Abstract
The chiR gene of Serratia marcescens 2170, encoding a LysR-type transcriptional activator, was identified previously as an essential factor for expression of chitinases and a chitin-binding protein, CBP21. To identify other genes that are essential for chitinase production, transposon mutagenesis with mini-Tn5Km1 was carried out, and 25 mutants that were unable to produce chitinases and CBP21 were obtained. Analysis of the mutated gene of one of the mutants, N22, revealed the presence of a pts operon in this bacterium, and a mutation was found in ptsI in the operon. In addition to its inability to produce chitinase, N22 did not grow well on N-acetyl-d-glucosamine (GlcNAc), (GlcNAc)2, and some other carbon sources, most of which were phosphotransferase system (PTS) sugars. Thus, the inability to produce chitinase was assumed to be caused by the defect in uptake of (GlcNAc)2 via the PTS, considering that (GlcNAc)2 is the minimal substrate for chitinase induction and the major product of chitin hydrolysis by chitinases of this bacterium. To confirm this assumption, the chb operon, encoding the (GlcNAc)2-specific enzyme II permease, was cloned by reference to its Escherichia coli counterpart, and the Serratia chb operon was shown to comprise chbB, chbC, bglA, chbR, and chbG. Disruption of chbC drastically reduced production of chitinases and CBP21 and impaired growth on colloidal chitin. These results indicate that uptake of (GlcNAc)2 is mediated by the PTS and that the (GlcNAc)2-specific enzyme II permease constitutes its major pathway. Since (GlcNAc)2 uptake is essential for induction of chitinases and CBP21 production, (GlcNAc)2 appears to be the key molecule in recognition and utilization of chitin by S. marcescens.
Chitin is the second-most-abundant biopolymer in nature and is composed of β-1,4-linked N-acetylglucosamine (GlcNAc). Serratia marcescens is an efficient biological degrader of chitin and one of the most extensively studied chitinolytic bacteria. This bacterium is a potential insect pathogen, and chitinase plays an important role in the virulence of this bacterium together with protease and lecithinase (9). From four different strains of this bacterium, namely, QMB1466 (13), BJL200 (5, 6), KCTC2172 (8), and 2170 (35), two chitinase genes (chiA and chiB) have been cloned, and a third chitinase gene, chiC, was recently identified in the KCTC2172 (8) and 2170 (28) strains. chiA, chiB, and chiC encode the family 18 chitinases A (ChiA), B (ChiB), and C1 (ChiC1). The three-dimensional structures of ChiA from QMB1466 and ChiB from BJL200 have been reported by Perrakis et al. (23) and van Aalten et al. (34), respectively. ChiA consists of an all-β-strand amino-terminal domain and a catalytic (β/α)8-barrel domain with a small α+β domain inserted between the seventh and eighth β-strands of the (β/α)8-barrel. ChiB consists of a catalytic domain that has folding similar to that of ChiA and a small carboxy-terminal putative chitin-binding domain. In addition to these three chitinases, CBP21 and chitinase C2 (ChiC2) have been detected in the culture supernatant of S. marcescens 2170. CBP21 is a chitin-binding protein of 21 kDa with a yet-unknown function, and production of this protein is coregulated with chitinases (27). ChiC2 is a proteolytic derivative of ChiC1 and corresponds to the catalytic domain of this chitinase. ChiC1 is the only S. marcescens chitinase which has a subfamily B-type catalytic domain (28) and a fibronectin type III domain. Therefore, ChiC1 is structurally unique compared with the other two chitinases. Among the three chitinases, ChiA has the highest hydrolyzing activity against insoluble chitinous substrates, while ChiC1 has the highest activity against soluble substrates (29). ChiA has exposed aromatic residues linearly aligned on the surface of the amino-terminal domain and the catalytic domain, and the importance of these residues in crystalline chitin hydrolysis has been demonstrated (33).
In contrast to the rapidly accumulating data on the structure-function relationship of chitinases and related proteins, little is known about the mechanisms of induction of chitinase production and of utilization of degradation products of chitin by S. marcescens cells. To identify the genes involved in chitinase production, transposon mutagenesis of strain 2170 was carried out and various mutants defective in chitinase production were isolated (35). Some of them were entirely defective in chitinase and CBP21 production. A detailed analysis of one such mutant, N1, resulted in identification of an essential gene for chitinase and CBP21 production. This gene, designated chiR, encodes a LysR-type transcriptional activator that is essential for the expression of chitinases and CBP21 (30). In this article, we describe the characterization of another mutant, N22, that is also defective in chitinase and CBP21 production, as N1 is. The mutation was found to be located in a gene homologous to ptsI, which encodes enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) carbohydrate transport system in Escherichia coli (25). The presence in 2170 of a chb operon similar to the E. coli counterpart encoding (GlcNAc)2-specific enzyme II permease is also described.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
S. marcescens 2170 was originally obtained from H. W. Ackermann (Department of Medical Biology, Faculty of Medicine, Laval University, Quebec, Canada). S. marcescens 2170 and its mini-Tn5Km1 mutants were grown at 30°C with shaking in a yeast extract-supplemented minimal (YEM) medium containing various carbon sources (22). YEM agar plates containing 0.2% (wt/vol) colloidal chitin and 50 μg of kanamycin per ml were used to test chitinase production. E. coli JM109 was used as the host organism and pUC119 was used as a vector for gene cloning. E. coli JM109 carrying pUC119 or its derivatives was grown in Luria-Bertani (LB) medium containing 100 μg of ampicillin per ml or 50 μg of kanamycin per ml. E. coli SM10(λpir) carrying pUTmini-Tn5Km1 (10, 18) was used for mini-Tn5Km1 mutation of S. marcescens 2170. E. coli S17-1(λpir) and the plasmid pFS200 were used to create insertion mutations in the chbC gene. pFS200 contains a chloramphenicol resistance (Cmr) cassette at the unique SmaI site of pGP704, a pir-dependent replication plasmid (20). E. coli S17-1(λpir) carrying pFS200 or its derivatives was grown at 30°C in LB medium containing 100 μg of ampicillin per ml and/or 50 μg of chloramphenicol per ml. pACPTS01, used in complementation tests of N22, carries the 2.3-kb DNA fragment from the S. marcescens genome containing the intact ptsI and ptsH genes.
