Abstract
The foodborne pathogen Campylobacter jejuni is the primary causative agent of gastroenteritis in humans. In the present study a whole genome microarray of C. jejuni was constructed and validated. These DNA microarrays were used to measure changes in transcription levels over time, as C. jejuni cells responded to a temperature increase from 37 to 42°C. Approximately 20% of the C. jejuni genes were significantly up- or downregulated over a 50-min period after the temperature increase. The global change in C. jejuni transcriptome was found to be essentially transient, with only a small subset of genes still differentially expressed after 50 min. A substantial number of genes with a downregulated coexpression pattern were found to encode for ribosomal proteins. This suggests a short growth arrest upon temperature stress, allowing the bacteria to reshuffle their energy toward survival and adaptation to the new growth temperature. Genes encoding chaperones, chaperonins, and heat shock proteins displayed the most dramatic and rapid upregulation immediately after the temperature change. Interestingly, genes encoding proteins involved in membrane structure modification were differentially expressed, either up- or downregulated, suggesting a different protein membrane makeup at the two different growth temperatures. Overall, these data provide new insights into the primary response of C. jejuni to surmount a sudden temperature upshift, allowing the bacterium to survive and adapt its transcriptome to a new steady state.
Since the foundation of the U.S. Foodborne Disease Active Surveillance Network in 1996, Campylobacter has been the most frequently diagnosed foodborne pathogen in human, followed by Salmonella and Shigella spp. (18, 35). Campylobacter infection is an acute diarrheal disease that ranges from a day of mild diarrhea to severe abdominal pain (30). Rarely, a Campylobacter infection results in the development of the Guillain-Barré syndrome, which is the primary cause of neuromuscular paralysis in the United States (30). Poultry is known to be the main food vehicle for Campylobacter, with up to 88% of broiler carcasses being contaminated with this microorganism (2, 26).
Campylobacter jejuni is able to grow at temperatures ranging from 30 to 47°C, with an optimal growth temperature of 42°C. Campylobacters are likely to encounter a wide range of temperatures during a contamination cycle and must therefore be able to sense, adapt, and respond to these temperature fluctuations. Temperature could constitute an important stimulus for Campylobacter; the organism may use temperature to sense that it has invaded the chicken reservoir (core temperature of 42°C) or the human host (core temperature of 37°C). Differential gene expression at these two temperatures may allow this organism to colonize its host efficiently, leading to commensalism or pathogenesis. With the complete genomic sequence of C. jejuni available (24), it is now possible to identify in silico the presence or absence of a particular gene. The search for chaperone homologues and heat shock proteins in the C. jejuni genome reveals up to 17 proteins, several of which have already been characterized, including GroEL, GroES, DnaJ, DnaK, GrpE, HrcA, and Lon (22). Although the function of chaperones and heat shock proteins might be significantly different than bacterial adaptation to host temperature, they should play a crucial role in the first line response. By surmounting a sudden temperature upshift, the bacteria should be able to survive and adapt to the new temperature. The importance of the heat shock response in intestinal tract colonization is supported by two observations: (i) the heat shock proteins GroEL and GroES are immunogenic in experimentally infected rabbits (40), and (ii) a DnaJ mutant is impaired in its ability to colonize chicken and is affected in its growth at 46°C (17). Hence, the heat shock response should play an essential role in intestinal tract colonization and bacterial survival at high temperature. Recently, C. jejuni has been shown to upregulate the expression of at least 24 proteins upon temperature upshift (17). Although that study identified DnaJ as being one of the temperature-regulated proteins, the remaining 23 proteins remain to be fully characterized.
DNA microarray has been recently used to compare Campylobacter interstrain variations at the genomic level by using a “low-cost” microarray technology (9) that utilized a C. jejuni genomic library previously constructed for the genome sequencing project (24). This microarray consists of PCR products amplified from the sequence-defined pUC18 clones by using universal vector primers. One major drawback of this low-cost microarray technology is the presence of more than one gene per element on the array, making this array extremely difficult to analyze and likely ineffective for transcriptome profiling. The construction of C. jejuni microarray (carrying a single gene per element) is described here and is used to investigate differential gene expression profile in response to a temperature upshift from 37 to 42°C over a period of 50 min.
Array construction and validation.
