Abstract
Animal cell viruses provide valuable model systems for studying many normal cellular processes, including membrane protein sorting. The focus of this study is an integral membrane protein encoded by the E3 transcription region of human adenoviruses called E3-13.7, which diverts recycling EGF receptors to lysosomes without increasing the rate of receptor internalization or intrinsic receptor tyrosine kinase activity. Although E3-13.7 can be found on the plasma membrane when it is overexpressed, its effect on EGF receptor trafficking suggests that the plasma membrane is not its primary site of action. Using cell fractionation and immunocytochemical experimental approaches, we now report that the viral protein is located predominantly in early endosomes and limiting membranes of endosome-to-lysosome transport intermediates called multivesicular endosomes. We also demonstrate that E3-13.7 physically associates with EGF receptors undergoing E3-13.7–mediated down-regulation in early endosomes. Receptor–viral protein complexes then dissociate, and EGF receptors proceed to lysosomes, where they are degraded, while E3-13.7 is retained in endosomes. We conclude that E3-13.7 is a resident early endocytic protein independent of EGF receptor expression, because it has identical intracellular localization in mouse cells lacking endogenous receptors and cells expressing a human cytomegalovirus-driven receptor cDNA. Finally, we demonstrate that EGF receptor residues 675–697 are required for E3-13.7–mediated down-regulation. Interestingly, this sequence includes a known EGF receptor leucine-based lysosomal sorting signal used during ligand-induced trafficking, which is also conserved in the viral protein. E3-13.7, therefore, provides a novel model system for determining the molecular basis of selective membrane protein transport in the endocytic pathway. Our studies also suggest new paradigms for understanding EGF receptor sorting in endosomes and adenovirus pathogenesis.
INTRODUCTION
Endocytosis is a dynamic process in which material originating from the plasma membrane is actively sorted through a series of intracellular compartments in close communication with the biosynthetic pathway (reviewed by Gruenberg and Maxfield, 1995). Many molecules, including ligand–receptor complexes, are rapidly internalized to the endocytic pathway and then either recycled to the plasma membrane or transported to lysosomes for disposal. Resident endocytic membrane proteins, on the other hand, are transported to their final destination immediately after their synthesis. Despite the continual flux of itinerant membrane cargo, endosomes remain morphologically and functionally distinct, a process made possible in part by precise mechanisms for retaining as well as retrieving endosome-specific proteins. In addition to their role as sorting stations, endosomes also serve to compartmentalize certain cellular responses, such as those mediated by signaling receptors that undergo ligand-induced internalization (reviewed by Baass et al., 1995). Hence, resident endosome-specific proteins likely participate in a variety of physiological processes ranging from protein sorting to cell signaling. In the absence of well-characterized integral membrane markers, however, the nature of the signals that distinguish resident from itinerant endocytic membrane proteins has been difficult to elucidate.
Animal cell viruses have provided valuable model systems for studying many normal cellular processes, including membrane protein sorting. Some of the best studied examples are enveloped RNA viruses that selectively bud from one side of polarized epithelial cells and that were used to establish membrane protein sorting routes in this cell type (Rodriguez-Boulan and Sabatini, 1978; Rodriguez-Boulan and Pendergast, 1980). Others include the human immunodeficiency virus-1 nef protein, which has provided insight into the molecular basis of CD4 down-regulation in infected T-cells (Aiken et al., 1994; Greenberg et al., 1997; Piguet et al., 1998), and the adenovirus-encoded E3-gp19 protein, which defined a novel class of endoplasmic reticulum retrieval signals recognized by COPI coatomer (Cosson and Letourneur, 1997). In addition to highlighting general principles of membrane protein sorting, elucidation of each of these host–virus interactions has also led to a better understanding of viral pathogenesis.
This laboratory studies another adenovirus protein called E3-13.7, which was initially identified because of its ability to specifically down-regulate the EGF receptor during an adenovirus infection (Carlin et al., 1989) or after retrovirus-mediated gene transfer (Hoffman et al., 1990). E3-13.7–mediated EGF receptor down-regulation occurs posttranslationally, resulting in the loss of unoccupied receptors from the cell surface and increased receptor degradation (Hoffman et al., 1992b). Like ligand, E3-13.7 causes EGF receptors to accumulate in internal vesicles of endosome-to-lysosome transport intermediates called multivesicular endosomes (MVEs) (Hoffman and Carlin, 1994). In contrast to ligand, E3-13.7–induced EGF receptor down-regulation occurs without an increase in the rate of receptor internalization or intrinsic tyrosine kinase activity (Hoffman and Carlin, 1994). To gain insight into the mechanism of E3-13.7 action, we sought to investigate its subcellular localization with the use of cell fractionation and immunocytochemical experimental approaches. Our data show that E3-13.7 is localized to early endosomes and MVEs independent of EGF receptor expression. In addition, our data suggest that E3-13.7 alters EGF receptor sorting behavior by interacting directly with receptors in early endosomes. Our data also suggest that leucine-based sorting signals conserved in both molecules act cooperatively to link receptors to lysosomal transport machinery. Interestingly, once recycling receptors have been diverted to lysosomes, the complex dissociates and E3-13.7 is retained in early endosomes, where it may then be reused.
MATERIALS AND METHODS
Cells, Viruses, and Antibodies
NR6 is an NIH 3T3–derived cell line lacking endogenous EGF receptors (Pruss and Herschman, 1977). ERwt is a clonal NR6-derived cell line stably expressing a wild-type human EGF receptor cDNA under regulation of a cytomegalovirus (CMV) promoter (Hoffman et al., 1993). Permanent NR6 cell lines expressing cytoplasmically truncated human EGF receptors have been described (Kil et al., 1999). Briefly, these cell lines were selected after transfection with EGF receptor cDNAs containing genetically engineered premature stop codons and are named based on the C-terminal amino acid residue in the EGF receptor coding region (i.e., c′-651 has a K652 stop substitution). A549 is a human lung carcinoma–derived epithelial cell line (Giard et al., 1973). All cells were maintained in DMEM supplemented with 10% FBS and 2 mM glutamine.
Adenovirus stocks were grown in spinner cultures of human KB cells maintained with Joklik's modified MEM supplemented with 5% horse serum and 2 mM glutamine, and titers were determined by plaque assay with the use of human embryonic kidney 293 cells. Two adenovirus mutants were used in this study: in724 is a splice-donor insertion mutant that overproduces the E3-13.7 protein, and dl753 has an internal deletion of 207 nucleotides in the E3-13.7 ORF and does not produce E3-13.7–related proteins (Tollefson et al., 1990; Hoffman et al., 1992b). Cells were acutely infected with 200–500 plaque-forming units per cell according to established protocols (Hoffman and Carlin, 1994).
The following antibodies were used in this study: adenovirus E3-13.7–specific rabbit peptide antiserum (Hoffman et al., 1990); cathepsin D mouse mAb (Transduction Laboratories, Lexington, KY); human-specific EGF receptor EGF-R1 mouse mAb (Waterfield et al., 1982); EGF receptor rabbit peptide antiserum (Santa Cruz Biotechnology, Santa Cruz, CA); furin convertase rabbit peptide antiserum (Affinity BioReagents, Golden, CO); human-specific interleukin-2 receptor mouse mAb (American Type Culture Collection, Rockville, MD); mouse-specific Lamp-1 1D4B rat mAb (Chen et al., 1985) (Developmental Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa, Iowa City, IA; maintained under National Institutes of Health contract NO1-HD-6-295); Rab 5 mouse mAb (Transduction Laboratories); Rab 7 goat peptide antiserum (Santa Cruz Biotechnology); Rho B rabbit peptide antiserum (Santa Cruz Biotechnology); human-specific Rho B mouse mAb (Santa Cruz Biotechnology); transferrin receptor H68.4 mouse mAb (White et al., 1990) (Zymed Laboratories, San Francisco, CA). Unless noted otherwise, these reagents cross-react with human and mouse proteins. All peptide antisera were affinity-purified.
Surface Reduction of Extracellular Disulfide Bonds
Cells were pulse-labeled for 1 h with l-[35S]cysteine (50 mCi/ml, 1075 Ci/mmol; New England Nuclear Life Sciences, Wilmington, DE) in cysteine-free MEM supplemented with 10% dialyzed FBS and 0.2% BSA and then incubated in chase medium (complete DMEM supplemented with 500 μM nonradioactive cysteine) for 3 h to allow proteins to achieve steady-state localization. To reduce external disulfide bonds of surface proteins, radiolabeled cells were incubated twice (25 min per incubation) with an ice-cold solution of 80 mM l-cysteine, 75 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 0.5 N NaOH, and 1% FBS (Low et al., 1992). Cells were rinsed twice with PBS containing iodoacetamide (1 mg/ml) and then lysed with RIPA detergent (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) in 50 mM Tris, pH 8.0, supplemented with 150 mM NaCl, 2 mM EDTA, 5 mM EGTA, 0.2 mM PMSF, 1 μM leupeptin, and 1 mg/ml iodoacetamide. Immunoprecipitations were carried out with the use of antibodies adsorbed to protein A–Sepharose CL-4B beads (Sigma Chemical, St. Louis, MO). Immunoprecipitates were solubilized in Laemmli buffer supplemented with iodoacetamide (1 mg/ml) and separated by SDS-PAGE (Laemmli, 1970) under reducing (0.1 M DTT) or nonreducing (no DTT) conditions. Gels were treated with En3Hance (New England Nuclear Life Sciences) for fluorography.
