Abstract
The 165-kb catabolic plasmid pAO1 enables the gram-positive soil bacterium Arthrobacter nicotinovorans to grow on the tobacco alkaloid l-nicotine. The 165,137-nucleotide sequence, with an overall G+C content of 59.7%, revealed, besides genes and open reading frames (ORFs) for nicotine degradation, a complete set of ORFs for enzymes essential for the biosynthesis of the molybdenum dinucleotide cofactor, as well as ORFs related to uptake and utilization of carbohydrates, sarcosine, and amino acids. Of the 165 ORFs, approximately 50% were related to metabolic functions. pAO1 conferred to A. nicotinovorans the ability to take up l-[14C]nicotine from the medium, with an Km of 5.6 ± 2.2 μM. ORFs of putative nicotine transporters formed a cluster with the gene of the d-nicotine-specific 6-hydroxy-d-nicotine oxidase. ORFs related to replication, chromosome partitioning, and natural transformation functions (dprA) were identified on pAO1. Few ORFs showed similarity to known conjugation-promoting proteins, but pAO1 could be transferred by conjugation to a pAO1-negative strain at a rate of 10−2 to 10−3 per donor. ORFs with no known function represented approximately 35% of the pAO1 sequence. The positions of insertion sequence elements and composite transposons, corroborated by the G+C content of the pAO1 sequence, suggest a modular composition of the plasmid.
The soil bacterium Arthrobacter nicotinovorans has the ability to grow on the tobacco plant alkaloid nicotine as a carbon and nitrogen source (15, 19). The metabolic pathway supporting this ability is linked to the presence in the bacterial cells of the catabolic plasmid pAO1 (8).
Catabolic plasmids are large DNA molecules, usually over 100 kb, which, although they are not essential for the viability of the host, carry genes that extend the metabolic versatility of bacteria and allow them to gain energy from the degradation of a large variety of natural or man-made organic compounds present in their surroundings. They can spread between different species of bacteria, and it is generally considered that catabolic plasmids are responsible for the horizontal dissemination of metabolic abilities among bacterial populations. The diverse biochemical features of their gene products make these plasmids also of interest from a biotechnological point of view. Of the many catabolic plasmids identified to date, only a few have been sequenced in their entirety (31, 35, 41; pWWO [TOL] plasmid EMBL/GenBank/DDBJ accession number AJ344068). pAO1 is the first from a member of the genus Arthrobacter, a large group of gram-positive coryneform bacteria.
The genes of l-nicotine degradation are clustered on pAO1 (3). They encode nicotine dehydrogenase (NDH), which hydroxylates C-6 of the pyridine ring of nicotine; 6-hydroxy-l-nicotine oxidase (6HLNO), which opens the pyrrolidine ring of nicotine; and ketone dehydrogenase, which hydroxylates C-2 of the pyridine ring. The enzyme that removes the γ-methylaminobutyrate site chain has not yet been characterized. 2,6-Dihydroxypyridine hydroxylase introduces one more hydroxyl group in position 3 of the pyridine ring. In the presence of oxygen, 2,3,6-trihydroxypyridine spontaneously forms the pigment nicotine blue (see reference 3 for an overview). When nicotine is provided as the sole carbon and nitrogen source, the bacteria are able to cleave and metabolize 2,3,6-trihydroxypyridine (15, 19). Utilization of nutrients from the environment by bacteria requires, besides the specific catabolic enzymes, uptake systems for the substrates. For nicotine, no transport system has been described thus far. In particular, it was not known for A. nicotinovorans whether the process is pAO1 dependent. In addition, missing from the nic cluster was the gene for 6-hydroxy-d-nicotine oxidase (6HDNO). This enzyme catalyzes the same oxidation reaction as does 6HLNO oxidase, but on the stereoisomer 6-hydroxy-d-nicotine (14). Nicotiana tabacum produces only l-nicotine and no d-nicotine, but l and d isomers of nornicotine have been detected in tobacco (26). Thus, the functional and genetic relationships of this d-nicotine-specific enzyme to the rest of the pAO1 nicotine pathway enzymes and genes remained uncertain.