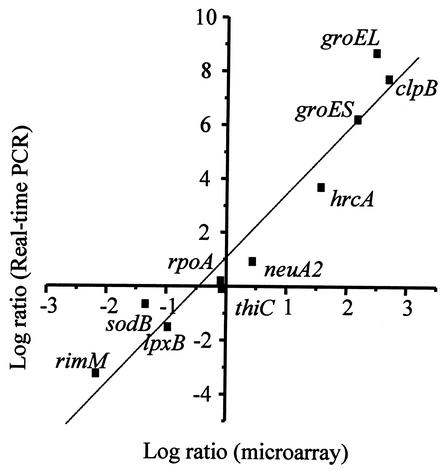
A C. jejuni array was constructed by PCR amplification of internal fragments of each open reading frame (ORF) from the annotated genomic sequence of C. jejuni NCTC 11168. Of all of the 1,654 predicted ORFs, 1,626 were successfully amplified; these represent 98.3% of the genome. Details of the methods used (microarray construction, RNA extraction, and labeling, as well as data collection and analysis) and a complete list of the genes represented on the microarrays are available online (http://www.cvm.okstate.edu/research/Facilities/CampyLab). The reliability of the data generated by using this microarray was assessed by cohybridization of two identical cDNA samples (prepared from total RNA isolated from two identical but independent bacterial cultures) that were labeled independently with Cy3 or Cy5 dyes. The pattern of hybridization revealed a linear correlation with no more than a 1.5-fold change in the relative gene expression level. This control experiment suggested that genes with an expression ratio beyond this range were either down- or upregulated. Therefore, only genes identified as being up- or downregulated by at least 1.5-fold were chosen for further study. Moreover, a 1.5-fold threshold has recently been reported as being biologically significant (15, 31). In addition, experiments were conducted independently twice (with dye swap) yielding at least four measurements per gene (representing two technical and two biological replicates, since each gene is present twice on each slide) to confirm the reproducibility of the gene expression data. Genes with highly reproducible expression ratios were selected. The Student t test was applied to the data, and genes with a P value of <0.01 were considered to have significant differential expression. By using a significance level of 0.01, we could anticipate a maximum of 17 false-positive results per time point (with 1,626 variables). However, this number is significantly reduced by the threshold of a 1.5-fold change in gene expression. In summary, genes were selected according to the following algorithm: (i) signal intensity more than three times the standard deviation of the background at least in one channel, (ii) a fold change in relative expression level of >1.5, and (iii) a P value of ≤0.01. Finally, the data generated were further validated by real-time quantitative reverse transcription-PCR (RT-PCR) by using an ABI Prism 7700 DNA analyzer (Applied Biosystems, Foster City, Calif.) and the QuantiTect SYBR green RT-PCR kit according to the manufacturer's protocol. Ten ORFs, exhibiting high, moderate, and low expression (as identified by microarray analysis), were selected for comparative real-time RT-PCR analysis (Fig. 1). The gene expression levels obtained by real-time quantitative RT-PCR analysis were normalized to that of the thiC gene, since its expression was found to be invariant under different temperature changes. The expression level of thiC was normalized to that of the rpoA gene, the expression of which was also found to be invariant. Quantitative values were obtained by using the comparative threshold cycle (ΔΔCT) method recommended by Applied Biosystems. The CT value corresponds to the PCR cycle at which occurs the first detectable increase in fluorescence associated with the exponential growth of PCR products. The relative expression of each gene was determined three times in each of the two experimental RNA samples and is expressed as the fold difference in quantity of cDNA molecules present at 42°C relative to that present at 37°C. The resulting gene expression ratio was log transformed and plotted against the average log ratio values obtained by microarray analysis (Fig. 1). A high level of concordance (r = 0.967) was observed between microarray and RT-PCR data despite quantitative differences in the level of change. Overall, RT-PCR and DNA microarray data differed by an average of fivefold. Consequently, microarray data could be calibrated to provide a quantitative estimate of differential fold expression. This underestimation of fold changes by DNA microarray analysis has been previously reported (41), suggesting a smaller dynamic range for microarray measurements compared to those of RT-PCR. Nevertheless, this validation study by real-time RT-PCR indicated that our microarray approach produced accurate fold change differences with sufficient sensitivity to identify differentially regulated transcripts.
FIG. 1.
Comparison of expression measurements by microarray and RT-PCR assays. The fold changes in gene expression in response to temperature upshift from 37 to 42°C were log transformed (in base 2). The real-time RT-PCR log2 values were plotted against the microarray data log2 values.
Experimental design.
We chose to study the C. jejuni response to temperature shift over a 50-min period, since the cell densities of the 37 and 42°C cultures were identical at the last time point. This suggests that the observed transcriptional profiles were not influenced by the growth phase or cell density differences of the two temperature conditions. Campylobacter sp. was grown microaerobically at 37°C in Mueller-Hinton medium to an optical density at 600 nm of 0.1. The Campylobacter broth was centrifuged (5 min, 8,000 × g), and the pellet was resuspended in 10 ml of fresh growth medium and then split equally in two flasks containing growth medium (200 ml) preconditioned at 37 or 42°C. Samples of 10 ml were collected at 5, 10, 20, 30, and 50 min after the temperature shift; rapidly mixed with one volume of RNALater solution (Ambion, Austin, Tex.); and placed on ice. Cells were immediately collected by centrifugation at 4°C (3 min, 8,000 × g) and resuspended in lysozyme-TE buffer (50 mM Tris-Cl [pH 8], 1 mM EDTA, 0.5 mg of lysozyme/ml). Total RNA was isolated by using a hot phenol-chloroform protocol (7). Total RNA was reverse transcribed by using random hexamers in the presence of aminoallyl-dUTP, followed by labeling with succinimidyl-ester monoreactive dyes (Cy3 or Cy5). The level of gene expression was monitored by competitive hybridization to the microarray with cDNA obtained from bacteria grown at 42 and 37°C for each time point. Each hybridization experiment was repeated two times by using total RNA isolated from two independent time course experiments. Microarray data were extracted by using GenPix Pro 3 software (Axon) and statistically analyzed as described earlier.