Conjugation and transposition of mini-Tn5Km1.
Mini-Tn5Km1 was delivered to wild-type S. marcescens 2170 via conjugation with E. coli SM10(λpir) carrying pUTmini-Tn5Km1 (10, 18). Fiftyfold-concentrated cultures of donor and recipient strains (both at an optical density at 660 nm of 0.6) were mixed, and 200 μl of the mixture was spotted on LB plates and incubated at 30°C for 6 h. Kanamycin-resistant transconjugants were obtained by spreading the mating mixture on LB plates containing kanamycin (50 μg/ml) and tetracycline (12.5 μg/ml), followed by incubation at 33°C for 24 h. Colonies that appeared on the plates were transferred onto agar plates of YEM medium containing 0.2% colloidal chitin and kanamycin (50 μg/ml) and incubated at 33°C. Clearing zones that formed around the mutant colonies were visually inspected, and mutants with altered clearing zones were selected.
Enzyme and protein assays.
Chitinase activity was measured by a modification of Schales' procedure (11) with colloidal chitin as the assay substrate. One unit of chitinase activity was defined as the amount of enzyme that produces 1 μmol of reducing sugar per min. The protein concentration was measured by the method of Lowry et al. (19) with bovine serum albumin as the standard.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12.5% slab gels was conducted as described by Ames (1) with the buffer system of Laemmli (17).
Cloning of the flanking DNA regions of inserted mini-Tn5Km1.
Chromosomal DNA of S. marcescens 2170 was extracted from the cells as described by Silhavy et al. (26), partially digested with Sau3AI, and separated by electrophoresis on a 0.5% agarose gel. The gel segment containing DNA fragments of between 7 and 12 kb in length was cut out, and DNA fragments in the gel were recovered by using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). The DNA fragments were ligated to BamHI-digested pUC119 and used to transform E. coli JM109 cells. Transformants carrying the plasmids containing various sizes of flanking DNA regions of the inserted mini-Tn5Km1 together with the kanamycin resistance gene were selected on LB agar plates containing 50 μg of kanamycin per ml.
Nucleotide sequence determination and sequence analysis.
DNA fragments inserted in the plasmids were sequenced by using an automated laser fluorescence sequencer (model 4000L; LI-COR, Lincoln, Nebr.). Sequencing reactions were done with a ThermoSequenase fluorescence-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) according to the supplier's instructions with a double-stranded template. Nucleotide sequence data were analyzed by using the GENETYX system (Software Kaihatsu Co., Tokyo, Japan). The deduced amino acid sequence was compared with those available in the translated GenBank, SWISS-PROT protein sequence, National Biomedical Research Foundation protein, and DDBJ databases.
Southern hybridization.
Southern hybridization was performed by using AlkPhos Direct (Amersham Pharmacia Biotech) as recommended by the manufacturer. A part of the kanamycin resistance gene of mini-Tn5Km1 was amplified by PCR with primers Km-F (5′-ATGAGCCATATTCAACGGGAAAC-3′) and Km-R (5′-TTACAAAAACTCATCGAGCATCAAATG-3′) and used as a hybridization probe. Chromosomal DNA of S. marcescens 2170 was digested with DraI and separated by agarose gel electrophoresis for hybridization.
Construction of plasmid pFS200.
The chloramphenicol resistance gene in the plasmid pACYC184 was amplified by PCR with primers Cmr-F (5′-GATCGGCACGTAAGAGGT-3′) and Cmr-R (5′-CGGTAAACCAGCAATAG). The amplified fragment (0.8 kb in size) was cloned into the unique SmaI site in plasmid pGP704 (20) to produce pFS200.
Construction of a mutant with a disrupted chbC gene.
Truncated chbC, corresponding to a 0.7-kb internal region of the gene, was amplified by PCR with primers ΔchbC-F (5′-TAGGAAAGCAGCCACACGT-3′) and ΔchbC-R (5′-CCATGAATACCGAAGAACCA-3′) and chromosomal DNA of S. marcescens 2170 as a template. The amplified fragment was ligated with EcoRV-cut pFS200 to generate plasmid pFSΔCHBC. Plasmid pFSΔCHBC was first introduced into E. coli S17-1(λpir) by electroporation and then transferred to S. marcescens 2170 by conjugation as previously described (30). Transconjugants were selected on LB agar plates containing chloramphenicol (100 μg/ml) and tetracycline (12.5 μg/ml).