Percoll Gradient Cell Fractionation
Cell homogenates were fractionated on Percoll (Pharmacia LKB Biotechnology, Piscataway, NJ) gradients essentially as described by Green et al. (1987). Briefly, cells were rinsed twice with PBS supplemented with 2 mM EDTA and 5 mM EGTA and then scraped in ice-cold homogenization buffer (HB) consisting of 10 mM HEPES, pH 7.5, 0.25 M sucrose, 1 mM EDTA, 0.2 mM PMSF, and 1 μM leupeptin. Cells were collected by centrifugation, resuspended in HB, and homogenized with 22 strokes of a Dounce homogenizer. The homogenate was diluted with an equal volume of fresh HB and centrifuged at 400 × g for 10 min at 4°C to precipitate unbroken cells and nuclei. Postnuclear supernatants were adjusted to a final concentration of 27% Percoll in 0.25 M sucrose with the use of a 90% Percoll stock solution and then layered over a 1-ml sucrose cushion consisting of 10× HB. Gradients were centrifuged for 90 min at 25,000 × g in an SS34 fixed-angle rotor (Sorvall Instruments, Newtown, CT) without braking. All fractions were collected manually starting from the top of the gradient.
Immunoblotting
Membranous organelles were solubilized with RIPA detergent in the presence of protease inhibitors. Solubilized membranes were centrifuged at 100,000 × g for 30 min at 4°C in a TL 100.3 fixed-angle rotor (Beckman Instruments, Palo Alto, CA) to precipitate Percoll and then concentrated in Centricon filters (10,000 molecular weight cutoff; Amicon, Beverly, MA). Equal aliquots from each fraction were resolved by SDS-PAGE and then transferred to nitrocellulose by standard techniques (Towbin et al., 1979). Blots were incubated with primary antibodies and appropriate HRP-conjugated secondary antibodies (Amersham Life Sciences, Arlington, IL; Jackson ImmunoResearch Laboratories, West Grove, PA) for detection by ECL (Amersham Life Sciences). For quantitation, immunoblots were incubated with fluorescein-conjugated secondary antibodies (Amersham Life Sciences), and fluorescent signals exposed in the linear range were scanned with the use of a Fluorimager SI scanner (Molecular Dynamics, Sunnyvale, CA). In some experiments, the fluorescent signal was amplified by a tertiary incubation with anti-fluorescein alkaline phosphatase and a fluorescent substrate (Amersham Life Sciences). Antibody dilutions were chosen experimentally for optimum specific staining.
β-Hexosaminidase Assays
β-Hexosaminidase activity was determined by incubating 10–20 mg of protein from individual Percoll gradient cell fractions diluted in a solution of 0.1 M 2-(N-morpholino)ethanesulfonic acid, pH 6.5, 1 mM p-nitrophenyl-β-d-glucosaminide, and 0.2% Triton X-100 for 90 min at 37°C. The reaction was quenched with 0.5 M glycine, pH 10, and absorbance was read at 405 nm with the use of a model 3550 automatic microplate reader (Bio-Rad Laboratories, Hercules, CA). Protein concentrations were determined by Bradford assay (Bio-Rad Laboratories).
Confocal Laser Scanning Microscopy
Cells grown on coverslips were prepared for staining essentially as described by Chavrier et al. (1990). Briefly, cells were permeabilized with 0.5% β-escin in a solution of 80 mM piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.8, supplemented with 5 mM EGTA and 1 mM MgCl2 for 5 min and then fixed with 3% paraformaldehyde–PBS for 15 min. Cells were stained with primary or secondary antibodies for 1 h or 30 min, respectively, at 37°C. Antibodies were diluted in a solution containing 0.5% β-escin and 3% radioimmunoassay-grade BSA and were blocked with a solution containing 5% normal serum from the host animal used to generate the secondary antibody between incubations with primary and secondary antibodies. Fluorophore-conjugated, species-specific secondary antibody Fab fragments that had been solid-phase absorbed to prevent cross-reactivity with primary antibodies made in other species were purchased from Jackson ImmunoResearch Laboratories. Cells were examined with a Zeiss (Göttingen, Germany) LSM 410 scanning laser confocal microscope with the use of the 488/568-nm wavelength lines of an argon–krypton laser. Some cells were optically sectioned every 0.5 μm. Image resolution with the use of a Zeiss 100x Plan-Neofluor oil objective and Zeiss LSM software was 512 × 512 pixels.
Cryosectioning and Immunoelectron Microscopy
Immunogold labeling of ultrathin cryosections was carried out essentially as described (McCaffery and Farquhar, 1995). Briefly, cells were fixed in either periodate-lysine-paraformaldehyde (75 mM phosphate buffer containing 2% formaldehyde, 70 mM lysine, and 10 mM sodium periodate, pH 6.2) for 4 h at room temperature or in 4–8% paraformaldehyde diluted in PBS, pH 7.4, for 1 h at room temperature. The cells were washed, scraped, and collected by low-speed centrifugation, and cell pellets were incubated in 2.3 M sucrose containing 20% polyvinylpyrrolidone for 1 h for cryoprotection. Cryoprotected pellets were mounted on aluminum cryopins and then frozen in liquid N2. Ultrathin cryosections were cut on a Reicherlt (Heidelberg, Germany) Ultracut T microtome equipped with an FCS cryostage, and sections were collected onto 300-mesh, formvar/carbon-coated nickel grids. Grids were washed by passing through several drops of PBS supplemented with 2.5% FCS and 0.01 M glycine, pH 7.4. Grids were blocked in 10% FCS and incubated overnight with mixed primary antibodies, each at ∼10 μg/ml. After washing, grids were incubated with mixed secondary antibody–gold conjugates (Jackson ImmunoResearch Laboratories) diluted 1:50 for 2 h. The grids were washed several times, first with PBS and then with double-distilled H2O. Grids were embedded in a mixture containing 3.2% polyvinyl alcohol (molecular weight 10,000), 0.2% methyl cellulose (400 cps), and 0.2% uranyl acetate. The sections were observed and photographed on a Philips (Eindhoven, the Netherlands) EM 410 transmission electron microscope. Some cells were preincubated with HRP (1 mg/ml; Sigma Chemical) for 30 min before fixation to facilitate identification of early endocytic compartments.
Determination of Metabolic Half-Lives
Cells were rinsed twice and then preincubated for 1 h with cysteine-free MEM. Cells were pulse-labeled for 30 min with l-[35S]cysteine and then incubated in chase medium exactly as described above. Cells were lysed with RIPA detergent at various times starting at 3 h for immunoprecipitation, SDS-PAGE, and phosphorstorage autoradiography. Digitized images were analyzed with the use of the ImageQuant software package (Molecular Dynamics), which averages five measurements of light emission for each pixel location, to give a pixel value proportional to the amount of stored radiation. The percentage of radioactivity remaining compared with the 3-h chase time was plotted as a function of time on a semilog scale, and protein half-lives were calculated by linear regression analysis. Serial dilutions of total radioactivity used for metabolic labeling were also quantitated by phosphorimaging to generate a standard curve for converting arbitrary phosphorimage units to microcuries of radioactivity. Phosphorimage units obtained from quantitation of radioactive immunoprecipitates were then converted to the total number of molecules labeled with the use of the following equations: microcuries of radioactivity (1)× specific activity of isotope = picomoles of labeled residues picomoles of labeled residues (2)÷ number of residues per molecule = picomoles of protein picomoles of protein (3) ÷ (1 × 1012) = total number of molecules labeled
Some cells were labeled with [35S]cysteine and others with 35S-Express Protein Labeling Mix (∼73% methionine and 27% cysteine). Specific activities of cysteine and methionine were 1075 and 1175 Ci/mmol, respectively.
Coimmunoprecipitation
Infected cells were metabolically labeled with l-[35S]cysteine from 15 to 18 h after infection and then fractionated on Percoll gradients exactly as described above. Membranous organelles enriched for E3-13.7 (fraction 2; see Figure 3) were solubilized with an equal volume of 2× CHAPS lysis buffer (40 mM 3-([3-chloramidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate, 20 mM Tris, pH 6.8, 1 M NaCl, 10 mM EDTA, 10 mM EGTA, 2 mM iodoacetamide) supplemented with protease inhibitors for 2 h on ice. Samples were clarified by centrifugation, precleared for 1 h at 4°C with formalin-fixed Staphylococcus aureus cells (Calbiochem-Novabiochem, La Jolla, CA) preadsorbed with 1% BSA, and then immunoprecipitated overnight at 4°C with the use of antibodies adsorbed to protein A–Sepharose CL-4B. Immunoprecipitates were washed five times with 1× CHAPS lysis buffer, solubilized with Laemmli buffer, and resolved by SDS-PAGE for fluorography and phosphorstorage autoradiography.