Global gene expression analysis.
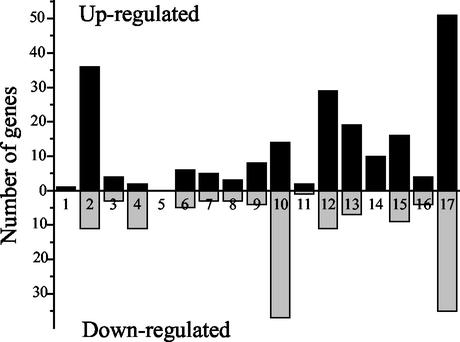
In total, 336 genes were identified to be differentially regulated by >1.5-fold (with 99% confidence) in at least one of the five time points. This represents ca. 20% of the 1,626 elements on the array. A total of 58% of the differentially transcribed genes were upregulated, and 42% were downregulated. The relative gene expressions in C. jejuni grown at 42°C versus C. jejuni grown at 37°C varied from 19.7- to 0.15-fold at different times after the temperature shift. Figure 2 provides a summary of the number of differentially expressed genes grouped by functional categories according to the C. jejuni NCTC 11168 genome annotation. This functional gene categorization provides information on the molecular mechanisms that allow C. jejuni to surmount a sudden temperature upshift. However, many genes whose expression was altered by temperature encode for proteins of unknown function (column 17, Fig. 2), thus displaying the limitations in understanding C. jejuni physiology. For genes of known or annotated function, those encoding proteins involved in energy metabolism (column 2, Fig. 2), cell wall and envelope constituents (column 12, Fig. 2), and transport and binding proteins (column 13, Fig. 2) are among the most upregulated genes. However, genes encoding proteins involved in synthesis and modification of macromolecules are among the most downregulated genes (column 10, Fig. 2).
FIG. 2.
Differentially expressed genes grouped by functional classification according to the Sanger Center C. jejuni genome database. Columns: 1, small molecules degradation; 2, energy metabolism; 3, central intermediary metabolism; 4, amino acid biosynthesis; 5, polyamine synthesis; 6, purines, pyrimidines, nucleosides, and nucleotides; 7, biosynthesis of cofactors, prosthetic groups, and carriers; 8, fatty acid biosynthesis; 9, regulatory functions; 10, synthesis and modification of macromolecules; 11, degradation of macromolecules; 12, cell envelope; 13, cell processes (transport and/or binding proteins); 14, cell processes (heat shock); 15, cell processes (other); 16, other; and 17, miscellaneous (unknown proteins). The solid or shaded bars represent the numbers of genes whose expression increased or decreased, respectively.
Cluster analysis of microarray data.
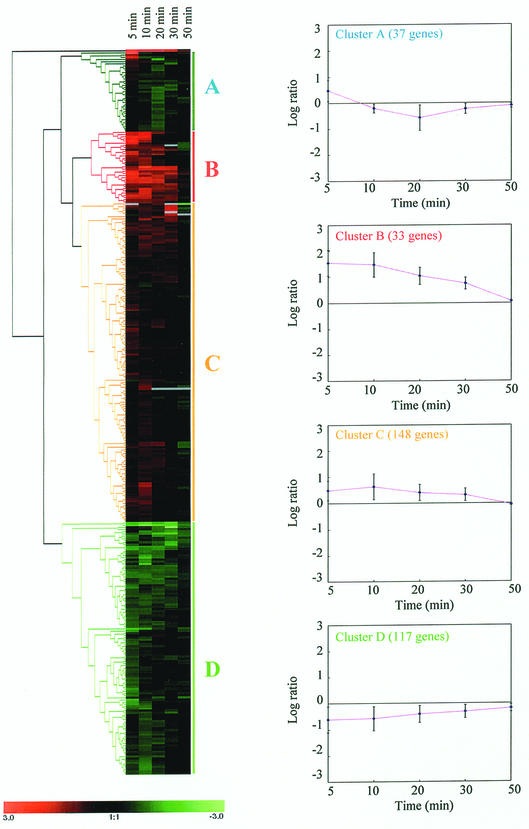
In order to identify and detect genes that were coregulated depending on the stage of temperature stress, a statistical analysis program was used to analyze the 336 genes previously selected. Briefly, the selected genes were subjected to hierarchical clustering by using Genesis software (version 1.0.2; Graz University of Technology [http://genome.tugraz.at]). This cluster analysis rearranges the data into new sets of better-ordered groups of genes sharing similar temporal expression pattern (10, 33). Four clusters were obtained, representing specific patterns of regulation (Fig. 3). Roughly, clusters B and C corresponded to genes that have their expression activated, whereas clusters A and D contained genes that have their expression repressed at 42°C relative to 37°C. One of the remarkable findings observed in this time course microarray analysis was that the global changes in gene expression upon temperature increase were essentially transient (Fig. 3). Immediately after the temperature stress, the bacterial cells responded with large changes in the expression level of 336 genes during the first 10 min, whereas <15% of these genes remained differentially expressed at 50 min. This gene expression pattern suggests that C. jejuni is able to adapt its transcript levels to a new steady state at 42°C over time with very few differentially expressed genes between the two growth temperature conditions. These initial global changes in transcript abundance at 5 and 10 min reflect the primary response of C.jejuni to surmount the sudden temperature increase, thereby allowing the bacterium to survive and ultimately adapt to the new temperature. This observation is in agreement with findings from Richmond et al. (27), who found very little differences in E. coli gene expression after heat shock (27). As exemplified here, time course experiments provide valuable information and are essential for deciphering the mechanism of bacterial response and adaptation to stresses. Temporal gene expression analysis can provide detailed insight into the mechanistic responses to stresses that are otherwise not easy to elucidate or identify using steady-state or single time point studies. Gene expression kinetics allow gene clustering and eventually putative gene function assignment based on a “guilt by association” principle. Steady-state or single time point studies of C. jejuni response to temperature changes would have missed ca. 85% of the genes whose expression is affected for the first 30 min.
