Abstract
A novel promoter in IS10R (OUTIIp) has been found in one of its ends in an inverted position relative to promoter pOUT. OUTIIp shows characteristics similar to those of rpoS-dependent promoters such as a gearbox expression pattern. It is under catabolite repression and positively regulated by ppGpp or conditioned media. This opens new challenges in IS10R transposition.
The nonessential gene nlpD, which codes for the NlpD lipoprotein (21), contains the promoter region for rpoS, a gene that codes for σS, an alternative sigma factor key in the expression during stationary phase and in the acquisition of resistance to environmental stress during this phase (22, 23). A copy of the insertion sequence IS10R has been found to interrupt the nlpD gene of Enterobacter cloacae strain CETC960, preventing the expression of rpoS from rpoSp (unpublished results). Nevertheless, this strain was not defective in σS (24); therefore, the coding gene was transcribed due to the presence of an unknown promoter located within IS10R. This element contains two well-characterized promoters that lie in opposite orientations near the boundary: pIN, a weak promoter required for transposase transcription, and pOUT, a strong promoter that sponsors transcription of an antisense RNA that, pairing with the tnp RBS region, represses transposase expression (7, 31).
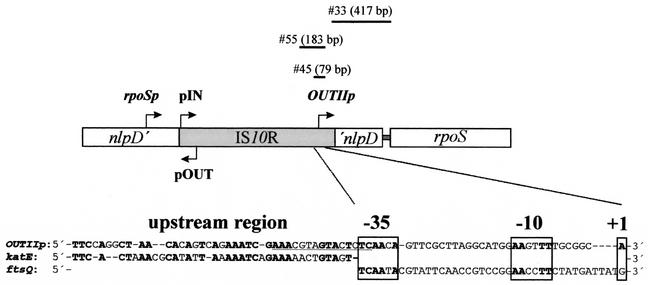
E. cloacae CECT960 rpoS transcriptional start site and identification of OUTIIp.
The presence of IS10R within nlpD did not prevent rpoS expression in E. cloacae CETC960, as there is evidence of a functional RpoS in this strain (24). However, IS10R prevented rpoS transcription from its own promoter, rpoSp, as shown by reverse transcriptase PCR (results not shown). These results suggest that a promoter within IS10R would drive rpoS transcription in CECT960.
The in vivo start site of E. cloacae rpoS RNA, identified by primer extension, indicated that rpoS transcription initiated at the same position in both exponential- and stationary-phase cells, although a weaker signal was obtained in the former as measured by densitometry with the Multi-Analyst 1.1 program from Bio-Rad (results not shown). The start point of transcription was identified 74 bp upstream of one border of IS10R in a position where no promoter has been described. We propose to name this novel promoter OUTIIp, since its location and orientation in one end of IS10R are similar to that of pOUT in the other end (7, 31).
The analysis of the promoter region did not show clear −35 and −10 sequences, presenting an overall identity of 50% with the σ70 consensus box (Fig. 1). The presence of certain nucleotides upstream of the −10 box (extended −10 sequences) increases the strength of promoters in gram-positive and -negative bacteria, making inessential the −35 recognition region (16, 19). We propose an extended −10 sequence (TG-AAGTTT) for OUTIIp that would facilitate transcription initiation in an otherwise poor promoter.
FIG. 1.
E. cloacae CECT960 rpoS organization, isolation of fragments no. 33, 45, and 55, and OUTIIp sequence. Several E. cloacae chromosomal sequences were PCR amplified. The sizes and locations with respect to the rpoS gene and the IS10R element are shown. Comparison of OUTIIp and katEp upstream sequences and of OUTIIp and the ftsQ promoters is also shown. −35 and −10 sequences and start points (+1) are shown in boxes. Identical nucleotides are shown in boldface type. A putative CRP-binding site is underlined in the OUTIIp sequence.