Chemicals.
Colloidal chitin was prepared from powdered chitin purchased from Funakoshi Chemical Co. (Tokyo, Japan) according to the methods described by Jeuniaux (12). The chitooligosaccharide mixture [(GlcNAc)3-4] was obtained from Pias Co. (Osaka, Japan). Restriction enzymes and modification enzymes were purchased from Takara Shuzo (Osaka, Japan), Toyobo Biochemicals (Osaka, Japan), and New England Biolabs (Beverly, Mass.).
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in GenBank/EMBL/DDBJ under accession no. AB085624 (ptsH, ptsI, and crr) and AB085625 (chbB, chbC, bglA, chbR, and chbG).
RESULTS
Isolation of mutants defective in chitinase production.
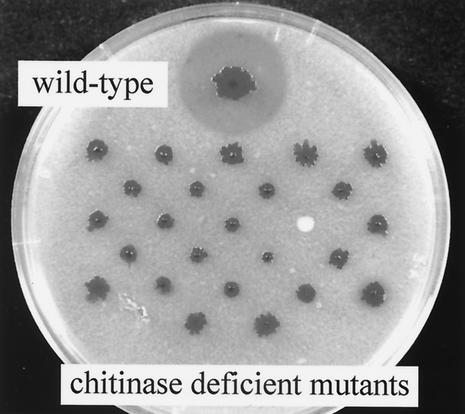
As described previously, the chiR gene, encoding a LysR-type transcriptional activator, was identified as an essential factor for the expression of all chitinases and the chitin-binding protein CBP21 of S. marcescens 2170 (30). In this study, we attempted to find other genes essential for the production of these proteins. Transposon mutagenesis of S. marcescens 2170 was carried out by using mini-Tn5Km1, and mutant colonies with either no clearing zones or altered clearing zones were screened on YEM agar plates containing colloidal chitin. Out of 2 × 104 kanamycin-resistant (Kmr) colonies, 25 formed no clearing zones (Fig. 1). Clearing zones were not observed around these mutant colonies even after 7 days of cultivation.
FIG. 1.
Plate assay for chitinase production by the wild-type strain and chitinase-deficient mutants. Mutants and wild-type 2170 were inoculated on YEM agar plates containing 0.2% (wt/vol) colloidal chitin and incubated at 33°C for 5 days.
In order to distinguish desirable mutants from the mutants with mini-Tn5Km1 insertions in chiR, Southern hybridization was carried out with part of the Kmr gene in the mini-Tn5Km1 as a probe. DraI digestion of the chromosomal DNA of S. marcescens 2170 normally gives a 3.5-kb DNA fragment containing the wild-type chiR gene. Mini-Tn5Km1 insertion will increase the size of the DNA fragment to 5.3 kb. Therefore, the mutants which gave a signal at 5.3 kb were considered to have insertion mutations in the chiR gene. Out of 25 Kmr mutants, 21 exhibited signals at the position corresponding to 5.3 kb. The other four mutants gave signals at positions not only different from 5.3 kb but also different from each other. In this study, we focused on one of the four mutants, designated mutant N22.
Phenotype of chitinase-deficient mutant N22.
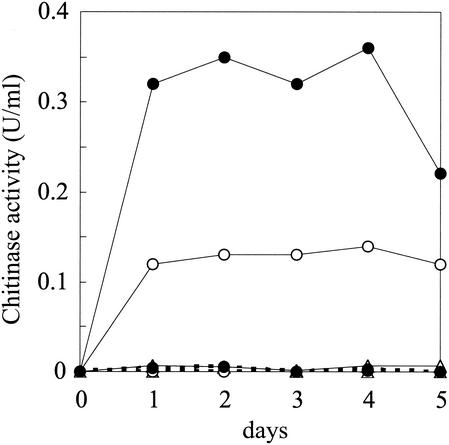
N22 and wild-type 2170 were cultivated in YEM liquid medium containing either colloidal chitin, (GlcNAc)3-4, or glycerol as a carbon source, and growth and chitinase production of the two strains were compared. The growth of N22 in the medium containing glycerol was as good as that of wild-type 2170; however, N22 did not grow well in the medium containing either colloidal chitin or (GlcNAc)3-4 (data not shown). As shown in Fig. 2, significant chitinase activity was detected in the culture supernatant of wild-type 2170 when it was grown in the medium containing colloidal chitin or (GlcNAc)3-4, while no activity was detected in the medium containing glycerol. On the other hand, no chitinase activity was detected in the culture supernatants of N22 grown in any tested media.
FIG. 2.
Chitinase activity detected in the culture supernatants of mutant N22 and wild-type 2170. Wild-type 2170 (solid lines) and N22 (dashed lines) were grown in YEM medium containing 0.1% colloidal chitin (•), (GlcNAc)3-4 (○), or glycerol (▵). Chitinase activity in the culture supernatant was measured by a modification of Schales' procedure with colloidal chitin as an assay substrate.