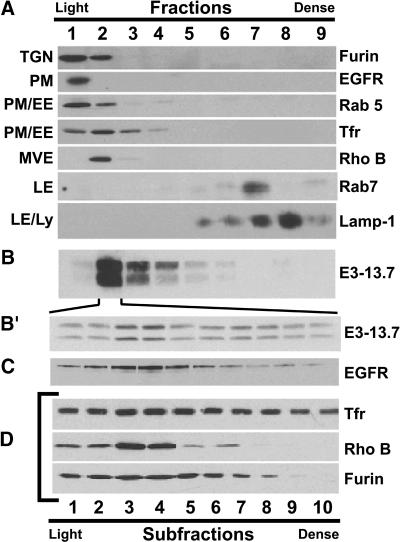
Figure 3.
E3-13.7 colocalizes with markers for early endosomes and MVEs. Postnuclear supernatants were fractionated on 27% Percoll density gradients, and fractions were immunoblotted for organelle-specific marker proteins listed in the figure. (A) Postnuclear supernatants from uninfected mouse cells were immunoblotted for organelle-specific marker proteins. (B and B′) Postnuclear supernatants from infected ERwt cells were immunoblotted for E3-13.7 in a total of 9 fractions from the original gradient (B) and in 10 subfractions obtained from fraction 2 (B′). (C and D) Postnuclear supernatants from infected ERwt cells were immunoblotted for EGF receptor (C) or organelle markers (D) in 10 subfractions obtained from fraction 2. TGN, trans-Golgi network; PM, plasma membrane; EE, early endosome; MVE, multivesicular endosome; LE, late endosome; Ly, lysosome.
RESULTS
Expression of Adenovirus E3-13.7 Proteins in Mouse Cell Models
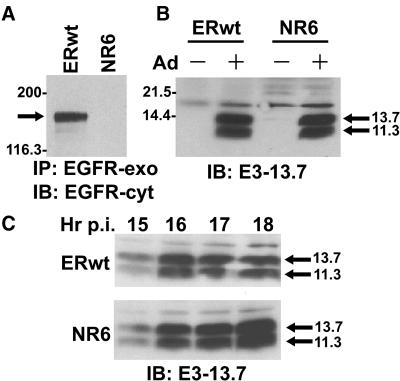
Two of the major goals of this study were to characterize E3-13.7 subcellular localization and to define the molecular requirements for E3-13.7–mediated EGF receptor down-regulation. Hence, many studies were carried out in mouse cell lines that either lack endogenous EGF receptors (NR6 cells) or that stably express human EGF receptor cDNAs under the control of a CMV promoter. For example, ERwt cells express wild-type human EGF receptors (Figure 1A). This cell line readily expresses early viral proteins after infection with human adenoviruses (Figure 1B), exhibiting peak E3-13.7 protein expression from 15 to 18 h after infection (Figure 1C). Because the CMV promoter is up-regulated by the adenovirus E1A protein (Gorman et al., 1989), steady-state EGF receptor protein levels are high in adenovirus-infected ERwt cells even though surface receptors undergo E3-13.7–mediated down-regulation (Hoffman et al., 1993). This is in contrast to cells expressing only endogenous receptors, in which steady-state EGF receptor levels are not maintained by replacement synthesis (Carlin et al., 1989). Hence, ERwt cells provide a comparatively rich source of EGF receptor for biochemical analysis during adenovirus infection.
Figure 1.
Characterization of mouse cell lines. (A) ERwt (EGF receptor-positive) and NR6 (EGF receptor-negative) cells were lysed and immunoprecipitated with a human-specific EGF receptor mAb recognizing an exocytic amino acid epitope (EGFR-exo), followed by immunoblotting with an EGF receptor rabbit peptide antiserum to a cytosolic epitope (EGFR-cyt). (B) Cells were mock-treated (−) or infected with adenovirus (+) and harvested 18 h later. (C) Adenovirus-infected cells were harvested 15–18 h after infection (p.i.). (B and C) Total cellular protein (∼80 μg) resolved by SDS-PAGE was immunoblotted with an E3-13.7–specific rabbit peptide antiserum. Molecular weight standards: myosin, 200,000; β-galactosidase, 116,300; soybean trypsin inhibitor, 21,500; lysozyme, 14,400.
E3-13.7 Is Concentrated in the Cell Interior at Steady State
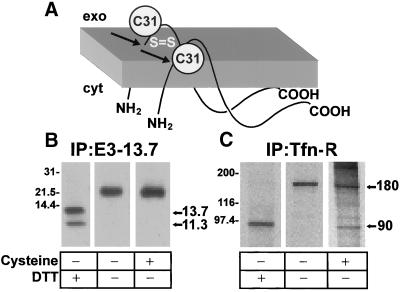
We first determined E3-13.7 steady-state distribution between the plasma membrane and the cell interior. E3-13.7 exists in two molecular weight forms: one corresponding to the full-length molecule, which has two membrane-spanning α-helices and cytosolic N and C termini (Figure 2A); and a lower-molecular-weight species lacking the N-terminal α-helix, which is cleaved by signal peptidase in the endoplasmic reticulum (Figure 2A, arrows) (Hoffman et al., 1992a; Krajcsi et al., 1992). Endoplasmic reticulum processing is incomplete, however, and both molecular weight species are routinely seen in adenovirus-infected cells and cells transfected with E3-13.7 expression vectors (Hoffman et al., 1990, 1992b; Tollefson et al., 1990). Sequences connecting the α-helices should be exposed at the cell exterior in either form of the molecule located at the plasma membrane. However, this region cannot be labeled by conventional surface biotinylation, because it lacks residues with free amino groups. Therefore, we took advantage of the fact that both E3-13.7 molecular weight species form disulfide-linked dimers at cysteine residue 31 (C31 in Figure 2A) (Hoffman et al., 1992a). Hence, disulfide bonds on proteins located at the cell surface should be reduced if cells are exposed to an external reducing agent such as cysteine, whereas intracellular proteins should be protected (Bretscher, 1989; Low et al., 1992). Infected cells were metabolically labeled, incubated with cysteine-containing medium, and then lysed in the presence of iodoacetamide to prevent de novo disulfide bond formation. When E3-13.7 immunoprecipitates were resolved under nonreducing conditions (i.e., in the absence of DTT), metabolically labeled proteins migrated as higher-molecular-weight dimers, regardless of whether or not cells were exposed to external cysteine (Figure 2B). This was in contrast to disulfide-linked transferrin receptors, which were partially reduced by external cysteine (Figure 2C), consistent with ∼20–45% of this molecule exhibiting plasma membrane localization, with the remainder in endocytic compartments (Klausner et al., 1983; Weiel and Hamilton, 1984). These data suggest that the majority of E3-13.7 is located intracellularly at steady state.
Figure 2.
The majority of E3-13.7 is protected from cell surface reduction at steady state. (A) Cartoon showing the membrane topology of E3-13.7 proteins (Hoffman et al., 1992a; Krajcsi et al., 1992). There are two distinct molecular mass species, a full-length 13.7-kDa protein and an 11.3-kDa protein cleaved cotranslationally at Ala-23 (arrows). Both species form disulfide-linked dimers at cysteine residue 31 (C31). (B and C) Infected cells were metabolically labeled for 1 h followed by a 3-h chase in nonradioactive medium to allow labeled proteins to achieve steady-state distribution. Some cells were incubated with a solution containing cysteine to reduce surface proteins with external disulfide bonds. Cell lysates were immunoprecipitated for E3-13.7 proteins (B) or the transferrin receptor (C). Immunoprecipitates were resolved by SDS-PAGE under reducing (+DTT) or nonreducing (−DTT) conditions. Sizes of reduced and nonreduced protein species are indicated to the right of each panel. E3-13.7 dimers have approximate molecular weights of 21,000–23,400 (Hoffman et al., 1992a). Additional molecular weight standards: phosphorylase B, 97,400; carbonic anhydrase, 31,000.
E3-13.7 Proteins Cosediment with Early Endosome and MVE Markers after Percoll Gradient Cell Fractionation
We sought to characterize the intracellular distribution of E3-13.7 by cell fractionation. This was achieved by first determining the location of organelle-specific markers after fractionation of postnuclear supernatants on a 27% isoosmotic Percoll gradient (Figure 3A). Fractions were immunoblotted for furin convertase (trans-Golgi network [TGN]) (Chapman and Munroe, 1994), the EGF receptor (plasma membrane), Rab 5 (plasma membrane and early endosomes) (Chavrier et al., 1990), transferrin receptor (plasma membrane and early endosomes) (Klausner et al., 1983; Lamb et al., 1983; Weiel and Hamilton, 1984), Rho B (MVEs) (Robertson et al., 1995), Rab 7 (late endosomes) (Feng et al., 1995), and Lamp-1 (late endosomes and lysosomes) (Lippincott-Schwartz and Fambrough, 1987) (Figure 3A). Fractions were also analyzed for β-hexosaminidase activity, a marker for lysosomes (our unpublished data). Based on marker distribution, we concluded that plasma membranes were concentrated in fraction 1, early endosomes in fractions 1–4, late endosomes in fractions 7 and 8, and lysosomes in fractions 8 and 9. With the exception of the EGF receptor (Figure 3C), marker protein distribution was identical in uninfected (Figure 3A) and infected (our unpublished data) cells.