A series of transcriptional lacZ fusions were constructed by PCR, obtaining fragments no. 33, 45, and 55 (Fig. 1), which were cloned into pRS551 (32), yielding plasmids pRS33, pRS45, and pRS55, which were transformed into Escherichia coli ZK126 (10). The copy number, measured as described previously (28), was in all cases between 80 and 90 plasmids per chromosome equivalent. The fragments were also cloned in monocopy into the E. coli RYC1000 chromosome as described earlier (32), rendering strains REM33, REM45, and REM55. β-Galactosidase measurements (25) conducted in rich and minimal media showed that pRS33 and REM33 failed to drive measurable LacZ levels, whereas the expression levels of pRS55 (8,935 ± 550 Miller units) and its corresponding lysogen REM55 (87 ± 5 Miller units) were similar to those measured in pRS45 (8,914 ± 500 Miller units) and REM45 (88 ± 5 Miller units). Therefore, the sequences required for OUTIIp activity must lie in the region defined by E4 and B5.
Influence of the C-terminal region of the α subunit of the RNA polymerase in OUTIIp activity.
The OUTIIp upstream region has a slightly high A/T content (Fig. 1) that could act as an upstream element interacting with the C-terminal region of the α subunit. Therefore, the requirement of the α C-terminal region in OUTIIp activity was studied. The REM strains were transformed with plasmids pLAD235 and pLAD256, which carry mutant rpoA genes under isopropyl-β-d-thiogalactopyranoside (IPTG) control (14). These mutant genes code for an α subunit deleted in their C-terminal region (235 residues in RpoAΔ235CTD and 256 in RpoAΔ2565CTD). In the presence of IPTG, transcription of mutant rpoA genes is turned on. In IPTG-induced wild-type cells harboring plasmid pLAD235 or pLAD256, 90% of RNA polymerases contains at least one mutant α subunit, while, in 50% of the polymerases, the two α subunits are mutant (34). Under these conditions, transcription starting in rrnB1p (a promoter that requires the C-terminal domain of the α subunit for initiation) is greatly reduced (29). As a control, plasmid pLAX181 (14) carrying the wild-type rpoA allele was used. LacZ activity was similar in the presence or absence of IPTG in the REM strains carrying pLAX181, pLAD235, and pLAD256 (results not shown). Thus, it can be concluded that the C-terminal region of the α subunit of the RNA polymerase is not required for OUTIIp activity. Moreover, the presence of upstream activating sequences did not seem to be required, as promoter activity of fragment no. 45 was the same as that of fragment no. 55 (which has a 104-bp upstream additional region).
Growth phase-dependent regulation of OUTIIp.
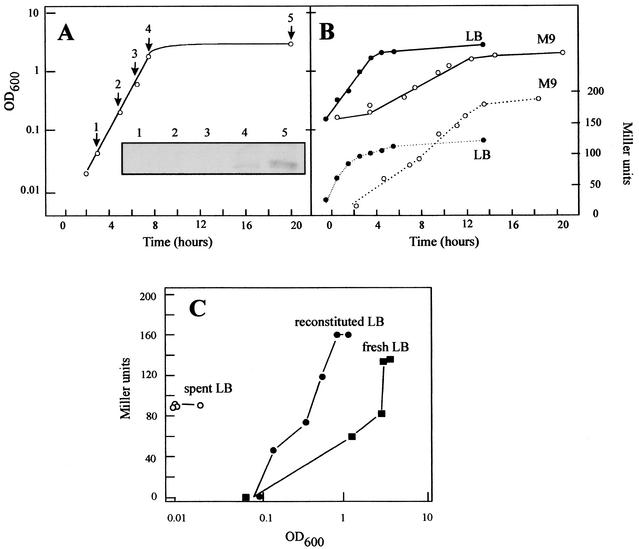
Lange and Hengge-Aronis (20) reported that E. coli rpoS expression follows a pattern similar to the growth curve. We studied whether OUTIIp presented a similar pattern of expression. RpoS, studied in E. cloacae CECT960 by Western blotting (Fig. 2A), began to be detected as a very faint band at the onset of stationary phase, reaching higher values at later times (measured by densitometry). Similar results were obtained when transcription driven by OUTIIp was examined in REM55 in both minimal and Luria-Bertani (LB) media (Fig. 2B). One of the signals that could be involved in this process is that of ppGpp, a transcriptional regulator that increases its levels in nutrient-limited cultures, its generation being a primary response to stress and nutritional deficiencies that are followed by the accumulation of poly(P) and RpoS (8, 12). However, the induction due to ppGpp could be limited to a basal expression (15) or could be modulated by the presence of other factors such as DksA (5). To verify if ppGpp regulates OUTIIp, β-galactosidase assays were performed with E. coli strain MG1655 and its derivative CF1693, ΔrelA, ΔspoT, a ppGpp-deficient strain (12) containing pRS55. β-Galactosidase expression during stationary phase decreased 50% in the absence of this alarmone, thus indicating the requirement of the alarmone in OUTIIp expression.