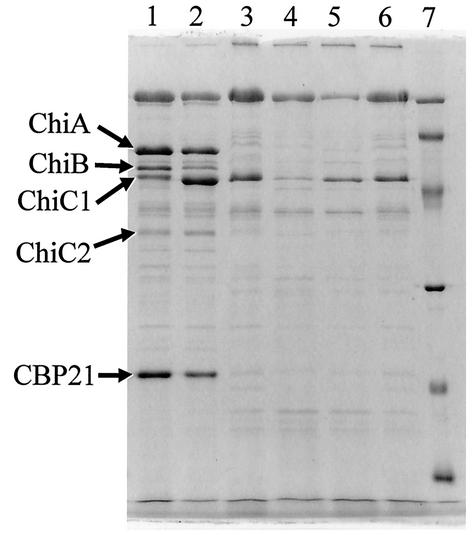
To confirm the absence of chitinases in the culture supernatant and to examine the effect of mutation on the production of chitin-binding protein CBP21 as well, proteins in the day 3 culture supernatants of N22 and wild-type 2170 were analyzed by SDS-PAGE. A previous study demonstrated that the production of chitinases and the production of CBP21 are regulated in parallel, in the sense that the substrate that induces production of chitinase also induces production of CBP21 (27). Equal aliquots (50 μg) of proteins prepared from the day 3 culture supernatants of N22 and 2170 grown in media with various carbon sources were separated by SDS-PAGE (Fig. 3). Wild-type 2170 produced CBP21 normally together with chitinases A, B, and C1 in the medium containing either colloidal chitin or (GlcNAc)3-4. On the other hand, neither CBP21 nor chitinases were detected in the culture supernatant of mutant N22, demonstrating that the mutant has lost the ability to produce not only chitinases but also CBP21.
FIG. 3.
SDS-PAGE analysis of CBP21 in the culture supernatants of N22 and wild-type 2170. Wild-type (lanes 1 to 3) and mutant (lanes 4 to 6) strains were grown in medium containing 0.1% colloidal chitin (lanes 1 and 4), (GlcNAc)3-4 (lanes 2 and 5), or glycerol (lanes 3 and 6), and the proteins (50 μg in each lane) in the day 3 culture supernatant were analyzed by SDS-PAGE.
Position of mini-Tn5Km1 insertion in mutant N22.
To identify the position of mini-Tn5Km1 insertion in N22, we cloned the inserted mini-Tn5Km1 with its flanking regions. Approximately 126,400 clones of the N22 genomic DNA library of E. coli were screened for Kmr, and 18 independent Kmr clones were obtained. Plasmids were isolated from these clones, and the nucleotide sequences of the inserted DNA regions were determined. The nucleotide sequence of a 3.2-kb region around the mini-Tn5Km1 insertion was finally obtained. Three open reading frames (ORFs), ORF1 (255 bp), ORF2 (1,725 bp), and ORF3 (507 bp), were found in the sequenced region, and mini-Tn5Km1 was within ORF2. All three ORFs are transcribed in the same direction. Proteins with sequences similar to those deduced from ORF1, ORF2, and ORF3 were searched for in the DDBJ database. It was found that the amino acid sequences deduced from ORF1, ORF2, and ORF3 were homologous to HPr (identity, 98%), enzyme I (identity, 86%), and enzyme IIAGlc (identity, 96%) in the E. coli PTS, respectively (25). In the E. coli genome, the genes for HPr (ptsH), enzyme I (ptsI), and enzyme IIAGlc (crr) are the components of the pts operon, and they are aligned in the same direction of transcription as ptsH-ptsI-crr from upstream to downstream (7). Therefore, it appears that S. marcescens has a pts operon that is very similar to its E. coli counterpart, and therefore, ORF1, ORF2, and ORF3 are hereafter referred to as the ptsH, ptsI, and crr genes, respectively, of S. marcescens. Several potential promoter sequences were observed, and one of them overlapped with a putative cyclic AMP receptor protein-binding site. Two potential promoters for crr, which are between ptsI and crr, were observed as well.
The phenotype of mutant N22 is, therefore, most likely caused by the disruption of ptsI by the mini-Tn5Km1 insertion. To confirm this, a complementation test using plasmid pACPTS01 carrying the 2.3-kb DNA fragment containing ptsI and ptsH was carried out. Introduction of the intact ptsI gene into N22 caused the production of chitinases to be restored, and clearing zones were formed around the colonies on the YEM agar plates containing colloidal chitin. SDS-PAGE analysis of the proteins in the culture supernatant demonstrated that N22(pACPTS01) secreted all chitinases and CBP21 in a ratio similar to that for wild-type 2170.
Growth of wild-type 2170 and N22 on various carbon sources.
The most probable explanation for the inability to produce chitinase and the growth defect in medium containing chitin or chitooligosaccharides is that disruption of ptsI makes the cells unable to take up (GlcNAc)2. (GlcNAc)2 is the minimal substrate able to induce chitinase production (35) and is the major hydrolysis product produced from chitin by S. marcescens chitinases (29).