Here we present the entire 165,137-nucleotide sequence of pAO1 of A. nicotinovorans and an analysis of its open reading frames (ORFs), and we provide evidence for the conjugal transfer of the plasmid and for a pAO1-dependent nicotine uptake system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
A. nicotinovorans harboring the catabolic plasmid pAO1 (8) and a cured derivative not harboring pAO1 were grown on citrate medium supplemented with mineral salts and vitamins (15, 21). pAO1 DNA was extracted as described previously (32). Plasmid pKDT542β (17), carrying a transposon with a chloramphenicol resistance gene, was propagated in Escherichia coli XL1-Blue grown on Luria-Bertani (LB) plates or in LB liquid medium in the presence of 10 μg of chloramphenicol/ml.
Determination of the pAO1 sequence.
The gene bank of the pAO1 DNA in the ZAP Express vector from Stratagene (Amsterdam, The Netherlands) was a kind gift of Karl Decker (Freiburg, Germany) and has been described before (3, 44). A total of 180 individual clones were sequenced. Gaps in the pAO1 DNA sequence were filled with the sequences determined from PCR products obtained with pAO1 DNA as the template and with primers derived from the ends of the known pAO1 sequences. Sequencing and PCR primers were the products of the Institute of Biology III Core Facility, synthesized with an ABI 3948 synthesizer (Applied Biosystems, Foster City, Calif.). DNA sequencing reactions (a total of 935 readings) were carried out with BigDye version 2 fluorescent terminator mixes (Applied Biosystems), purified by Sephadex G50 gel filtration over MultiScreen plates (Millipore), and analyzed with ABI 310 genetic analyzers. DNA not providing information under standard conditions due to GC or AT accumulation, homopolymeric or dinucleotide repeat regions, or secondary structures could be analyzed by using either BigDye-dGTP, the addition of Thermofidelase (Fidelity Systems, Gaithersburg, Md.)-10% dimethyl sulfoxide, or a combination of these. Compressions during electrophoresis could often be overcome by increasing the capillary temperature to 60°C. The Staden package containing the GAP4 software (6) was used for sequence assembly, primer design, and editing. FASTA files of ORFs and codon usage tables were generated by SPIN. Coding regions were identified with BLAST (1), using BLASTCL3 for batch searches of ORFs encoding more than 60 amino acids. Coding sequences with associated ribosome binding sites were predicted with GENEMARK (7) and GLIMMER (http://www.tigr.org/software/glimmer/) and compared to the BLAST alignments to establish the coding sequence start. Regions lacking identified ORFs and giving no BLAST results but revealing, on visual inspection, coding sequences with ribosomal binding sites were included in the annotation. Unidentified ORFs were scanned with PROSITE (http://www.expasy.org/prosite/) to obtain functional information. Annotation and GC content analysis were performed with ARTEMIS (http://www.sanger.ac.uk/Software/Artemis/), from which the graphic depiction was captured with HARDCOPY (http://www.hardcopy.de/).
Electroporation of an A. nicotinovorans derivative lacking pAO1 with pKGT452β
A. nicotinovorans electrocompetent cells were prepared and transformed as described by Gartemann and Eichenlaub (17), with 1 μg of pKGT452β DNA per 200 μl of bacterial suspension, with the aid of a Bio-Rad Gene Pulser. The electroporated bacteria were plated on LB plates with 10 μg of chloramphenicol per ml for the selection of Cmr bacterial cells. The pKGT452β DNA does not replicate in A. nicotinovorans, and Cmr bacteria appear following transposition to the chromosome of the cmx gene-carrying transposon present on the plasmid (17). No Cmr colonies were obtained with the same bacterial suspensions when they were not electroporated or when they were electroporated without plasmid DNA.