FIG. 2.
(A) E. cloacae rpoS expression. Samples were withdrawn at the times indicated in the growth curve (LB medium, 37°C), and total protein was isolated and normalized for equal total concentration. The inset shows the results of a Western blot by using polyclonal serum against σS. OD600, optical density at 600 nm. (B) OUTIIp activity versus growth rate in LB medium and in M9 supplemented with 0.4% glycerol (vol/vol) media. β-Galactosidase activity (dashed line) and optical density at 600 nm (solid line) were measured at different times during the growth of E. coli strain REM55. (C) Effect of spent LB medium (sterilized supernatant obtained after centrifugation of a 24-h-grown culture) on OUTIIp activity. REM55 was inoculated in LB broth and in spent LB medium and in reconstituted LB medium (spent LB medium plus 20× LB medium to a final concentration of 1×, pH adjusted to 7.5). The optical density at 600 nm and β-galactosidase activities were measured at different incubation times. β-Galactosidase activity was plotted against the optical density at 600 nm. The data shown are the average of at least three independent cultures, with duplicate measurements. The overall variation was below 15%.
Slow-growing cells (M9 medium supplemented with 0.4% [vol/vol] glycerol) showed a level of expression almost twice that of fast-growing cells (LB medium). The inverse relationship between promoter strength and growth rate showed by OUTIIp is characteristic of gearbox promoters such as katEp and ftsQp (2). The OUTIIp upstream sequence presents around 60% similarity to E. coli katEp (35), and its −35 and −10 sequences have a 75% identity (Fig. 1) with those of E. coli ftsQp (1). As ftsQ and katE are σS dependent, the influence of σS on OUTIIp was studied. E. coli strains ZK918 and rpoS::Kmr (4) and its isogenic strain ZK126 were transformed with pRS55. A 63% decrease in OUTIIp strength was observed in the absence of RpoS. Although OUTIIp is partially σS dependent, it does not bear two important characteristics of σS-dependent promoters (11): it does not present upstream intrinsic curvature (results not shown), as katEp and most of σS-dependent promoters do, and it bears a poor similarity with the −10 σS consensus sequence (AAGTTT versus CTATACT). Moreover, OUTIIp lacks the proposed features for an optimally σS-selective promoter: no −35 region, a TC motif at the −14/−13 positions, and a TATACT −10 hexamer (3). In fact, OUTIIp presents a relatively well-conserved −35 sequence (TCAACA). Perhaps all these features could be responsible for the low activity of this promoter. As OUTIIp activity was not completely abolished in an rpoS strain, it is possible that the presence of a −35 sequence allows σ70 RNA polymerase to recognize it. In this sense, the presence of an extended −10 sequence, a feature more common in gram-positive bacteria than in gram-negative bacteria (16, 19), is probably important. This fact supports the idea of a gram-positive origin of Tn10, as its G+C content is closer to that of Bacillus subtilis than to that of Enterobacteriaceae (9).
Catabolite repression.