When gradient fractions from adenovirus-infected cells were analyzed for E3-13.7, viral proteins were highly enriched in the low-density fraction 2, with smaller amounts in fractions 3–6 (Figure 3B). The absence of E3-13.7 in fraction 1 enriched for plasma membrane is consistent with the surface reduction results shown in Figure 2. Because low-density fraction 2 is enriched for several distinct organelle markers (Figure 3A), it was further resolved into 10 subfractions to better characterize E3-13.7 steady-state localization. As shown in Figure 3B′, E3-13.7 was detected in all 10 of the subfractions, but it was clearly enriched in subfractions 3 and 4. EGF receptors undergoing down-regulation in the same cell showed a similar distribution (Figure 3C). Based on the distribution of marker proteins (Figure 3D), transferrin receptor–positive early endosomes were distributed throughout all 10 subfractions, with modest enrichment in subfractions 3–6; Rho B–positive MVEs were distributed in subfractions 1–6, with substantial enrichment in subfractions 3 and 4; and furin-positive TGN was evenly distributed in subfractions 1–6, with smaller amounts present in subfractions 7 and 8. Together, these data suggest that E3-13.7 is localized to transferrin receptor–positive endosomes and Rho B–positive MVEs. Because MVEs originate from early sorting endosomes (Aniento et al., 1993), Rho B–positive MVEs may represent distinct vesicles or regions still attached to early endosomes.
E3-13.7 Proteins Localize to Early Endosomes and MVEs by Immunocytochemistry
Although the cell fractionation results suggested that E3-13.7 was localized primarily in early endocytic compartments, we could not exclude the possibility that it was also a resident TGN protein because of its cosedimentation with the TGN marker furin (Figure 3). Therefore, we also determined steady-state intracellular location by single-label confocal laser scanning microscopy (CLSM) analysis. Because of the host animals used to raise primary antibodies to organelle-specific marker proteins, these studies were carried out in adenovirus-infected human A549 cells to facilitate the dual-label studies described below. In contrast to the mouse cell lines, A549 cells do not replenish EGF receptors after E3-13.7–mediated down-regulation (Carlin et al., 1989). Cells were either mock-treated or infected with an E3-13.7–positive adenovirus and then fixed and stained 8 h later. Some cells were also treated with cycloheximide from 5.5 to 8 h after infection. Although resident Golgi/TGN membrane proteins will remain at that location in the presence of cycloheximide, itinerant proteins en route to other organelles will not (Luzio et al., 1990). E3-13.7 exhibits a punctate cytoplasmic staining pattern in the cycloheximide-treated infected cells (our unpublished data). Infected cells also exhibit punctate cytoplasmic staining in the absence of cycloheximide, but with additional Golgi/TGN staining, as would be expected during the early stages of an adenovirus infection (our unpublished data). Mock-infected cells were negative for E3-13.7 staining with or without (our unpublished data) cycloheximide treatment. These results indicate that E3-13.7 is not a resident Golgi/TGN protein.
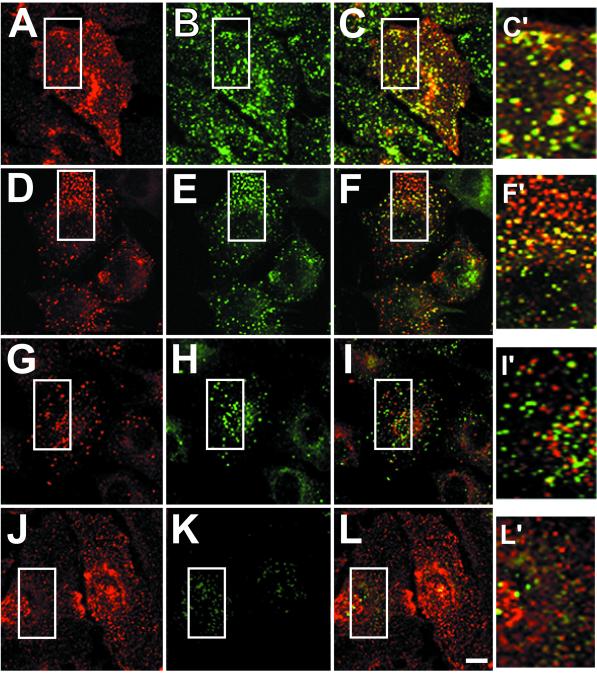
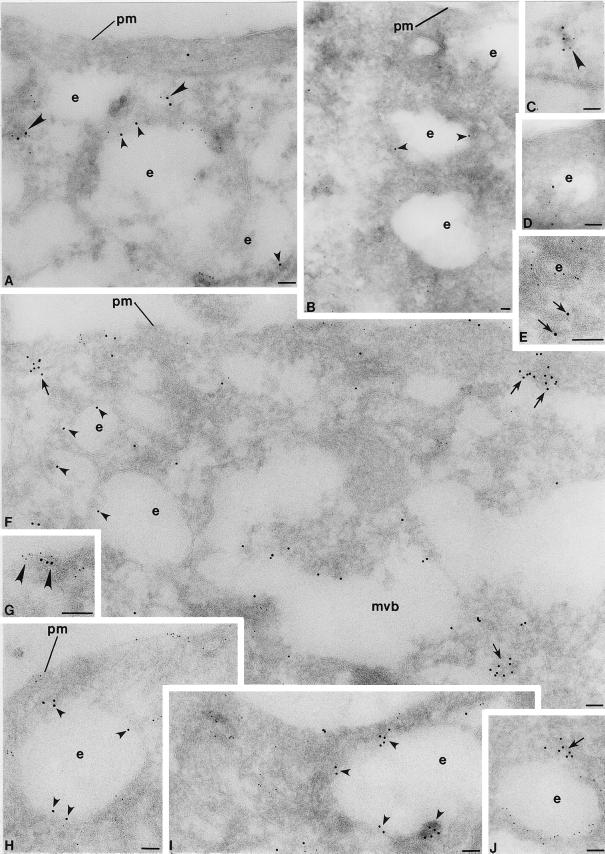
Infected cells that had not been treated with cycloheximide were then costained for E3-13.7 and endocytic markers (Figure 4). E3-13.7 exhibits extensive colocalization with both the transferrin receptor and Rho B (see “yellow” superimposed images in Figure 4, C′ and F′, respectively). In contrast, E3-13.7 has very limited overlap with Rab 7 (Figure 4I′) and essentially no overlap with the soluble lysosomal hydrolase cathepsin D (Figure 4L′), a major ligand for the cation-independent mannose 6-phosphate receptor (CI-M6PR), which is also a marker for mature MVEs (Hirst et al., 1998). Steady-state localization of E3-13.7 to early endocytic compartments was confirmed by immunoelectron microscopy analysis of ultrathin cryosections (Figure 5). Some of the infected cells were preincubated for 30 min with HRP to facilitate identification of early endosomes (Figure 5, A–D). Ultrathin cryosections were then costained for E3-13.7 and HRP, followed by colloidal gold–conjugated secondary antibodies. E3-13.7 was clearly localized to peripheral endosomes (Figure 5, A–D, small arrowheads), some of which were also positive for HRP taken up during the 30-min pulse (Figure 5, A and C, large arrowheads). Figure 5F (discussed below) shows a clear example of E3-13.7 localization to an MVE-limiting membrane. Together with the cell fractionation data, multiple immunocytochemical analyses show that E3-13.7 proteins are located primarily in peripheral early endosomes and Rho B–positive, CI-M6PR–negative MVEs at steady state.
Figure 4.
E3-13.7 colocalization with endocytic marker proteins. Adenovirus-infected A549 cells were permeabilized and fixed at 8 h after infection and then dual-stained for E3-13.7 proteins with a rabbit peptide antiserum and antibodies to transferrin receptor (A–C), Rho B (D–F), Rab 7 (G–I), or cathepsin D (J–L). E3-13.7 is red in A, D, G, and J. Transferrin receptor, Rho B, Rab 7, and cathepsin D are green in B, E, H, and K, respectively. Red and green channels were merged after both fluorescent signals were adjusted to similar levels (C, F, I, and L), and “yellow” indicates the overlap of red and green fluorescence. Insets from merged images are enlarged on the right (C′, F′, I′, and L′). Areas corresponding to the enlarged insets are boxed in all of the panels. All images constitute individual sections from the middle of the cell. Bar, 10 μm.
Figure 5.
Analysis of E3-13.7 distribution by ultrathin cryosectioning and immunoelectron microscopy. (A–D) Infected ERwt cells were incubated with HRP (1 mg/ml) for 30 min before fixation and cryopreservation. Sections were costained for E3-13.7 (10-nm gold particles) and HRP (5-nm gold particles). (E–J) Infected ERwt cells were costained for E3-13.7 (10-nm gold particles) and EGF receptor (5-nm gold particles). Large arrowheads, colocalized 5-nm and 10-nm gold particles; small arrowheads, E3-13.7 localized to peripheral endosomes. In F–J, arrows indicate lack of E3-13.7 colocalization with EGF receptor. e, endosome; mvb, multivesicular body; pm, plasma membrane. Bars, 0.1 μm.