FIG. 3.
Hierarchical clustering of 336 genes that varied significantly (P < 0.01 and a fold change > 1.5) in their expression profiles in response to a temperature change from 37 to 42°C. Each row represents a single gene expression, and each column represents an individual time point after the temperature upshift. An increasing green intensity indicates genes whose expression decreased, whereas an increasing red intensity indicates genes whose expression increased in response to the temperature upshift. The gray color indicates missing data. The mean of the gene expression patterns of each cluster is represented by plotting the log2 value of the expression ratio versus time. The error bars represent the standard deviation of the gene expression ratio.
Cluster A is composed of genes that are significantly downregulated at 20 min and are back near the baseline level at 50 min after the temperature increase. The most notable subgroup of genes with repressed expression includes a number of genes involved in ribosomal protein synthesis and modification (e.g., rplR, rplO, rplX, rpsH, rplL, rlpN, rpsQ, rpsN, rplF, and rpsI) and one gene involved in ribosome maturation and modification (ksgA, a putative dimethyladenosine transferase). In several gene expression studies with Saccharomyces cerevisiae, it has been observed that the expression level of ribosomal genes is similarly affected by various environmental stresses (13, 16). This repression of ribosomal genes suggests a brief growth arrest that allows the cell to reshuffle energy devoted to an increased expression of genes involved in protective response and adaptation to the new growth condition. This reduction in ribosomal gene expression, together with the upregulation of genes encoding proteins involved in energy metabolism, reflects the energy-starved condition of the cell and the necessity for saving and reshuffling energy for the increased expression of proteins involved in repairing damages caused by the temperature upshift. In addition, this transient decrease in ribosomal proteins probably resulted in a modest repression of de novo protein synthesis, allowing the bacterium to surmount heat shock damage and adapt its transcriptome to the new growth temperature. At 50 min after the temperature shift, the expression of the ribosomal genes returned to a basal level, indicating that C. jejuni maintains a constant level of ribosomal protein and RNA between 37 and 42°C. Since 42°C is the optimal growth temperature for C. jejuni, this bacterium probably responds, like Escherichia coli, to temperature augmentation by increasing the rate of peptide chain elongation.