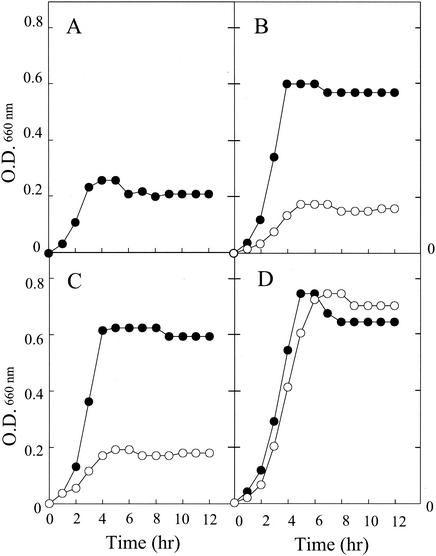
If the ptsI gene of S. marcescens identified in this study is essential for uptake of (GlcNAc)2, N22 would show a growth defect on (GlcNAc)2 as well as on other known PTS carbohydrates. Therefore, the growth of N22 and wild-type 2170 on various carbohydrates was examined. The carbohydrates tested in these experiments included GlcNAc, (GlcNAc)2, glycerol, glucose, and fructose. Figure 4 shows the growth of wild-type 2170 and N22 in medium containing either GlcNAc, (GlcNAc)2, or glycerol as examples. Since YEM medium contains a small amount of yeast extract, a low level of growth occurs even in the absence of supplemented carbohydrate, as demonstrated by a control experiment shown in Fig. 4A. Wild-type 2170 grew well on all carbohydrates examined. On the other hand, N22 grew well only on glycerol. The growth on GlcNAc, (GlcNAc)2, and fructose was not better than that without supplementary carbohydrate. The growth of N22 on glucose was retarded significantly, but the cell density finally reached the same level as for wild-type 2170 after longer incubation.
FIG. 4.
Growth of wild-type 2170 and mutant N22 on YEM medium supplemented with various carbon sources. Wild-type (•) and mutant (○) strains were grown in the medium without supplementary a carbon source (A) or with (GlcNAc)2 (B), GlcNAc (C), or glycerol (D). The concentration of all carbon sources in YEM medium was 0.05% (wt/vol). O.D., optical density.
Glycerol is well known as a non-PTS carbohydrate, while uptake of GlcNAc, (GlcNAc)2, and fructose is mediated by the PTS in the E. coli system. Considering the close taxonomic relationship between E. coli and S. marcescens, these results strongly suggested that the ptsI gene of S. marcescens is essential for the uptake and utilization of (GlcNAc)2 as well as other PTS carbohydrates and GlcNAc and that the inability of N22 to produce chitinases and CBP21 is due to the defect in (GlcNAc)2 uptake.
Presence of the chb operon in S. marcescens.
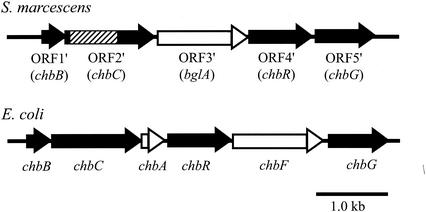
The observation that (GlcNAc)2, like other PTS carbohydrates, was not utilized by N22 does not necessarily mean that the uptake of (GlcNAc)2 is directly mediated by the PTS. Indeed, E. coli ptsI mutants do not grow on a number of non-PTS carbon sources (24). Therefore, to study whether S. marcescens uses the PTS for (GlcNAc)2, a homolog of the E. coli chb operon was searched for in S. marcescens. It has been recently reported that in E. coli the uptake of (GlcNAc)2 is mediated by the PTS, and the (GlcNAc)2-specific enzyme II permease is encoded in the chb operon (14, 15). The operon consists of chbB (encodes the IIB domain of enzyme II permease), chbC (IIC domain), chbA (IIA domain), chbR (possibly a repressor for the chb operon), chbF (possibly phosphochitobiase), and chbG (function unknown).
To detect a chbC homolog in S. marcescens, PCR amplification of part of a chbC homolog was attempted by using primers based on the E. coli chbC sequence. An amplified fragment of 0.7 kb was obtained by PCR, and the fragment was cloned into pUC119 and sequenced. The nucleotide sequence of the amplified fragment exhibited 83% identify with the corresponding region of E. coli chbC, suggesting the presence of a chbC homolog in S. marcescens. To obtain the entire nucleotide sequence of the S. marcescens chb operon, the flanking regions of the determined sequence were cloned as DNA fragments containing a Cmr gene from the chbC disruption mutant described below by shotgun cloning. Finally, the nucleotide sequence of a 5.6-kb region was obtained, and five ORFs were identified in the sequenced region (Fig. 5). ORF1′ (315 bp), encodes a polypeptide of 105 amino acids which is very similar to E. coli (GlcNAc)2-specific enzyme IIB (identity 83%) and was thus designated Serratia chbB. The GTG initiation codon of ORF2′ (1,359 bp) is found 12 bp downstream from the chbB termination codon. This ORF encodes a polypeptide of 453 amino acids which is very similar to E. coli (GlcNAc)2-specific enzyme IIC (identity of 80%) and was thus designated Serratia chbC. ORF3′ (1,383 bp) encodes a polypeptide of 461 amino acids which shows similarity to β-glucosidases, including a putative β-glucosidase (YdhP) from Bacillus subtilis (58% identity), β-glucosidase A (BglA) from Clostridium thermocellum (34% identity), and a putative 6-phospho-β-glucosidase (BglA) from E. coli (31% identity). Thus, the gene of ORF3′ was designated bglA. The GTG initiation codon of ORF4′ (846 bp) is found 15 bp downstream from the bglA termination codon. The ORF encodes a polypeptide of 282 amino acids that is very similar to ChbR (67% identity), a putative repressor of the E. coli chb operon, and was thus designated Serratia chbR. ORF5′ (816 bp) encodes a polypeptide of 272 amino acids that is very similar to ChbG (67% identity) and was thus designated Serratia chbG.
FIG. 5.