Conjugation of A. nicotinovorans/pAO1 Nic+ Cms with the A. nicotinovorans Nic− Cmr derivative not carrying pAO1.
The A. nicotinovorans strains were grown in LB medium to an A578 of 1.0. One milliliter of donor A. nicotinovorans/pAO1 Nic+ Cms and 1 ml of recipient pAO1-negative A. nicotinovorans Nic− Cmr were mixed, pelleted by centrifugation at 10,000 × g, resuspended in 100 μl, and applied to a sterile 0.22-μm-pore-size, 3-cm-diameter Millipore GS membrane placed on LB medium. Following incubation of the plate for 20 h at 30°C, the membrane was removed and the bacteria from the membrane were suspended in 20 ml of LB medium. Serial dilutions of the bacterial suspension were plated on citrate medium supplemented with 0.05% (wt/vol) nicotine and 10 μg of chloramphenicol per ml and incubated at 30°C for 48 h. Cmr colonies grown on nicotine plates from bacteria which have taken up pAO1 turn dark and present a halo of nicotine blue secreted into the medium. In control platings with donor strain only, no Cmr colonies were observed.
Determination of l-[14C]nicotine uptake by A. nicotinovorans and E. coli XL1-Blue.
l-[14C]nicotine (1.25 mCi/mmol) was prepared as described previously (13) and was the kind gift of Karl Decker (Freiburg, Germany). The free base was obtained from the pure dipicrate of [14C]CH3-l-nicotine by exhaustive ether extraction and gave a chromatographically homogenous spot on thin-layer plates. The A. nicotinovorans strains carrying and not carrying pAO1 were grown on citrate medium for 30 h at 30°C to an A578 of 1.0. To 5 ml of the culture were added 1.5 μCi of l-[14C]nicotine (which gave an approximately 80 μM nicotine solution), and 1-ml samples were removed at various time points. The bacteria were collected by centrifugation, the pellet was suspended in 1 ml of double-distilled water, the suspension was pelleted, and the bacteria were resuspended in 50 μl of 50 mM Tris-HCl (pH 8.0)-10 mM EDTA, lysed with 100 μl of 200 mM NaOH-1% (wt/vol) sodium dodecyl sulfate, and neutralized with 75 μl of 2 M Tris-HCl (pH 7.0)-0.5 M NaCl. The samples were centrifuged at 15,000 × g in order to remove precipitated material, and the protein concentration was determined with Roti-Quant according to the instructions of the supplier (Roth, Karlsruhe, Germany). Two hundred microliters of the cleared supernatant (30 μg of protein) was added to 5 ml of scintillation solution (Roth), and the radioactivity was counted in a Packard liquid scintillation counter for 2 min each. E. coli XL1-Blue bacteria were grown under the same conditions as A. nicotinovorans in citrate medium supplemented with 0.8% (wt/vol) tryptone and 0.1% (wt/vol) yeast extract. Uptake of l-[14C]nicotine was determined as described above. The effect of l- and d-amino acids on nicotine uptake was determined with bacterial cultures supplemented before the addition of nicotine with various concentrations of the corresponding amino acid as indicated in the legends to the figures.
Determination of the Km of nicotine uptake by A. nicotinovorans carrying pAO1.
A. nicotinovorans/pAO1 grown on citrate medium for 30 h as described above was induced with unlabeled l-nicotine (2 μM) for 20 min, and the bacteria were pelleted by centrifugation at 12,000 × g for 1 min, washed, recentrifuged for 1 min, and resuspended in citrate medium. The time dependence of nicotine uptake was determined at 5, 10, 15, and 20 min for each of the concentrations of 2, 10, 20, 40, and 80 μM l-[14C]nicotine (specific activity, 1.25 mCi/mmol). The initial velocity of nicotine uptake determined for each nicotine concentration was used to calculate the Km and Vmax of nicotine uptake with the aid of the program Microcal Origin (Microcal Software, Inc, Northampton, Mass.).