Cluster B contains 33 genes and displays the most highly and rapidly upregulated genes. The genes in this cluster were immediately upregulated after the increase in temperature. After 10 min of exposure at 42°C, their expression gradually decreased with time to reach a baseline level at 50 min. Cluster B is dominated by genes encoding chaperones, chaperonins, and heat shock proteins. Specifically, groEL, groES, dnaK, dnaJ, hspR, cbpA, hrcA, lon, clpB (protease), hslU, and Cj1034c transcripts all increased (Table 1). Proteins belonging to the heat shock family have been intensively studied and have been shown to be induced in response to stress, in particular as a result of a sudden temperature upshift (42). These proteins act by repairing and preventing damages caused by an accumulation of unfolded proteins. Several of the heat shock proteins (e.g., DnaK and GroEL) also play a crucial role under normal physiological conditions by assisting in the proper folding of newly synthesized proteins. Recently, C. jejuni has been shown to mount a heat shock response, with the preferential synthesis of 24 proteins upon temperature increase (17). Of these 24 proteins, only DnaJ has been further characterized. Our study confirms the upregulation of dnaJ upon temperature stress and demonstrates the temperature-responsive regulation of many other heat shock proteins (Table 1). The molecular mechanism of heat shock regulation in C. jejuni is poorly understood. Many studies have demonstrated that heat shock regulation differs among bacterial species. In E. coli, the alternative sigma activator σ32 (encoded by rpoH) mediates the expression of most heat shock proteins (42). Analysis of the C. jejuni genome reveals the absence of a σ32 homologue, suggesting a regulation mechanism of the heat shock response different than those in other gram-negative bacteria such as E. coli. In Bacillus subtilis and other gram-positive bacteria, the heat shock response is regulated by diverse regulatory strategies depending on the specific heat shock genes (42). In B. subtilis, class I heat shock genes are negatively regulated by HrcA, class II genes are positively regulated by the transcriptional activator σB, and class III genes are negatively regulated by CtsR. The HrcA repressor acts at the DNA level by binding to conserved palindromic sequence, named CIRCE (for controlling inverted repeat of chaperone expression) (43). Upon heat shock, HrcA dissociates from its operators, leading to the transcriptional expression of class I (groE and dnaK) heat shock genes. Interestingly, the C. jejuni genome contains a homologue of the hrcA gene arranged in a cluster with two heat shock genes, grpE and dnaK. Consequently, HrcA might serve as a negative regulator of grpE and dnaK in C. jejuni just as it does in B. subtilis. This hypothesis is supported by the identification of a cis-acting regulatory element (CIRCE) upstream of the hrcA gene (36). The C. jejuni CIRCE sequence exhibits a putative stem-loop structure with some similarities to the consensus CIRCE sequence. The C. jejuni CIRCE sequence is 5′-CTAGCAATC-N8-GAGTGCTAG-3′, with unmatching bases underlined (the loop of the putative hairpin structure is composed of eight nucleotides instead of nine in the consensus sequence). This campylobacter CIRCE sequence was also identified upstream of the groESL operon translation start site (37). However, this CIRCE sequence could not be identified in front of the other genes from cluster B, suggesting that these other genes might be regulated differently than the groESL and hrcA-grpE-dnaK operons. The C. jejuni genome lacks homologues of the other two B. subtilis heat shock regulators, σB and CtsR. Interestingly, the C. jejuni genome contains a homologue of Streptomyces albus HspR regulator (4). HspR is organized in an operon with the cbpA gene, which belongs to the heat shock protein DnaJ family. HspR is contained within cluster B, and thus HspR might likely regulate several genes from cluster B. Indeed, recent investigations have shown that HspR regulates the expression of dnaK, groESL, and cbpA genes in C. jejuni. Therefore, C. jejuni may use more than one strategy to simultaneously regulate distinct sets of heat shock genes. This complex regulatory network may allow the bacterium to fine tune heat shock gene expression in response to a temperature upshift or other stimuli (e.g., oxidative shock, acid shock, and osmotic shock).
TABLE 1.
Fold change in the expression level of C.jejuni heat shock genes in response to a temperature upshift from 37 to 42°C
| Gene | Gene product or function | Induction ratioa |
|---|---|---|
| groEL | 60-kDa chaperonin | 5.5 |
| groES | 10-kDa chaperonin | 3.8 |
| grpE | Heat shock protein | 19.7 |
| dnaK | Heat shock protein | 3.5 |
| dnaJ | Chaperone DnaJ | 1.4 |
| hspR | Putative heat shock transcriptional regulator | 2.6 |
| cbpA | Putative curved DNA-binding protein | 5.1 |
| hrcA | Putative heat shock regulator | 3.7 |
| lon | ATP-dependent protease | 1.4 |
| clpB | Protease | 5.1 |
| hslU | Putative heat shock protein | 1.4 |
| Cj1034c | Possible DnaJ-like protein | 1.6 |
The induction ratio is the maximum value obtained over five time points.
Another set of genes in cluster B contains napABGH, Cj1358c and Cj1357c genes, which encode a nitrate reductase of the periplasmic nap type, a NapC/NirT/NrfH homologue, and a putative periplasmic cytochrome c (homologous to the nitrate reductase NfrA of E. coli), respectively. This set of genes could be predicted to allow the bacterium to carry out respiration with nitrate and nitrite as electron acceptors. In fact, it has recently been reported that nitrate sustains C. jejuni growth under oxygen-limited conditions, indicating its role in energy conservation (28). Thus, the upregulation of these genes may be caused by a decreased pO2 of the culture medium at 42°C compared to that which occurs at 37°C (since gas solubility decreases with increasing temperature) allowing C. jejuni to respire on nitrate. This hypothesis was supported by the downregulation of genes encoding for proteins involved in aerobic metabolism (constituting the cluster D) upon temperature increase. Analysis of the C. jejuni genome revealed the presence of two other reductases that are predicted to allow the use of fumarate (FrdCAB homologues) and N- or O-oxide reductase (DorA/TorA homologue) as alternative electron acceptors to oxygen. In contrast to the nitrate reductase, these two other reductases were not differentially expressed at 42°C relative to 37°C. One explanation for their absence of upregulation could be the lack of sufficient electron acceptors (fumarate and N- or O-oxide compounds) in Mueller-Hinton broth. Likewise, Mueller-Hinton broth should contain only traces of nitrate, and yet genes involved in nitrate reduction were differentially expressed. The homologue E. coli Nap enzymes have been shown to exhibit a high affinity for nitrate, making it ideal for scavenging a low nitrate concentration as encountered in vivo (32). Thus, the trace amount of nitrate within the broth medium might be sufficient to induce the nap genes expression at a low oxygen concentration.