E3-13.7 and EGF Receptors Are Transiently Located in the Same Early Endocytic Compartments, Where They Form a Physical Complex
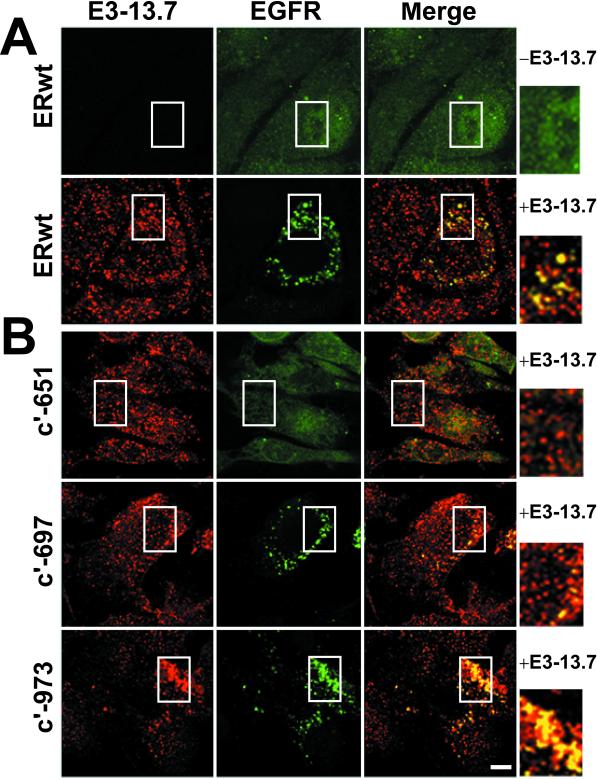
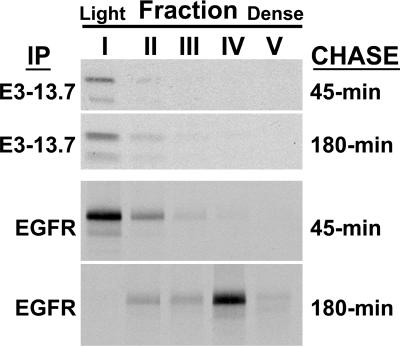
Cell fractionation data suggesting that E3-13.7 colocalizes with EGF receptors undergoing down-regulation (Figure 3, B′ and C) were confirmed in ERwt cells with the use of dual-label CLSM (Figure 6A; note that results in Figure 6B are discussed in a later section). Whereas EGF receptors were located primarily at the plasma membrane in cells infected with an E3-13.7–negative virus (Figure 6A, top three panels), a portion of the receptors undergoing down-regulation in infected cells colocalized with E3-13.7–positive intracellular vesicles (see “yellow” superimposed images in bottom panels). Not all of the EGF receptor vesicles colocalized with E3-13.7, however, consistent with the idea that colocalization is transient. Similar results were obtained when cells were costained for E3-13.7 and EGF receptors in ultrathin cryosections (Figure 5, E–J). Although the molecules were colocalized in some images (Figure 5G, large arrowheads), often they were not, even when they were apparently present in the same endocytic compartment (Figure 5J, arrow). These same studies also confirmed that in contrast to EGF receptors, E3-13.7 frequently was not present at the plasma membrane (Figure 5H). The hypothesis that E3-13.7 and EGF receptors colocalize transiently was further tested with the use of Percoll gradient cell fractionation to track metabolically labeled immunoprecipitates. ERwt cells were pulse-labeled for 15 min, incubated with excess nonradioactive amino acids for 45 or 180 min, and harvested for Percoll gradient cell fractionation. We found it necessary to collect five fractions (labeled I to V in Figure 7) instead of nine, as we had in previous experiments, to detect low-abundance pulse-labeled molecules. Preliminary studies with organelle markers showed that plasma membrane and early endosomes were concentrated in fractions I and II and that late endosomes and lysosomes were concentrated in fractions IV and V (our unpublished data). EGF receptors were concentrated in fraction I, corresponding to plasma membrane/early endosomes at both time points in mock-treated cells (our unpublished data), as was E3-13.7 in adenovirus-infected cells (Figure 7, top panels). In contrast, metabolically labeled EGF receptors were located in fraction I at the 45-min time point but had shifted to fraction IV, enriched for late endosomes/lysosomes, by the 180-min time point in adenovirus-infected cells (Figure 7, bottom panels).
Figure 6.
Intracellular EGF receptors colocalize with E3-13.7. (A) ERwt cells infected with E3-13.7–negative or E3-13.7–positive adenoviruses were stained with a rabbit polyclonal antibody to E3-13.7, a mouse mAb to an external EGF receptor epitope, and corresponding secondary Fab fragments conjugated to rhodamine (donkey anti-rabbit) or fluorescein (goat anti-mouse). Red and green channels were merged after both fluorescent signals were adjusted to similar levels, and “yellow” indicates the overlap of red and green fluorescence. All images constitute individual sections from the middle of the cell. (B) NR6 cell lines expressing cytoplasmically truncated human EGF receptors listed to the left were infected with an E3-13.7–positive adenovirus and then costained for E3-13.7 and EGF receptor exactly as described in A. Insets from merged images are enlarged on the right. Areas corresponding to the enlarged insets are boxed in all of the panels. Bar, 10 μm.
Figure 7.
Metabolically labeled E3-13.7 proteins and EGF receptors follow divergent pathways subsequent to early endosomes. ERwt cells that had been infected with in724 were pulse-labeled for 15 min at 15 h after infection, incubated in chase medium for 45 min or 180 min, and then processed for Percoll gradient cell fractionation. Five fractions (I–V) were collected starting from the top of the gradient. Membranes were solubilized with RIPA detergent and sequentially immunoprecipitated with antibodies to E3-13.7 and the EGF receptor. Immunoprecipitates were resolved by SDS-PAGE and detected by fluorography.
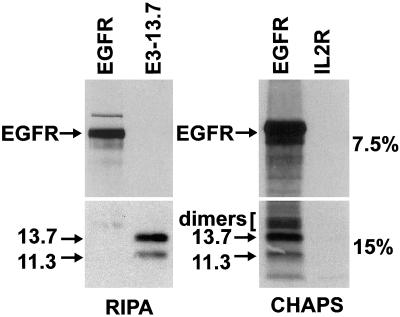
To evaluate the likelihood that E3-13.7 physically interacts with the EGF receptor in an endocytic compartment, we carried out coimmunoprecipitation studies with the use of solubilized membrane fractions enriched for both molecules isolated from metabolically labeled ERwt cells. Membrane fractions were solubilized with mildly denaturing RIPA detergent (Figure 8, left panels) or the zwitterionic detergent CHAPS, which is often used to preserve membrane protein interactions without promoting nonspecific hydrophobic aggregation (Figure 8, right panels). Samples were prepared with the use of buffers containing 500 mM NaCl to prevent nonspecific electrostatic interactions. Cell lysates were then immunoprecipitated with EGF receptor and E3-13.7–specific antibodies, and immunoprecipitates were resolved on 7.5% gels for EGF receptor detection or 15% gels to detect E3-13.7 proteins. Neither of the antibodies coimmunoprecipitated a protein partner from the RIPA detergent cell lysates (Figure 8) or from lysates made with nonionic detergents such as Triton X-100 or NP-40 (our unpublished data). However, both molecular weight forms of the viral protein were coimmunoprecipitated from CHAPS detergent lysates with an antibody to the EGF receptor, but not with an irrelevant isotype-matched antibody (to the interleukin-2 receptor) (Figure 8). Some of the coprecipitating viral protein also migrated as disulfide-linked dimers, suggesting inefficient reduction by DTT. Although EGF receptor coimmunoprecipitation with E3-13.7 was not feasible because of low E3-13.7 immunoprecipitation efficiency from CHAPS lysates (our unpublished data), these data nevertheless suggest that colocalized EGF receptor and E3-13.7 form a specific protein complex. Quantitation of coimmunoprecipitating proteins by phosphorimaging, taking into account the number of radiolabeled cysteine residues per molecule, suggests a molar ratio of E3-13.7 to EGF receptor of ∼1:4. This ratio must be interpreted with caution, because the E3-13.7/EGF receptor interaction affinity may be low and not well preserved, or a majority of E3-13.7 may be insoluble, in CHAPS detergent. Nevertheless, these data suggest that only a fraction of E3-13.7 is associated with the EGF receptor at any given time, consistent with transitory protein–protein interactions.
Figure 8.
E3-13.7 enriched in early endosomes coimmunoprecipitates with the EGF receptor. Postnuclear supernatants from adenovirus-infected ERwt cells that had been metabolically labeled from 15 to 18 h after infection were fractionated on 27% Percoll gradients. Fraction 2 membranes enriched for E3-13.7 and the EGF receptor were solubilized with RIPA or CHAPS detergent, and lysates were immunoprecipitated with the use of antibodies to the proteins listed at the top of the figure. One-half of each immunoprecipitate was resolved on a 7.5% (top panels), and the other on a 15% (bottom panels), polyacrylamide gel for detection by fluorography. Note that autoradiograms of EGF receptor immunoprecipitates were exposed for 2 d, compared with 14 d for E3-13.7 immunoprecipitates.