Schematic diagrams of the S. marcescens chb operon and the E. coli chb operon. Closed arrows indicate the genes common to the operons of the two bacteria. Open arrows indicate the genes found only in the operon of one of the two bacteria. The hatched box in chbC indicates the region amplified by PCR and used in targeted mutagenesis.
In the E. coli chb operon, chbF is located between chbR and chbG, but a gene homologous to chbF was not found in the S. marcescens chb operon. In addition, a counterpart of bglA in the S. marcescens chb operon was not found in the E. coli chb operon, and instead chbA is located downstream of chbC. The S. marcescens chb operon has no gene corresponding to chbA in the E. coli chb operon.
Targeted mutagenesis of S. marcescens chbC.
The presence of a chbC gene in S. marcescens 2170 strongly suggests that the uptake of (GlcNAc)2 is mediated by the PTS. Therefore, we attempted targeted mutagenesis of chbC to study the phenotypic change caused by a disruption of this gene. A part of the chbC gene corresponding to an internal 0.7-kb region was amplified by PCR and cloned into EcoRV-cut pFS200 to generate plasmid pFSΔCHBC. pFS200 is a pir-dependent replication plasmid containing a Cmr gene. pFSΔCHBC was transferred from E. coli S17-1(λpir) to S. marcescens 2170 by conjugation, and transconjugants were selected based on chloramphenicol resistance. Chloramphenicol-resistant transconjugants should contain pFSΔCHBC integrated into the chromosome by homologous recombination. The integration occurred between the chbC gene and the plasmid, leading to two incomplete copies of the chbC gene. Integration of pFSΔCHBC into the chromosomal DNA of S. marcescens 2170 was confirmed by Southern hybridization using the chbC fragment of 0.7 kb as a probe.
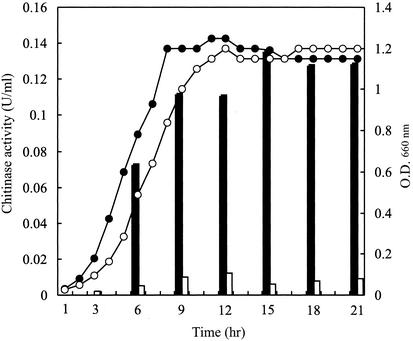
The phenotype of the chbC disruption mutant thus obtained was first examined by testing formation of clearing zones on YEM agar plates containing colloidal chitin. The mutant colony did form a clearing zone, but it was much smaller than that formed by wild-type 2170. The clearing zone of the wild-type strain was visible on the day after inoculation, while the clearing zone of the mutant strain became visible more than 3 days after inoculation. Next, chitinase production was compared under conditions in which the chbC disruptant and the wild-type 2170 grow almost equally. The two strains were grown in medium containing both glycerol and (GlcNAc)2, and the growth of the cells and chitinase activity in the culture supernatant were monitored. As shown in Fig. 6, the mutant and wild-type strains grew almost equally in this medium. Since glycerol does not repress chitinase production, (GlcNAc)2 induced chitinase production by wild-type 2170 normally. On the other hand, the level of chitinase activity detected in the culture supernatant of the mutant strain was less than 1/10 of that of wild-type 2170.
FIG. 6.
Growth of wild-type 2170 and the chbC disruption mutant and chitinase activity detected in the culture supernatants. Closed and open bars indicate the chitinase activities of the wild-type and mutant strains, respectively. The growth of wild-type (•) and mutant (○) strains was monitored by measuring the optical density (O.D.) at 660 nm. Chitinase activity in the culture supernatant was measured by a modification of Schales' procedure with colloidal chitin as the assay substrate. The concentrations of (GlcNAc)2 and glycerol in the YEM medium were 0.1% (wt/vol).
From these results, we concluded that uptake of (GlcNAc)2 into S. marcescens cells is mediated by the PTS and that (GlcNAc)2-specific enzyme IIC, encoded by the chb operon, is involved in the major pathway of (GlcNAc)2 uptake. Uptake of (GlcNAc)2 into cells is essential for the production of chitinases and CBP21 and for the utilization of chitin outside cells by this bacterium.
DISCUSSION
Mutation of the ptsI gene, which is a member of the pts operon in S. marcescens found in this study, resulted in a lack of chitinase production and almost no growth on either colloidal chitin or chitooligosaccarides. The phenotype of N22 can be explained by the inability of the cells to take up (GlcNAc)2 through the PTS. (GlcNAc)2 is the major product of chitin hydrolysis by S. marcescens chitinases and the smallest substrate for induction of chitinase production as well (35). The chbC gene, which is homologous to E. coli chbC, encoding (GlcNAc)2-specific enzyme IIC, was identified in this bacterium. Disruption of this gene drastically reduced the production of chitinases and CBP21 and impaired growth on colloidal chitin. However, chbC disruption did not abolish chitinase production completely. Although the level was much lower than that of the wild-type strain, some chitinase activity was detected when the mutant was cultivated in medium containing either colloidal chitin or (GlcNAc)2. This probably means that the chbC disruption mutant cells still take up a small amount of (GlcNAc)2 through some other PTS transport system. It has been reported that the E. coli mannose-specific enzyme II component encoded by the man operon mediates the uptake of not only mannose but also GlcNAc, glucosamine, fructose, glucose, and trehalose (24). A small amount of (GlcNAc)2 could be taken up by the mutant cells if such an enzyme II component with low specificity is present in S. marcescens. The small amount of (GlcNAc)2 taken up into the cells through such a component would induce a low level of chitinase production.