Cluster C contains 148 genes. The gene expression in cluster C resembled that of cluster B. The level of gene expression in this cluster was upregulated between 1.5- to 2-fold at 5 and 10 min after a temperature increase from 37 to 42°C. As with genes from clusters A and B, cluster C gene expression gradually decreased over time until it reached basal level at 50 min after the temperature change. Interestingly, many genes encoding proteins known or presumed to be involved in chemotaxis, flagellum biosynthesis, and flagellar motility are part of this cluster. A similar effect on the expression of flagellar and chemotaxis genes in response to heat shock has also been recently demonstrated in E. coli (27). This set of genes includes fliI (flagellum-specific ATP synthase), fliQ (flagellar biosynthetic protein), fliD (flagellar hock-associated protein), flgH (flagellar L-ring protein precursor), flgD (flagellar hock assembly protein), flgE2 (flagellar hock subunit protein), flgI (flagellar P-ring protein precursor), flaA (flagellin), flaC (flagellin), Cj1312 (possible flagellum protein), cheV (chemotaxis protein), cheW (chemotaxis protein), Cj0262c (putative methyl-accepting chemotaxis signal transduction protein), and neuB3 (N-acetylneuraminic acid synthetase, known to be involved in flagellin glycosylation). This observation is in agreement with the findings of Alm et al. (1) that the flaB gene expression is induced 2.5-fold at a growth temperature of 42°C compared to induction at 37°C, suggesting that flagellar gene expression is temperature regulated. Although flaB was not selected as differentially expressed by using the gene selection algorithms described above, the microarray data show that the flaB gene expression is in fact upregulated by 1.4-fold (P < 0.01) at 5, 10, 20, and 30 min and reaches a basal expression level at 50 min. This microarray data definitely demonstrates that a growth temperature of 42°C induces flagellar biosynthesis and consequently motility. The molecular mechanism of Campylobacter chemotaxis and flagellar gene regulation are ill defined. The present study reveals that these genes are upregulated upon a temperature increase. Undoubtedly, flagellum and chemotaxis proteins should play an essential role in Campylobacter gut colonization. Indeed, nonmotile mutants are unable to colonize intestinal tracts (19, 20, 25, 38). Therefore, the upregulation of chemotaxis and flagellar genes at 42°C, which is the core temperature of the chicken reservoir, might condition C. jejuni in order to spread rapidly through large broiler flocks and to colonize efficiently the human intestinal tract. This hypothesis is supported by the observation that flagellin proteins are overexpressed when Campylobacter is maintained within chicken implants compared to in vitro growth observed at 37°C (6) and that the chick colonization potential of C. jejuni could be increased 10,000-fold after a single passage in chickens (5).
Interestingly, several genes encoding for restriction and modification enzymes are included in cluster C. These genes are Cj0031 (putative type IIS restriction-modification enzyme, N-terminal half), Cj0032 (putative type IIS restriction-modification enzyme, C-terminal half), Cj1549c (putative type I restriction enzyme R protein), and Cj1551c (putative type I restriction enzyme S protein). The role of the DNA restriction and modification systems in C. jejuni is unclear at present. These restriction-modification enzymes might be involved in the breakdown of foreign DNA. These enzymes might also be necessary for stimulating the formation of DNA fragmentation and recombination resulting in antigenic diversity and variation, such as the homologous recombination observed for the virulence-associated flagellin locus of C. jejuni (14, 21, 39). Interestingly, the presence of restriction-modification systems in Helicobacter pylori has been recently associated with the bacterial ability to infect its host (3), implying that these enzymes might affect the virulence gene expression. Based on this observation, it is tempting to suggest a similar function in C. jejuni, where these enzymes would control the expression of genes involved in chicken gut colonization.
Another group of genes from cluster C encodes for proteins involved in energy metabolism. Representative genes from this functional group encode for Ni/Fe hydrogenase (hydAB), Ni/Fe hydrogenase B-type cytochrome (hydC), formate dehydrogenase (fdhABC), and cb-type cytochrome c oxidase (ccoO, ccoN, and ccoQ). The upregulation of these genes at 42°C clearly correlates with the upregulation of the nitrate reductase napABGH, Cj1358c and Cj1357c from cluster B. Undoubtedly, C. jejuni shifted from aerobic metabolism to anaerobic metabolism upon temperature increase; this was probably caused by a decrease in the oxygen concentration in the broth medium at the higher temperature.