Determination of 6HLNO activity.
Enzyme activity in the supernatant of lysed bacteria was determined spectrophotometrically, at each time point analyzed for l-[14C]nicotine uptake, as described previously (9).
Nicotine blue formation by A. nicotinovorans carrying pAO1 in the presence of l- and d-amino acids.
A. nicotinovorans/pAO1 cultures grown on citrate medium were supplemented with 100 μM each l- or d-amino acids or with increasing concentrations of d-Arg and l-Pro as indicated in the legends to the figures. Following 10 min of incubation, 50 μM l-nicotine was added. Samples were removed after 3 h and centrifuged, and the blue pigment present in the supernatant was measured spectrophotometrically at 600 nm. Alternatively, to cultures supplemented with 100 μM l-Pro or d-Arg, 1.5 μCi of l-[14C]nicotine was added and the radioactivity taken up by the bacteria was determined at 1, 5, 15, and 30 min as described above.
The DNA sequence of pAO1 has been deposited in the EMBL database under accession number AJ507836.
RESULTS
Nucleotide composition.
The circular plasmid is comprised of 165,137 bp (an irresolvable compression in the region of bp 163480 may increase this size by up to 5 bp). This includes a 376-bp direct repeat region. The overall G+C content is 59.7%, which is somewhat less than the G+C content of 62% of A. nicotinovorans itself (27) but in the range of 59 to 66% for the genus Arthrobacter (24). The G+C content is also reflected by the lack of restriction sites for PacI (TTAATTAA), PmeI (GTTTAAAC), and SmiI (ATTTAAAT). The bias towards G and C in the third codon position is a consequence of this structural feature (not shown).
With no obvious origin of replication (see below), position 1 of the sequence was arbitrarily assigned to a BamHI site. A total of 83.2% of the plasmid genome appears to have a coding function, and 126 ORFs either correspond to biochemically characterized proteins or can be correlated with functions by high degrees of similarity to sequences in the databases. Additionally, 39 ORFs predicted by GENEMARK, GLIMMER, or visual inspection but having no significant similarity to sequences in the databases may be functionally relevant (Table 1). The ORFs are equally distributed between the strands, with 79 on one and 85 on the other.
TABLE 1.
Summary of ORFs identified by significant homology (BLAST search) or prediction or previously experimentally verified
| ORF | Length (amino acids) | CDS position (start codon-stop codon)a | Gene or function of closest relative (source) | Data bank reference | Identity (%) | E valueb |
|---|---|---|---|---|---|---|
| 1 | 133c | 1- 296c | GENEMARK prediction; no homology | |||
| 2 | 264 | 344-138c | Probable hydrolase (M. leprae), related to LipG (M. tuberculosis H37Rv) | emb|CAC30853.1| | 91/282 (32) | 3e-031 |
| emb|CAB07104.1| | 83/281 (29) | 2e-023 | ||||
| 3 | 317 | 1140-2093c | Sucrose hydrolase (E. coli) | emb|CAA57219.1| | 66/255 (25) | 2e-009 |