E3-13.7 and EGF Receptor Exhibit Distinct Metabolic Half-Lives in Adenovirus-infected Cells
The data presented thus far suggest that E3-13.7 is a resident early endocytic membrane protein that is not transported efficiently to lysosomes. This hypothesis was tested by comparing rates of E3-13.7 and EGF receptor protein degradation. Infected cells that had been labeled with 35S-labeled amino acids for 1 h were incubated in chase medium for 3 h to allow for steady-state localization and then harvested at intervals during the next 4.5 h for recovery of radiolabeled proteins by immunoprecipitation. In addition to the E3-13.7–positive in724 virus, some cells were infected with the E3-13.7–negative dl753 virus to verify E3-13.7–mediated EGF receptor degradation. Immunoprecipitates were quantitated by phosphorimaging to determine metabolic half-lives by linear regression analysis (our unpublished data). We determined relative rates of protein turnover (Table 1) after converting arbitrary units from the phosphorimage analysis to total number of molecules labeled (see MATERIALS AND METHODS) to normalize for differences in the initial amount of each protein. These calculations assume similar labeling and immunoprecipitation efficiencies for E3-13.7 and the EGF receptor. As expected, EGF receptors in in724-infected cells exhibited faster turnover kinetics than receptors in dl753-infected cells (249 versus 106 molecules/min, respectively, in the experiment described in Table 1). The metabolic turnover of E3-13.7 proteins was substantially slower than that of EGF receptors undergoing down-regulation in the same cell (59 versus 249 molecules/min, respectively). Hence, the viral protein does not appear to be transported to lysosomes for degradation at the same rate as the EGF receptor.
Table 1.
E3-13.7 and EGF receptor protein turnover kinetics
| E3-13.7 | EGF receptor | EGF receptor | |
|---|---|---|---|
| Infecting virusa | in724 | in724 | dl753 |
| Protein half-life (h)b | 5.3 | 2.9 | 21.1 |
| Turnover rate (molecules/min)c | 59 | 249 | 106 |
ERwt cells were infected with in724, which overexpresses E3-13.7, or dl753, which does not produce E3-13.7.
From linear regression analysis of metabolically labeled protein decay plots (our unpublished data). Values in the table were derived from a single experiment. Similar values were obtained for each molecule in at least two independent experiments.
Calculation made by converting arbitrary phosphorimage units to total number of molecules labeled (see MATERIALS AND METHODS).
Effect of EGF Receptor on E3-13.7 Steady-State Localization
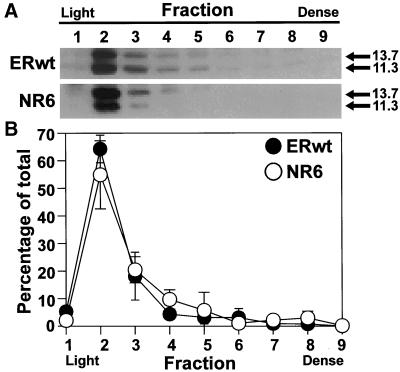
Our data also led us to hypothesize that E3-13.7 should localize to early endosomes independent of EGF receptor expression. This hypothesis was tested by comparing the steady-state distribution of E3-13.7 on Percoll gradients in ERwt cells versus NR6 cells. The E3-13.7 proteins exhibited the same relative distribution in both of the mouse cell lines (Figure 9A), consistent with our hypothesis. When E3-13.7 signals were quantitated by enhanced chemifluorescence, 64.2 ± 3.5% and 54.9 ± 8.7% of total E3-13.7 protein was found in fraction 2 in ERwt cells and NR6 cells, respectively (Figure 9B). In contrast, only 2–5% of total E3-13.7 was detected in fraction 1 enriched for plasma membrane in either of the mouse cell lines.
Figure 9.
Effect of EGF receptor expression on the steady-state distribution of E3-13.7 proteins. Postnuclear supernatants from in724-infected ERwt or NR6 cells were fractionated on 27% Percoll density gradients, and fractions were processed for immunoblotting of E3-13.7 immunoprecipitates. Immunoblots were developed by ECL (A) or by chemifluorescence (B). Fluorescence in the linear range was scanned and quantitated with the use of ImageQuant software from Molecular Dynamics. Data in B are presented as SEM ± SD, n = 3; symbols obscure some SEM bars. ●, ERwt cells; ○, NR6 cells.
EGF Receptor Cytoplasmic Residues 675–697 Are Required for E3-13.7–mediated Down-Regulation
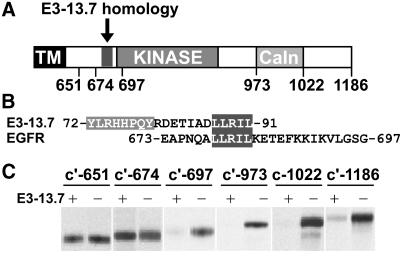
To determine the molecular basis of E3-13.7–mediated EGF receptor down-regulation, stable NR6 cell lines expressing cytoplasmically truncated EGF receptors (Figure 10A) were infected with E3-13.7–positive or E3-13.7–negative viruses and then labeled metabolically to determine receptor protein stability. As shown in Figure 10C, receptors with truncations in the C terminus (c′-1022 and c′-973) as well as near the juxtamembrane domain–kinase catalytic core border (c′-697) underwent E3-13.7–mediated down-regulation. In contrast, the protein stability of receptors with truncations in the juxtamembrane domain (c′-674 and c′-651) was unaffected by E3-13.7 expression. Similar results were obtained by dual-label CLSM (Figure 6B), in which c′-697 and c′-973 but not c′-651 receptors colocalized with E3-13.7 in internal vesicles. These data suggest that EGF receptor residues 675–697 are essential for E3-13.7–mediated down-regulation. Interestingly, this region contains a leucine-based sorting signal (679-LL) implicated in ligand-induced endosome-to-lysosome transport of EGF receptors (Kil et al., 1999; Kil and Carlin, 2000) that is also part of a motif conserved in the cytosolic tail of the viral protein (highlighted in Figure 10B).
Figure 10.
EGF receptor cytoplasmic residues 675–697 are required for E3-13.7–mediated down-regulation. (A) Cartoon showing the EGF receptor transmembrane (TM) domain and the locations of a region bearing homology to E3-13.7, the kinase catalytic core, and the calcium response and internalization (CaIn) domain in the cytoplasmic tail. Locations of cytoplasmic truncations encoded by cDNAs with premature stop codons are also indicated. (B) Homologous sequences located at the E3-13.7 C terminus and the EGF receptor juxtamembrane domain are shown in the single-letter code and highlighted in dark gray. Locations of additional consensus tyrosine-based sorting signals in E3-13.7 (72-YLRH and 76-HPQY) are highlighted in light gray. (C) NR6 cell lines expressing each of the EGF receptor proteins indicated in the figure were infected with adenoviruses with (+) or without (−) an intact E3-13.7 ORF. Cells were metabolically labeled for 1 h, incubated in chase medium for 6 h, and harvested for immunoprecipitation with a human EGF receptor antibody specific for an external epitope. Immunoprecipitates were resolved by SDS-PAGE for fluorography.
DISCUSSION
The model that best fits our data is summarized as follows. In the absence of EGF receptor expression, E3-13.7 is selectively retained in early endosomes and MVE-limiting membranes. In cells expressing the EGF receptor, E3-13.7 physically engages receptors undergoing constitutive recycling in an early endocytic compartment. This either links EGF receptors to an intrinsic MVE-sorting signal located in the viral protein or induces a conformational change exposing a cryptic MVE-sorting signal in the EGF receptor. Viral protein–EGF receptor complexes then dissociate, perhaps as they encounter progressively lower luminal pH. Whereas EGF receptors proceed to lysosomes, where they are degraded, E3-13.7 is either retained or retrieved to early endosomes, where it may then be reused.
What, then, is the nature of the putative MVE-sorting signal proposed by this model? Data obtained with cells expressing cytoplasmically truncated EGF receptors suggest that leucine-based motifs conserved in both molecules act cooperatively to link inactive EGF receptors to the lysosomal sorting machinery. Besides the critical leucine residues (Figure 10B), both molecules have N-terminal acidic residues (Glu-673 in the EGF receptor and Arg-82 in E3-13.7) that are also thought to play a key role in leucine motif–dependent sorting events (reviewed by Sandoval and Bakke, 1994). In addition to the fact that the E3-13.7 C terminus containing such a signal is necessary for its activity (Carlin et al., 1989), a corresponding signal located in the EGF receptor juxtamembrane domain is also necessary for the efficient transport of ligand–receptor complexes to lysosomes (Kil et al., 1999; Kil and Carlin, 2000). Because E3-13.7 is a disulfide-linked dimer (Hoffman et al., 1992a), the leucine motif could be presented as a higher-order oligomer in viral protein–EGF receptor protein complexes. Hence, as in other leucine-type signals, sorting activity may be regulated by signal multimerization (Arneson and Miller, 1995). Leucine motifs located in other molecules have also been reported to bind selectively to the clathrin adaptor complexes AP-1 and AP-3 (Rapoport et al., 1998; Vowels and Payne, 1998), providing a possible molecular basis for regulation of some of transport steps mediated by this type of signal.
The fact that E3-13.7 and the EGF receptor ultimately follow divergent sorting pathways suggests that each molecule has additional unique sorting signals. For example, our model predicts that E3-13.7 has signals that mediate endosome retention and/or retrieval. Although their identity is not currently known, consensus tyrosine-based signals also located in the E3-13.7 cytoplasmic tail (Figure 10B) are reasonable candidates (Vinogradova et al., 1998 ). These same signals probably mediate TGN-to-early endosome biosynthetic transport as well. Although our data suggest that newly synthesized E3-13.7 is delivered directly to early endosomes, we cannot exclude the possibility that biosynthetic transport involves an obligatory cell surface intermediate. Such an intermediate would be extremely difficult to detect if it is rapidly internalized and then efficiently retained in endosomes. The existence of transitory cell surface intermediates for other integral membrane proteins targeted to the endocytic pathway, such as lysosomal glycoproteins, usually requires high levels of protein expression. In those cases, the possibility that cell surface expression is an artifact of saturation of other specific transport steps has yet to be resolved (Williams and Fukada, 1990; Harter and Mellman, 1992; Mathews et al., 1992). Hence, although it is possible to detect small amounts of E3-13.7 at the cell surface when it is substantially overexpressed (Hoffman et al., 1992a), the physiological significance of this observation remains an open question.