Finally, another worthwhile functional set of genes from cluster C encodes proteins involved in surface structure biosynthesis and modification, UDP-glucose 4-epimerase (galE), phosphoheptose isomerase (gmhA2), 3-deoxy-d-manno-octulosonic acid transferase (kdtB), putative transferase (Cj1321), acylneuraminate cytidylyltransferase (neuA2), N-acetylneuraminic acid synthetase (neuB3), putative ADP-heptose synthase (waaE), and a putative glycosyltransferase (wlaE). It has been previously reported that the protein encoded by galE plays an essential role in the synthesis of C. jejuni lipopolysaccharide (LPS), probably by catalyzing the interconversion of UDP-glucose into UDP-galactose (11). A galE mutant expresses a truncated lipid A core molecule and is affected in its ability to invade and adhere to human intestinal cells, thus demonstrating the involvement of GalE in LPS biosynthesis and the significance of LPS as a virulence factor (11). GalE gene is part of a 16-kb region, named pgl/wla locus (12), which contains 13 genes encoding proteins involved in LPS and lipooligosaccharide biosynthesis. Genes from this locus (galE and wlaBCDEFGHIKLM) have also been shown to be involved in a mechanism of general protein glycosylation in C. jejuni (34). Surprisingly, the microarray data show that several genes from this cluster are expressed differently. Whereas wlaE and galE are upregulated upon temperature increase, wlaK is downregulated (and is part of cluster D). These results suggest that the genes from the pgl/wla locus are transcribed as multiple operons. In support of this conclusion is the absence of a polar mutation by insertion of a chloramphenicol or kanamycin cassette in either orientation within several genes from the wlaB-M region (12). From these microarray data, it is apparent that the expression of several genes from the protein glycosylation locus is temperature regulated in C. jejuni. This differential regulation of glycosylases genes should lead to differential glycosylation patterns of surface structures between 37 and 42°C. This complex mechanism of protein glycosylation may allow the bacterium to efficiently colonize different ecological niches, such as the intestinal tract of birds (core temperature of 42°C) and mammals (core temperature of 37°C), and/or to modify the protein membrane “make-up” in order to evade the host immune system.
Cluster D contains 117 genes. The expression level of the genes in this cluster is decreased by 1.5- to 3-fold at 5 and 10 min after the temperature upshift. A subgroup of noteworthy genes encode for the succinate dehydrogenase (sdhABC). This enzyme complex is a membrane component of the tricarboxylic acid cycle. It catalyzes the oxidation of succinate to fumarate and participates in the aerobic respiratory chain by reducing the ubiquinone pool in the membrane. The gene expression of E. coli succinate dehydrogenase has been studied and appears to be under the control of both oxygen concentration and carbon sources (23, 29). The E. coli enzyme is subjected to anaerobic repression via the two regulators, ArcA and Fnr (23, 29). Whereas the C. jejuni genome lacks homologues of both ArcA and Fnr, Campylobacter sdhABC genes seem to be similarly repressed under a low oxygen concentration as a result of a temperature increase to 42°C. The downregulation of this set of genes is in agreement with the upregulation of genes encoding proteins involved in the anaerobic metabolism (genes from cluster B and C). Given that ArcA and Fnr homologues are absent in the Campylobacter genome, this microorganism should regulate gene expression in response to oxygen deprivation via a novel and uncharacterized mechanism. E. coli ArcA has also been shown to repress other enzymes involved in aerobic metabolism, including manganese superoxide dismutase (encoded by the sodA gene) (8). Interestingly, C. jejuni iron superoxide dismutase gene (named sodB) is part of cluster D, suggesting that sodB and sdhABC might be coregulated via an anaerobic responsive repressor.
One other major functional group of genes in cluster D encodes proteins involved in surface structure biosynthesis and modification, including WlaK (putative aminotransferase), LpxB (lipid A-disaccharide synthase), and Fcl (putative fucose synthase). The downregulation of these proteins, together with the upregulation of other proteins belonging to the same functional group, clearly suggests a differential surface structure pattern between 42 and 37°C.
Conclusion.
The present study provides a genome-wide expression profile of C. jejuni in response to a temperature change from 37 to 42°C. A complete repertoire of genes are either induced or repressed at 42°C relative to 37°C. This time course study of gene expression reveals that a large number of genes are differentially expressed during the first 10 min, whereas only a few of them remain differentially expressed 50 min after temperature upshift. This report identifies up to 336 genes that have their expression affected by the temperature change. Whereas clustering analysis was used as a first step to identify coregulated genes, a genetic analysis of the regulatory pathways needs to be undertaken in order to fully characterize the major regulators.
Acknowledgments
This study was supported by NIH grant number RR15564 from the COBRE program of the National Center for Research Resources and by OCAST grant number HR02-012RS.
The technical assistance of D. Smalley (microarray printing) is gratefully acknowledged. I am grateful to all of the staff from OSU and the OU microarray core facilities. I also thank B. Barrow, J. Andrus, and I. Turcot for providing helpful comments on the manuscript.