| 4 | 322 | 2124-3092c | Lipase (esterase) (B. halodurans) | dbj|BAB05967.1| | 92/241 (38) | 1e-039 |
| 5 | 444 | 3148-4482c | Inositol transport protein IolF (B. subtilis) | emb|CAB16007.1| | 94/339 (27) | 5e-032 |
| 6 | 337 | 4567-5580 | Putative LacI family transcriptional regulator (S. coelicolor) | emb|CAB62677.1| | 96/301 (31) | 2e-022 |
| 7 | 206 | 5603-6223 | Chromosome partitioning protein-like protein ParB (R. equi) | gb|AAG21765.1| | 39/81 (48) | 1e-011 |
| 8 | 153 | 6349-6810 | GENEMARK prediction, related to chromosome partitioning protein (R. equi) | dbj|BAB16671.1| | 38/130 (29) | 0.011 |
| 9 | 162 | 6807-7295 | Chromosome partitioning protein-like protein (R. equi) | gb|AAG21765.1| | 49/144 (34) | 1e-012 |
| 10 | 88 | 8256-8522 | Redoxin homolog NrdH (C. glutamicum) | gb|AAD25054.1| | 34/72 (47) | 2e-012 |
| 11 | 170 | 8879-9391 | Single-stranded DNA-binding protein (A. aurescens) | gb|AAM74937.1| | 135/171 (78) | 1e-071 |
| 12 | 103 | 9742-10053c | GENEMARK prediction; no homology | |||
| 13 | 221 | 10125-10790c | Manual prediction; no homology | |||
| 14 | 164 | 10930-11424 | MC37 (Micrococcus sp. strain 28) | gb|AAK62511.1| | 41/108 (37) | 4e-007 |
| 15 | 549 | 11429-13078 | MC38 (Micrococcus sp. strain 28) | gb|AAK62512.1| | 112/275 (40) | 9e-050 |
| 16 | 395 | 13155-14342 | GENEMARK prediction; related to AviX4 (S. viridochromogenes) | gb|AAK83164.1| | 33/97 (34) | 0.013 |
| 17 | 165 | 14600-15097c | GLIMMER prediction; no homology | |||
| 18 | 226 | 15604-16284 | KfrA protein, plasmid RK2 | gb|AAK73376.1| | 32/107 (29) | 2e-005 |
| 19 | 300 | 16726-17628 | Hypothetical protein (P. aeruginosa), related to Van WB2 (E. faecium) | gb|AAG04138| | 99/263 (37) | 5e-040 |
| gb|AAG34689.1| | 71/233 (30) | 7e-031 | ||||
| 20 | 406 | 17879-19099 | GENEMARK prediction; no homology | |||
| 21 | 70 | 19130-19342 | Manual prediction; no homology | |||
| 22 | 168 | 19585-20091 | Putative RNA polymerase sigma factor (S. coelicolor) | emb|CAB63191.1| | 49/166 (29) | 4e-008 |
| 23 | 287 | 20094-120957 | GENEMARK prediction; related to SAM-dependent methyltransferases (C. glutamicum) | dbj|BAB99995.1| | 20/50 (40) | 0.11 |
| 24 | 249 | 21370-22119c | Hypothetical protein (D. radiodurans), related to cellulase precursor (C. thermocellum) | gb|AAF09927.1| | 52/211 (25) | 6e-011 |
| 25 | 165 | 22231-22728 | Manual prediction; no homology | |||
| 26 | 116 | 22743-23093 | Manual prediction; no homology | |||
| 27 | 666 | 23110-25110 | Putative secreted protein (S. coelicolor), related to putative | emb|CAB51964.1| | 100/270 (37) | 4e-044 |
| ATP/GTP-binding protein (S. coelicolor) | emb|CAD30941.1| | 92/201 (45) | 1e-037 | |||
| 28 | 99 | 25144-25443c | Manual prediction; no homology | |||
| 29 | 97 | 25440-25733c | Manual prediction; no homology | |||
| 30 | 95 | 25874-26161 | Unknown (R. etli) | gp|U80928 | 30/90 (33) | 0.001 |
| 31 | 458 | 26161-27537 | Hypothetical protein (E. amylovora) | gb|AAG31049.1| | 88/331 (26) | 8e-013 |
| 32 | 226 | 27948-28628c | Putative transcriptional regulator PdhR (S. seoulensis) | gb|AAF37157.1| | 76/192 (39) | 9e-029 |