Although the continued transport of the EGF receptor to lysosomes after dissociation of the viral protein–receptor complex could occur by a default mechanism, in all likelihood it is also signal-mediated. In addition to the leucine signal described above, two additional lysosomal sorting signals have been mapped to the carboxyl half of the EGF receptor cytoplasmic tail, including a region that binds the SNX-1–sorting nexin (Kornilova et al., 1996; Kurten et al., 1996). According to our model, one of these signals may mediate transport of EGF receptors following their incorporation into forming MVEs. MVEs undergo a remarkable maturation process in response to a variety of stimuli, resulting in an overall increase in size as well as in the number of internal vesicles (reviewed by Gu and Gruenberg, 1999). Membrane trafficking in MVEs is clearly highly regulated, because membrane proteins are differentially distributed within these compartments. The transferrin receptor, for example, is found on the limiting membrane of relatively immature MVEs (Hopkins, 1983), the CI-M6PR is enriched in MVEs with an intermediate degree of maturation (Hirst et al., 1998), and Lamp I is found predominantly in mature MVEs (van Deurs et al., 1993). In addition, selected cargo, including ligand-occupied EGF receptors (Miller et al., 1986), is sequestered in internal vesicles. Hence, EGF receptor down-regulation likely depends on several discrete sorting events activated during MVE maturation. Because ligand-occupied EGF receptors also accumulate in the internal vesicles of MVEs en route to lysosomes (Miller et al., 1986), we hypothesize that E3-13.7 has usurped a mechanism normally used during ligand-induced receptor down-regulation. Our model predicts that although some EGF receptor lysosomal sorting signals, including the leucine signal usurped by E3-13.7, may be activated by ligand-induced oligomerization, others acting distally may function in the context of monomers. The major argument against this model is that cytoplasmically truncated c′-697 receptors lacking the distal signals undergo E3-13.7–mediated down-regulation. However, sorting signals are likely to operate quite differently in cytoplasmically truncated receptors versus conformationally constrained, full-length receptors.
Although we believe that the model described here best explains how E3-13.7 alters EGF receptor trafficking, other scenarios are certainly possible. For example, a single E3-13.7 molecule may promote formation of a large oligomeric complex with multiple receptor molecules and undergo cotransport to lysosomes. It is also possible that E3-13.7 competes with recycling EGF receptors for an unknown “recycling” component in early endosomes, causing default receptor routing to lysosomes. Both of these alternative models predict that E3-13.7 should behave differently in cells expressing EGF receptors versus those that do not. For example, the first model predicts that E3-13.7 should be distributed to late endocytic compartments in ERwt but not NR6 cells. The second model, on the other hand, predicts that E3-13.7 should exhibit increased surface expression in ERwt cells relative to NR6. Hence, although difficult to rule out, each alternative makes at least one prediction that is not well supported by our data.
Although a few membrane-associated early endocytic proteins have been identified, notably the peripheral membrane protein EEA1 (Mu et al., 1995), an endosome-specific COPI coat protein (Daro et al., 1997; Gu et al., 1997), and the small G-binding protein Rho B (Robertson et al., 1995), relatively few integral membrane proteins are known that reside exclusively in early endosomes or MVEs. E3-13.7, therefore, represents a novel model system for understanding the molecular basis of membrane protein sorting within these early endocytic compartments. The possibility that E3-13.7–EGF receptor complexes promote MVE maturation may also have implications for viral pathogenesis, because similar phenomena have been reported for a number of intracellular pathogens (Sanfridson et al., 1997). Several roles for MVE expansion in E3-13.7–expressing cells can be imagined. First, MVE expansion may serve to degrade specific host cell proteins that would otherwise be detrimental to host virus defense. In addition to the EGF receptor and select related tyrosine kinase receptors (Kuivinen et al., 1993), E3-13.7 recently was reported to be involved in down-regulating the Fas ligand, thereby preventing Fas-induced apoptosis (Shisler et al., 1997). Fas ligand degradation could be selective or a by-product of an expanded MVE compartment. Second, viral components may be accumulated and processed in an expanded MVE compartment. This could include secondary infecting viral particles whose degradation prevents superinfection, maturation of virally encoded proteins, or destruction of superfluous viral proteins. Third, the expanded MVE compartment may promote functions other than protein degradation, because MVEs intersect secretory as well as biosynthetic pathways (Gu and Gruenberg, 1999). Finally, localization of specific proteins or lipids to MVEs may alter the integrity of the cell membrane, helping to set the stage for cell lysis. It is important to keep in mind that in each of these scenarios E3-13.7 is a relatively low-abundance early viral protein in a natural infection (Chow et al., 1979). The E3 promoter is also regulated by NF-κB and therefore implicated in viral persistence (Shurman et al., 1989). Hence, E3-13.7 likely mediates a change in homeostatic balance rather than the complete clearance of target proteins. Continued investigation of E3-13.7 cells, therefore, promises to provide new insights into the role of MVE expansion in viral pathogenesis.
ACKNOWLEDGMENTS
We thank Maryanne Pendergast for help with CLSM. This paper is dedicated to the memory of Leila Diamond, Ph.D. This work was supported by National Institutes of Health grants CA49540 and DK57306 (to C.C.)
REFERENCES
- Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneson LS, Miller J. Efficient endosomal localization of major histocompatibility complex class II-invariant chain complexes requires multimerization of the invariant chain targeting sequence. J Cell Biol. 1995;129:1217–1228. doi: 10.1083/jcb.129.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baass PC, DiGuglielmo GM, Authier F, Posner BI, Bergeron JJM. Compartmentalized signal transduction by receptor tyrosine kinases. Trends Cell Biol. 1995;5:465–470. doi: 10.1016/s0962-8924(00)89116-3. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes in A431 cells induced by EGF. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin CR, Tollefson AE, Brady HA, Hoffman BL, Wold W. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell. 1989;57:135–144. doi: 10.1016/0092-8674(89)90179-7. [DOI] [PubMed] [Google Scholar]
- Chapman RE, Munroe S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- Chen JW, Murphy TL, Willingham MC, Pastan I, August JT. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Broker TR, Lewis JB. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979;134:265–287. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer (COP1)-coated vesicles: role in intracellular transport and protein sorting. Curr Opin Cell Biol. 1997;9:484–487. doi: 10.1016/s0955-0674(97)80023-3. [DOI] [PubMed] [Google Scholar]
- Daro E, Sheff D, Gomez M, Kreis T, Mellman I. Inhibition of endosome function in CHO cells bearing a temperature-sensitive defect in the coatomer (COPI) component ε-COP. J Cell Biol. 1997;139:1747–1759. doi: 10.1083/jcb.139.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. In vitro cultivation of human tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- Gorman CM, Gies D, McCray G, Huang M. The human cytomegalovirus major immediate early promoter can be transactivated by adenovirus early proteins. Virology. 1989;171:371–385. doi: 10.1016/0042-6822(89)90605-3. [DOI] [PubMed] [Google Scholar]
- Green SA, Zimmer KP, Griffiths G, Mellman I. Kinetics of intracellular transport and sorting of lysosomal membrane and plasma membrane proteins. J Cell Biol. 1987;105:1227–1240. doi: 10.1083/jcb.105.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Bronson S, Lock M, Neumann M, Pavlakis GN, Skowronski J. Co-localization of HIV-1 nef with the AP-2 adaptor protein complex correlates with nef-induced CD4 down-regulation. EMBO J. 1997;16:6964–6976. doi: 10.1093/emboj/16.23.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Maxfield MR. Membrane transport in the endocytic pathway. Curr Opin Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- Gu F, Aniento F, Parton RG, Gruenberg J. Functional dissection of COP-I subunits in the biogenesis of multivesicular bodies. J Cell Biol. 1997;139:1183–1195. doi: 10.1083/jcb.139.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Gruenberg J. Biogenesis of transport intermediates in the endocytic pathway. FEBS Lett. 1999;452:61–66. doi: 10.1016/s0014-5793(99)00561-x. [DOI] [PubMed] [Google Scholar]
