrastructural differences in the exosporium of the Sterne and Vollum strains of Bacillus anthracis. Can. J. Microbiol. 14:1297-1299. [DOI] [PubMed] [Google Scholar]
- 23.Matz, L., and P. Gerhardt. 1965. Chemical ultrastructure of the exosporium enveloping spores of Bacillus cereus. Bacteriol. Proc., p. 31.. [DOI] [PMC free article] [PubMed]
- 24.Matz, L. L., T. C. Beaman, and P. Gerhardt. 1970. Chemical composition of exosporium from spores of Bacillus cereus. J. Bacteriol. 101:196-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., West Sussex, United Kingdom.
- 28.Ohye, D. F., and W. G. Murrell. 1973. Exosporium and spore coat formation in Bacillus cereus T. J. Bacteriol. 115:1179-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 178:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Priest, F. G. 1993. Systemics and ecology of Bacillus, p. 3-16. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. Biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 31.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 32.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 33.Turnbough, C. L., Jr. Discovery of phage-displayed peptide ligands for species-specific detection of Bacillus spores. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 34.Vellanoweth, R. L., and J. C. Rabinowitz. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6:1105-1114. [DOI] [PubMed] [Google Scholar]
- 35.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611-615. [DOI] [PubMed] [Google Scholar]
- 36.Young, D., and J. F. Kearney. 1995. Sequence analysis and antigen binding characteristics of Ig SCID Ig+ mice. Int. Immunol. 7:807-819. [DOI] [PubMed] [Google Scholar]