REFERENCES
- 1.Alm, R. A., P. Guerry, and T. J. Trust. 1993. The Campylobacter sigma 54 flaB flagellin promoter is subject to environmental regulation. J. Bacteriol. 175:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berndtson, E., M. L. Danielsson-Tham, and A. Engvall. 1996. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int. J. Food Microbiol. 32:35-47. [DOI] [PubMed] [Google Scholar]
- 3.Bjorkholm, B. M., J. L. Guruge, J. D. Oh, A. J. Syder, N. Salama, K. Guillemin, S. Falkow, C. Nilsson, P. G. Falk, L. Engstrand, and J. I. Gordon. 2002. Colonization of germ-free transgenic mice with genotyped Helicobacter pylori strains from a case-control study of gastric cancer reveals a correlation between host responses and HsdS components of type I restriction-modification systems. J. Biol. Chem. 277:34191-34197. [DOI] [PubMed]
- 4.Bucca, G., Z. Hindle, and C. P. Smith. 1997. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J. Bacteriol. 179:5999-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cawthraw, S., T. M. Wassenaar, R. Ayling, and D. G. Newell. 1996. Increased colonization potential of Campylobacter jejuni strain 81116 after passage through chickens and its implication on the rate of transmission within flocks. Epidemiol. Infect. 117:213-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chart, H., D. Conway, J. A. Frost, and B. Rowe. 1996. Outer membrane characteristics of Campylobacter jejuni grown in chickens. FEMS Microbiol. Lett. 145:469-472. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compan, I., and D. Touati. 1993. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J. Bacteriol. 175:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry, B. N., S. Feng, Y. Y. Chen, D. G. Newell, P. J. Coloe, and V. Korolik. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry, B. N., N. J. Oldfield, V. Korolik, P. J. Coloe, and J. M. Ketley. 2000. Genetics of Campylobacter lipopolysaccharide biosynthesis, p. 381-403. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 13.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 16.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konkel, M. E., B. J. Kim, J. D. Klena, C. R. Young, and R. Ziprin. 1998. Characterization of the thermal stress response of Campylobacter jejuni. Infect. Immun. 66:3666-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morooka, T., A. Umeda, and K. Amako. 1985. Motility as an intestinal colonization factor for Campylobacter jejuni. J. Gen. Microbiol. 131(Pt. 8):1973-1980. [DOI] [PubMed] [Google Scholar]
- 20.Nachamkin, I., X. H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 59:1269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuijten, P. J., A. J. van den Berg, I. Formentini, B. A. van der Zeijst, and A. A. Jacobs. 2000. DNA rearrangements in the flagellin locus of an flaA mutant of Campylobacter jejuni during colonization of chicken ceca. Infect. Immun. 68:7137-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, S. F. 2000. Environmental regulatory genes, p. 423-440. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 23.Park, S. J., C. P. Tseng, and R. P. Gunsalus. 1995. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15:473-482. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. V. Vliet, S. Whitehead, and B. G. Barell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 25.Pavlovskis, O. R., D. M. Rollins, R. L. Harberberger, A. E. Green, L. Habash, S. Stroko, and R. I. Walker. 1991. Significance of flagella in colonization resistance of rabbits immunized with Campylobacter spp. Infect. Immun. 66:938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescott, J. F., and C. W. Bruin-Mosch. 1981. Carriage of Campylobacter jejuni in healthy and diarrheic animals. Am. J. Vet. Res. 42:164-165. [PubMed] [Google Scholar]
- 27.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen, J., and R. P. Gunsalus. 1997. Role of multiple ArcA recognition sites in anaerobic regulation of succinate dehydrogenase (sdhCDAB) gene expression in Escherichia coli. Mol. Microbiol. 26:223-236. [DOI] [PubMed] [Google Scholar]
- 30.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 66-88. In M. J. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 31.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart, V., Y. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207-208. [DOI] [PubMed] [Google Scholar]
- 34.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 35.Tauxe, R. V. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thies, F. L., H. Karch, H. P. Hartung, and G. Giegerich. 1999. Cloning and expression of the dnaK gene of Campylobacter jejuni and antigenicity of heat shock protein 70. Infect. Immun. 67:1194-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thies, F. L., A. Weishaupt, H. Karch, H. P. Hartung, and G. Giegerich. 1999. Cloning, sequencing, and molecular analysis of the Campylobacter jejuni groESL bicistronic operon. Microbiology 145:89-98. [DOI] [PubMed] [Google Scholar]
- 38.Wassenaar, T. M., B. A. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139(Pt. 6):1171-1175. [DOI] [PubMed] [Google Scholar]
- 39.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141(Pt. 1):95-101. [DOI] [PubMed] [Google Scholar]
- 40.Wu, Y. L., L. H. Lee, D. M. Rollins, and W. M. Ching. 1994. Heat shock- and alkaline pH-induced proteins of Campylobacter jejuni: characterization and immunological properties. Infect. Immun. 62:4256-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wurmbach, E., T. Yuen, B. J. Ebersole, and S. C. Sealfon. 2001. Gonadotropin-releasing hormone receptor-coupled gene network organization. J. Biol. Chem. 276:47195-47201. [DOI] [PubMed] [Google Scholar]
- 42.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Henggee-Aronis (ed.), Bacterial stress responses. Amercian Society for Microbiology, Washington, D.C.
- 43.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]