Several interesting differences were observed between the S. marcescens chb operon and the E. coli chb operon. Most importantly, the S. marcescens chb operon had no chbA homolog which would encode (GlcNAc)2-specific enzyme IIA (Fig. 5). PTS enzyme II comprises three domains, the so-called IIA, IIB, and IIC. In some cases, IIA, IIB, and IIC, or IIB and IIC, are in a single polypeptide and are encoded by one gene. In the E. coli chb operon, chbA (encoding IIA) is located immediately downstream of chbB (encoding IIB), which is preceded by chbC (encoding IIC). IIA receives a phosphate group from another PTS component, HPr, and transfers it to IIB. Because of the indispensable role of IIA, chbA may be located in some other region of the S. marcescens genome, although there is an example in which the gene for the IIA component specific for a particular PTS sugar was not identified: trehalose is taken up by E. coli cells through trehalose-specific enzyme II, but only treB, encoding IICB, has been identified (3). In this case, it has been presumed that there is no IIA component specific for trehalose and, instead, the gene product of crr, which is a IIA component of glucose-specific enzyme II, substitutes for the role of trehalose-specific IIA. However, this is a rather exceptional case and probably is not applicable to the Serratia chb operon.
As demonstrated in the present study, uptake of (GlcNAc)2 is essential to induce chitinases and to support the growth of S. marcescens on chitin. ChiA, ChiB, and ChiC1 secreted by this bacterium release (GlcNAc)2 as the major hydrolysis product from chitin (29). These observations indicate that (GlcNAc)2 is the key molecule in the degradation and utilization of chitin by S. marcescens. In another words, S. marcescens cells recognize chitin outside the cells through uptake of (GlcNAc)2 and utilize it as a form of (GlcNAc)2. It is well known that a number of chitinases produced by a variety of chitinolytic bacteria release (GlcNAc)2 as the major hydrolysis product from chitin. The importance of (GlcNAc)2 in the induction of chitinase production has been reported not only for S. marcescens but also for some other chitinolytic bacteria. For example, Miyashita et al. reported that (GlcNAc)2 is the smallest substance that induces production of chitinases from Streptomyces lividans (21). GlcNAc and glucose repressed chitinase production by this bacterium, just as in the case of S. marcescens (22). (GlcNAc)2 also induces production of chitin hydrolases by a marine bacterium, Vibrio furnissii (2). However, unlike the PTS-mediated (GlcNAc)2 uptake by S. marcescens, V. furnissii takes up (GlcNAc)2 through a specific transporter independent of the PTS (16), while the uptake of GlcNAc is mediated by the PTS, including GlcNAc-specific enzyme II (nagE) (4). On the other hand, GlcNAc was the smallest substance to induce chitinase production by a Pseudoalteromonas strain, and in addition, glucose did not act repressively in this case (31). The most interesting exception reported is that of an Alteromonas strain reported by Tsujibo et al. (32). In that case, β-(1-6)-(GlcNAc)2, which was enzymatically converted from (GlcNAc)2, was the most effective inducer of chitinase production. (GlcNAc)2 did not act as an inducer of chitinase production. In addition, (GlcNAc)2 as well as GlcNAc actually repressed the production of chitinases, while glucose did not act repressively.
These observations imply that (GlcNAc)2 is the key molecule in the degradation and utilization of chitin by many chitinolytic bacteria; however, there is great diversity among various chitinolytic bacteria regarding the mechanisms of regulation of chitinase production as well as the mechanism for uptake of degradation products derived from chitin.
Acknowledgments
We thank Haruyoshi Seino (Pias Co.) for generously providing the chitooligosaccharide mixture.
This work was partly supported by a grant-in-aid for scientific research (12660070) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
REFERENCES
- 1.Ames, G. F. 1974. Resolution of bacterial proteins by polyacrylamide gel electro-phoresis on slabs. Membrane, soluble, and periplasmic fractions. J. Biol. Chem. 249:634-644. [PubMed] [Google Scholar]
- 2.Bassler, B. L., C. Yu, Y. C. Lee, and S. Roseman. 1991. Chitin utilization by marine bacteria: degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276-24286. [PubMed] [Google Scholar]
- 3.Boos, W., U. Ehmann, H. Forkl, W. Klein, M. Rimmele, and P. Postma. 1990. Trehalose transport and metabolism in Escherichia coli. J. Bacteriol. 172:3450-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouma, C. L., and S. Roseman. 1996. Sugar transport by the marine chitinolytic bacterium Vibrio furnissii: molecular cloning and analysis of the glucose and N-acetylglucosamine permeases. J. Biol. Chem. 271:33457-33467. [DOI] [PubMed] [Google Scholar]
- 5.Brurberg, M. B., V. G. Eijsink, and I. F. Nes. 1994. Characterization of a chitinase gene (chiA) from Serratia marcescens BJL200 and one-step purification of the gene product. FEMS Microbiol. Lett. 124:399-404. [DOI] [PubMed] [Google Scholar]
- 6.Brurberg, M. B., V. G. Eijsink, A. J. Haandrikman, G. Venema, and I. F. Nes. 1995. Chitinase B from Serratia marcescens BJL200 is exported to the periplasm without processing. Microbiology 141:123-131. [DOI] [PubMed] [Google Scholar]