The expression of some operons is inhibited in a coordinated process known as catabolite repression when glucose is available. Growth in 0.4% (wt/vol) glucose inhibited OUTIIp transcription (6 ± 0.2 Miller units versus 87 ± 5 Miller units) in the REM55 strain. Catabolite repression is set in train by the ability of glucose to reduce the level of cyclic AMP (cAMP) in the cell. This metabolite is bound by CRP, a cAMP receptor protein that binds to more or less conserved DNA sequences and whose presence is necessary to initiate transcription at many promoters (18). A putative CRP-binding site was found overlapping the −35 sequence of OUTIIp (Fig. 1). To determine if OUTIIp catabolite repression observed in the presence of glucose was CRP dependent, pRS55 was introduced in a crp strain, SBS688 (27). A 41% drop in OUTIIp activity was observed. These results indicate that OUTIIp is subject to catabolite repression. OUTIIp is probably a class II CRP-dependent promoter because, as shown in Fig. 1, the putative CRP-binding site (73% identity with the left half of the consensus sequence) overlaps the −35 determinant for the binding of RNA polymerase (6). Class II CRP-dependent promoters have a single DNA site for CRP overlapping the DNA site for RNA polymerase. Transcription at class II CRP-dependent promoters requires two mechanistic components: an overcoming of an inhibitory effect of the α C-terminal domain of RNA polymerase and a direct activation involving the α N-terminal domain (6). Therefore, absence of the α C-terminal domain would not have an effect on CRP-mediated OUTIIp activity in agreement with our results. The decrease in expression in the crp strain was not as dramatic as in the presence of glucose, probably due in part to the lower growth rate of the mutant that would increase expression of OUTIIp as it follows a gearbox pattern. The observed expression in the crp strain could also be explained by the presence of proteins involved in catabolite regulation other than CRP and such as Cra (30).
Quorum sensing.
This process is a regulatory mechanism involved in the control of cell density-dependent expression of many bacterial genes in response to high intracellular concentrations of N-acylhomoserine lactones (33). We have studied the effect of spent E. cloacae LB media in the activity of OUTIIp to find the presence of a possible signal secreted by bacteria during their growth or during nutrient starvation that would cause the induction of OUTIIp. Growth and OUTIIp activity in strain REM55 in fresh LB, spent LB, and reconstituted spent LB media were measured. The results (Fig. 2C) suggested that a signal molecule should appear during late exponential phase or at the onset of stationary phase in order to induce OUTIIp even in the absence of growth.
Effect of BarA.
Mukhopadhyay et al. (26) have recently described the inductor effect of protein BarA upon exponential-phase rpoS expression. To check the possible dependence of BarA during the exponential phase, a barA::λplacMu53 strain, HS703 (26), and wild-type MC4100 were transformed with pRS55. β-Galactosidase levels measured along the growth curve showed a 65.7% decrease in the barA strain.
There are some common features in RpoS expression in E. coli and E. cloacae CECT960, such as catabolite repression and dependence on both growth phases and ppGpp. RpoS expression is BarA dependent during exponential growth both in E. coli (via nlpDp) (26) and E. cloacae CECT960 (via OUTIIp). Thus, OUTIIp mimics the combination of nlpDp and rpoSp in the regulation of RpoS expression.
Readthrough transcription across an IS10 terminus probably inhibits its ability to bind transposase (17). A similar mechanism has been proposed in phage Mu in which transcription proceeding towards the left end from the Pc promoter could interfere with the binding of protein A to the left end of the genome (13). It is tempting to suggest a role of OUTIIp in preventing transposition of free IS10R, thus favoring transposition of the composite Tn10 transposon. The regulation observed in promoter OUTIIp suggests a link between cellular physiology and transposition. Increase in cAMP and ppGpp and the presence of an unknown signal at the onset of stationary phase activate OUTIIp, and thus, interaction between transposase and the end under transcription of IS10R could be reduced.
Transcription of IS10R neighboring regions may occur due to the presence of promoter pOUT or OUTIIp. The presence of all IS10R elements might not be dispensable, transcription of some genes being dependent on these promoters, as it occurs in E. cloacae CECT960 with the expression of the rpoS gene. This opens an exciting challenge now that more genomes are known and analyzed.
Acknowledgments
This work was supported by project BIO097-1246. E. Martínez was a recipient of a scholarship associated with it.
We acknowledge R. Hengge-Aronis for σS antibodies and M. Espinosa, G. del Solar, P. López, H. E. Schellhorn, M. Vicente, P. Palacios, J. L. García, and F. Moreno for their supply of strains, phages, and plasmids and for discussion.