- Harter C, Mellman I. Transport of the lysosomal glycoprotein lgp120 (lgp-A) to lysosomes does not require appearance on the plasma membrane. J Cell Biol. 1992;117:311–325. doi: 10.1083/jcb.117.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Futter CE, Hopkins CR. The kinetics of mannose 6-phosphate receptor trafficking in the endocytic pathway in HEp-2 cells: the receptor enters and rapidly leaves multivesicular endosomes without accumulating in a prelysosomal compartment. Mol Biol Cell. 1998;9:809–816. doi: 10.1091/mbc.9.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BL, Takishima K, Rosner MR, Carlin C. Adenovirus and protein kinase C have distinct molecular requirements for regulating epidermal growth factor receptor trafficking. J Cell Physiol. 1993;157:535–543. doi: 10.1002/jcp.1041570313. [DOI] [PubMed] [Google Scholar]
- Hoffman BL, Ullrich A, Wold W, Carlin C. Retrovirus-mediated transfer of an adenovirus gene encoding an integral membrane protein is sufficient to down regulate the receptor for epidermal growth factor. Mol Cell Biol. 1990;10:5521–5524. doi: 10.1128/mcb.10.10.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P, Carlin C. Adenovirus E3 protein causes constitutively internalized EGF receptors to accumulate in a prelysosomal compartment, resulting in enhanced degradation. Mol Cell Biol. 1994;14:3695–3706. doi: 10.1128/mcb.14.6.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P, Yaffe MB, Hoffman BL, Yei S, Wold WSM, Carlin C. Characterization of the adenovirus E3 protein that down-regulates the epidermal growth factor receptor. J Biol Chem. 1992a;267:13480–13487. [PubMed] [Google Scholar]
- Hoffman PH, Rajakumar P, Hoffman B, Heuertz R, Wold WSM, Carlin CR. Evidence for intracellular down-regulation of the epidermal growth factor receptor during adenovirus infection by an EGF-independent mechanism. J Virol. 1992b;66:197–203. doi: 10.1128/jvi.66.1.197-203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C. Intracellular routing of transferrin and transferrin receptors in epidermoid carcinoma A431 cells. Cell. 1983;35:321–330. doi: 10.1016/0092-8674(83)90235-0. [DOI] [PubMed] [Google Scholar]
- Kil S, Carlin C. EGF receptor residues Leu679, Leu680 mediate selective sorting of ligand-receptor complexes in early endocytic compartments. J Cell Physiol. 2000;185:47–60. doi: 10.1002/1097-4652(200010)185:1<47::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Kil SJ, Hobert ME, Carlin C. A leucine-based determinant in the EGF receptor juxtamembrane domain is required for the efficient transport of ligand-receptor complexes to lysosomes. J Biol Chem. 1999;274:3141–3150. doi: 10.1074/jbc.274.5.3141. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Van Renswoude J, Ashwell G, Kempf C, Schecter AN, Dean A, Bridges KR. Receptor-mediated endocytosis of transferrin in K562 cells. J Biol Chem. 1983;258:4715–4724. [PubMed] [Google Scholar]
- Kornilova E, Sorkina T, Beguinot L, Sorkin A. Lysosomal targeting of epidermal growth factor receptors via a kinase-dependent pathway is mediated by the receptor carboxyl-terminal residues 1022–1123. J Biol Chem. 1996;271:30340–30346. doi: 10.1074/jbc.271.48.30340. [DOI] [PubMed] [Google Scholar]
- Krajcsi P, Tollefson AE, Anderson CW, Stewart AR, Carlin CR, Wold WSM. The E3–10.4K protein of adenovirus is an integral membrane protein that is partially cleaved between Ala22 and Ala23 and has a Ccyt orientation. Virology. 1992;187:131–144. doi: 10.1016/0042-6822(92)90302-6. [DOI] [PubMed] [Google Scholar]
- Kuivinen E, Hoffman BL, Hoffman PA, Carlin CR. Structurally related class I and class II receptor protein tyrosine kinases are down-regulated by the same E3 protein coded by human group C adenoviruses. J Cell Biol. 1993;120:1271–1279. doi: 10.1083/jcb.120.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamb J, Ray F, Ward J, Kushner J, Kaplan J. Internalization and subcellular localization of transferrin and transferrin receptors in HeLa cells. J Biol Chem. 1983;258:8751–8758. [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Fambrough DM. Cycling of the integral glycoprotein LEP100 between the plasma membrane and lysosomes: kinetic and morphological analysis. Cell. 1987;49:669–677. doi: 10.1016/0092-8674(87)90543-5. [DOI] [PubMed] [Google Scholar]
- Low SH, Tang BL, Wong SH, Hong W. Selective inhibition of protein targeting to the apical domain of MDCK cells by brefeldin A. J Cell Biol. 1992;118:51–62. doi: 10.1083/jcb.118.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Brake B, Banting G, Howell KE, Braghetta P, Stanley KK. Identification, sequencing, and expression of an integral membrane protein of the trans-Golgi network (TGN-38) Biochem J. 1990;270:97–102. doi: 10.1042/bj2700097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews PM, Martinie JB, Fambrough DM. The pathway and targeting signal for delivery of the integral membrane glycoprotein LEP100 to lysosomes. J Cell Biol. 1992;118:1027–1040. doi: 10.1083/jcb.118.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffery JM, Farquhar MG. Localization of GTPases by indirect immunofluorescence and immunoelectron microscopy. Methods Enzymol. 1995;257:259–279. doi: 10.1016/s0076-6879(95)57031-4. [DOI] [PubMed] [Google Scholar]
- Miller K, Beardmore J, Kanely H, Schlessinger J, Hopkins CR. Localization of the EGF receptor within the endosome of EGF-stimulated A431 cells. J Cell Biol. 1986;102:500–509. doi: 10.1083/jcb.102.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EP, Toh BH. EEA1, an early endosome-associated protein: EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Piquet V, Chen Y, Mangasarian A, Foti M, Carpentier J, Trono D. Mechanism of nef-induced CD4 endocytosis: nef connects CD4 with the mu chain of adaptor complexes. EMBO J. 1998;17:2472–2481. doi: 10.1093/emboj/17.9.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss RM, Herschman HR. Variants of 3T3 cells lacking mitogenic response to epidermal growth factor. Proc Natl Acad Sci USA. 1977;74:3918–3921. doi: 10.1073/pnas.74.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport I, Chen YC, Cupers P, Shoelson SE, Kirchausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D, Paterson HF, Adamson P, Hall A, Monaghan P. Ultrastructural localization of Ras-related proteins using epitope-tagged plasmids. J Histochem Cytochem. 1995;43:471–480. doi: 10.1177/43.5.7537292. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Pendergast M. Polarized distribution of viral envelop proteins in the plasma membrane of infected epithelial cells. Cell. 1980;20:45–54. doi: 10.1016/0092-8674(80)90233-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Sabatini DD. Asymmetric budding of viruses in epithelial cell monolayers: a model system for study of epithelial polarity. Proc Natl Acad Sci USA. 1978;75:5071–5075. doi: 10.1073/pnas.75.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval IV, Bakke O. Targeting of membrane proteins to endosomes and lysosomes. Trends Cell Biol. 1994;4:292–297. doi: 10.1016/0962-8924(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Sanfridson A, Hester S, Doyle C. Nef proteins encoded by human and simian immunodeficiency viruses induce the accumulation of endosomes and lysosomes in human T cells. Proc Natl Acad Sci USA. 1997;94:873–878. doi: 10.1073/pnas.94.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisler J, Yang C, Walter B, Ware CF, Gooding LR. The adenovirus E3–10.4/14.5 complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J Virol. 1997;71:8299–8306. doi: 10.1128/jvi.71.11.8299-8306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurman L, Sen R, Bergman Y. Adenovirus E1A products activate the Ig k-chain enhancer in fibroblasts. J Immunol. 1989;143:3806–3812. [PubMed] [Google Scholar]
- Tollefson AE, Krajcsi P, Yei S, Carlin CR, Wold W. A 10,400-molecular weight membrane protein is coded by region E3 of adenovirus. J Virol. 1990;64:794–801. doi: 10.1128/jvi.64.2.794-801.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deurs B, Holm PK, Kayser L, Sandvig K, Hansen SH. Multivesicular bodies in HEp-2 cells are maturing endosomes. Eur J Cell Biol. 1993;61:208–224. [PubMed] [Google Scholar]
- Vinogradova O, Carlin CR, Sönnichsen FD, Sanders CR. Membrane interaction and structure of the cytoplasmic domian of the E3-13.7 kDa adenovirus protein that disrupts normal intracellular trafficking of the EGF receptor. J Biol Chem. 1998;273:17343–17350. doi: 10.1074/jbc.273.28.17343. [DOI] [PubMed] [Google Scholar]
- Vowels JJ, Payne GS. A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 1998;17:2482–2493. doi: 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield MD, Mayes ELV, Stroobant P, Bennet PLP, Young S, Goodfellow PN, Banting GS, Ozanne B. A monoclonal antibody to the human epidermal growth factor receptor. J Cell Biochem. 1982;20:149–161. doi: 10.1002/jcb.240200207. [DOI] [PubMed] [Google Scholar]
- Weiel JE, Hamilton TA. Quiescent lymphocytes express intracellular transferrin receptors. Biochem Biophys Res Commun. 1984;119:598–603. doi: 10.1016/s0006-291x(84)80291-0. [DOI] [PubMed] [Google Scholar]
- White S, Taetle R, Seligman PA, Rutherford M, Trowbridge IS. Combinations of anti-transferrin receptor monoclonal antibodies inhibit human tumor cell growth in vitro and in vivo: evidence for synergistic antiproliferative effects. Cancer Res. 1990;50:6295–6301. [PubMed] [Google Scholar]
- Williams MA, Fukada M. Accumulation of membrane glycoproteins in lysosomes requires a tyrosine residue in a particular position in the cytoplasmic tail. J Cell Biol. 1990;111:955–966. doi: 10.1083/jcb.111.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]