- 7.De Reuse, H., and A. Danchin. 1988. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J. Bacteriol. 170:3827-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gal, S. W., J. Y. Choi, C. Y. Kim, Y. H. Cheong, Y. J. Choi, S. Y. Lee, J. D. Bahk, and M. J. Cho. 1998. Cloning of the 52-kDa chitinase gene from Serratia marcescens KCTC2172 and its proteolytic cleavage into an active 35-kDa enzyme. FEMS Microbiol. Lett. 160:151-158. [DOI] [PubMed] [Google Scholar]
- 9.Grimont, P. A., and F. Grimont. 1987. The genus Serratia. Annu. Rev. Microbiol. 32:221-248. [DOI] [PubMed] [Google Scholar]
- 10.Herrero, M., V. D. Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imoto, T., and K. Yagishita. 1971. A simple activity measurement of lysozyme. Agric. Biol. Chem. 35:1154-1156. [Google Scholar]
- 12.Jeuniaux, C. 1966. Chitinase. Methods Enzymol. 8:644-650. [Google Scholar]
- 13.Jones, J. D. G., K. L. Grady, T. V. Suslow, and J. R. Bedbrook. 1986. Isolation and characterization of genes encoding two chitinase enzymes from Serratia marcescens. EMBO J. 5:467-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keyhani, N. O., and S. Roseman. 1997. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc. Natl. Acad. Sci. USA 94:14367-14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keyhani, N. O., L. Wang, Y. C. Lee, and S. Roseman. 2000. The chitin disaccharide, N,N′-diacetylchitobiose, is catabolized by Escherichia coli and is transported/phosphorylated by the phosphoenolpyruvate:glycose phosphotransferase system. J. Biol. Chem. 275:33084-33090. [DOI] [PubMed] [Google Scholar]
- 16.Keyhani, N. O., Wang. L.-X., Y. C. Lee, and S. Roseman. 1996. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Characterization of an N,N′-diacetyl-chitobiose transport system. J. Biol. Chem. 271:33409-33413. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzo, V. D., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 20.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyashita, K., T. Fujii, and A. Saito. 2000. Induction and repression of a Streptomyces lividans chitinase gene promoter in response to various carbon sources. Biosci. Biotechnol. Biochem. 64:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Monreal, J., and E. T. Reese. 1969. The chitinase of Serratia marcescens. Can. J. Microbiol. 15:689-696. [DOI] [PubMed] [Google Scholar]
- 23.Perrakis, A., I. Tews, Z. Dauter, A. B. Oppenheim, I. Chet, K. S. Wilson, and C. E. Vorgias. 1994. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2:1169-1180. [DOI] [PubMed] [Google Scholar]
- 24.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saffen, D. W., K. A. Presper, T. L. Doering, and S. Roseman. 1987. Sugar transport by the bacterial phosphotransferase system: molecular cloning and structural analysis of the Escherichia coli ptsH, ptsI, and crr genes. J. Biol. Chem. 262:16241-16253. [PubMed] [Google Scholar]
- 26.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Suzuki, K., M. Suzuki, M. Taiyoji, N. Nikaidou, and T. Watanabe. 1998. Chitin binding protein (CBP21) in the culture supernatant of Serratia marcescens 2170. Biosci. Biotechnol. Biochem. 62:128-135. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki, K., M. Taiyoji, N. Sugawara, N. Nikaidou, B. Henrissat, and T. Watanabe. 1999. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 343:587-596. [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, K., N. Sugawara, M. Suzuki, T. Uchiyama, F. Katouno, N. Nikaidou, and T. Watanabe. 2002. Chitinases A, B, and C1 from Serratia marcescens 2170: enzymatic properties and synergism on chitin degradation. Biosci. Biotechnol. Biochem. 66:1075-1083. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki, K., T. Uchiyama, M. Suzuki, N. Nikaidou, M. Regue, and T. Watanabe. 2001. LysR-type transcriptional regulator ChiR is essential for production of all chitinases and a chitin-binding protein, CBP21, in Serratia marcescens 2170. Biosci. Biotechnol. Biochem. 65:338-347. [DOI] [PubMed] [Google Scholar]
- 31.Techkarnjanaruk, S., S. Pongpattanakitshote, and A. E. Goodman. 1997. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl. Environ. Microbiol. 63:2989-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujibo, H., K. Kondo, K. Tanaka, K. Miyamoto, N. Baba, and Y. Inamori. 1999. Molecular analysis of the gene encoding a novel transglycosylative enzyme from Alteromonas sp. strain O-7 and its physiological role in the chitinolytic system. J. Bacteriol. 181:5461-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiyama, T., F. Katouno, N. Nikaidou, T. Nonaka, J. Sugiyama, and T. Watanabe. 2001. Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170. J. Biol. Chem. 276:41343-41349. [DOI] [PubMed] [Google Scholar]
- 34.van Aalten, D. M. F., B. Synstad, M. B. Brugerg, E. Hough, B. W. Piise, V. G. H. Eijsink, and R. K. Wierenga. 2000. Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9-Å resolution. Proc. Natl. Acad. Sci. USA 97:5842-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe, T., K. Kimura, T. Sumiya, N. Nikaidou, K. Suzuki, M. Suzuki, M. Taiyoji, S. Ferrer, and M. Regue. 1997. Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179:7111-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]