REFERENCES
- 1.Aldea, M., T. Garrido, J. Pla, and M. Vicente. 1990. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 9:3787-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldea, M., T. Garrido, and A. Tormo. 1993. Gearbox gene expression and growth rate. World J. Microbiol. Biotechnol. 9:414-420. [DOI] [PubMed] [Google Scholar]
- 3.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoter σS dependent? Role of the -13/-14 nucleotide promoter positions and region 2.5 of σS. Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 4.Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel, M. M. Zambrano, and R. Kolter. 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of σ70. J. Bacteriol. 173:4482-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, L., D. Gentry, T. Elliott, and M. Cashel. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184:4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busby, S., and R. H. Ebright. 1997. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 7.Case, C. C., S. M. Roels, J. E. González, E. L. Simons, and R. W. Simons. 1988. Analysis of the promoters and transcripts involved in IS10 anti-sense RNA control. Gene 72:219-236. [DOI] [PubMed] [Google Scholar]
- 8.Cashel, M., and K. E. Rudd. 1987. The stringent response, p. 1410-1438. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 9.Chalmers, R., S. Sewitz, K. Lipkow, and P. Crellin. 2000. Complete nucleotide sequence of Tn10. J. Bacteriol. 182:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connell, N., Z. Han, F. Moreno, and R. Kolter. 1987. An E. coli promoter induced by the cessation of growth. Mol. Microbiol. 1:195-201. [DOI] [PubMed] [Google Scholar]
- 11.Espinosa-Urgel, M., C. Chamizo, and A. Tormo. 1996. A consensus structure for σS-dependent promoters. Mol. Microbiol. 21:657-659. [DOI] [PubMed] [Google Scholar]
- 12.Gentry, D. A., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor, σS, is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goosen, N., and P. van de Putte. 1986. Role of the Ner protein in bacteriophage Mu transposition. J. Bacteriol. 167:503-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayward, R. S., K. Igarashi, and A. Ishihama. 1991. Functional specialization within the α-subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 221:23-29. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch, M., and T. Elliott. 2002. Role of ppGpp in rpoS stationary-phase regulation in Escherichia coli. J. Bacteriol. 184:5077-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keilty, S., and M. Rosenberg. 1987. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 262:6389-6395. [PubMed] [Google Scholar]
- 17.Kleckner, N. 1989. Transposon Tn10, p. 227-268. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 18.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749-795. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The −35 recognition region of E. coli sigma 70 is inessential for initiation of transcription at an “extended −10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 20.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in E. coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 21.Lange, R., and R. Hengge-Aronis. 1994. The nlpD gene is located in an operon with rpoS on the E. coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13:733-743. [DOI] [PubMed] [Google Scholar]
- 22.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σs subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1996. Environmental regulation of virulence gene expression in Escherichia coli, Salmonella, and Shigella spp., p. 2803-2812. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 24.Martínez-García, E., A. Tormo, and J. M. Navarro-Llorens. 2001. Further studies on RpoS in Enterobacteria: identification of rpoS in Enterobacter cloacae and Kluyvera cryocrescens. Arch. Microbiol. 175:395-404. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Mukhopadhyay, M., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in E. coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 27.Prieto, M. A., and J. L. García. 1997. Identification of a novel positive regulator of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. Biochem. Biophys. Res. Commun. 232:759-765. [DOI] [PubMed] [Google Scholar]
- 28.Projan, S. J., S. Carleton, and R. P. Novick. 1983. Determination of plasmid copy number by fluorescence densitometry. Plasmid 9:182-190. [DOI] [PubMed] [Google Scholar]
- 29.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 30.Saier, M. H., Jr., and T. M. Ramseier. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 178:3411-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons, R. W., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1983. Three promoters near the termini of IS10: pIN, pOUT and pIII. Cell 34:673-682. [DOI] [PubMed] [Google Scholar]
- 32.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 33.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. Macintyre, and G. S. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, H., K. Severinov, A. Goldfarb, D. Fenyo, B. Chait, and R. H. Ebright. 1994. Location, structure, and function of the target of a transcriptional activator protein. Genes Dev. 8:3058-3067. [DOI] [PubMed] [Google Scholar]
- 35.Von Ossowski, I., M. R. Mulvey, P. A. Leco, A. Borys, and P. C. Loewen. 1991. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J. Bacteriol. 173:514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]