| 33 | 496 | 28858-30348 | ABC transporter protein, ATP-binding component (S. coelicolor) | emb|CAB66285.1| | 228/484 (47) | e-113 |
| 34 | 347 | 30345-31388 | Probable ribose ABC transporter (C. perfringens) | dbj|BAB81335.1| | 52/222 (23) | 3e-015 |
| 35 | 347 | 31385-32428 | Ribose/xylose/arabinose/galactoside ABC-type transport systems, permease components (T. tengcongensis) | gb|AAM24023.1| | 83/297 (27) | 7e-021 |
| 36 | 392 | 32506-33684 | Probable sugar ABC transporter (periplasmic) (R. etli) | gb|AAM54922.1| | 81/309 (26) | 1e-020 |
| 37 | 175 | 33752-34279 | GENEMARK, GLIMMER prediction; no homology | |||
| 38 | 522 | 34276-35844 | 2-Keto-gluconate dehydrogenase (X. axonopodis pv. Citri) | gb|AAM36981.1| | 166/516 (32) | 4e-053 |
| 39 | 458 | 35897-37273 | Succinate-semialdehyde dehydrogenase (M. tuberculosis) | gb|AAK44465.1| | 255/454 (56) | e-143 |
| 40 | 388 | 37349-38515 | Putative oxidoreductase (S. coelicolor) | emb|CAB66291.1| | 242/382 (63) | e-139 |
| 41 | 402 | 38515-39723 | Conserved hypothetical protein SCC57A.22c. (S. coelicolor) | emb|CAB66290.1| | 210/403 (52) | e-108 |
| 42 | 389 | 39724-40893 | Putative transferase (S. coelicolor), related to glycerate kinase | emb|CAA22714.1| | 133/346 (38) | 1e-057 |
| (X. campestris pv. Campestris) | gb|AAM43442.1| | 133/346 (38) | 1e-057 | |||
| 43 | 890 | 41260-43932c | DNA helicase (T. volcanium) | dbj|BAB59970.1| | 101/453 (22) | 3e-015 |
| 44 | 332 | 43929-44927c | GENEMARK, GLIMMER prediction; no homology | |||
| 45 | 615 | 45283-47130c | GENEMARK prediction; no homology | |||
| 46 | 203 | 47346-47957 | GENEMARK prediction; no homology | |||
| 47 | 224 | 48405-49079 | GENEMARK prediction, related to integrase (C. glutamicum) | dbj|BAB98812.1| | 25/85 (29) | 0.074 |
| 48 | 368 | 49039-50145 | Transposase homolog (M. gordonae) | gb|AAB54012.1| | 212/368 (57) | e-114 |
| 49 | 385 | 50142-51299 | Unknown (M. gordonae) | gb|AAB54013.1| | 117/349 (33) | 7e-033 |
| 50 | 309 | 51455-52384 | Transposase B of transposon Tn554 (S. aureus) | dbj|BAB47606.1| | 110/284 (38) | 2e-050 |
| 51 | 137 | 52381-52794 | Transposase C of transposon Tn554 (S. aureus) | dbj|BAB47607.1| | 36/112 (32) | 2e-011 |
| 52 | 72 | 52823-53041 | Manual prediction; no homology | |||
| 53 | 101 | 53083-53388 | GENEMARK prediction; no homology | |||
| 54 | 204 | 53903-54517c | Hypothetical protein (M. loti), related to NADPH:quinone | dbj|BAB48001.1| | 102/181 (56) | 2e-054 |
| oxidoreductase (M. loti) | dbj|BAB51303.1| | 96/170 (56) | 5e-051 | |||
| 55 | 114 | 54899-55243c | Hypothetical protein (M. loti) | dbj|BAB49953.1| | 34/99 (34) | 2e-011 |
| 56 | 421 | 55240-56505c | Amine oxidase (Synechocystis sp. strain PCC 6803) | dbj|BAA10142.1| | 64/245 (26) | 1e-009 |
| 57 | 176 | 56912-57442 | GLIMMER prediction; no homology | |||
| 58 | 450 | 57584-58936 | NAD-dependent aldehyde dehydrogenases (C. glutamicum ATCC 13032) | dbj|BAB97443.1| | 278/50 (61) | e-157 |
| 59 | 146 | 59275-59715 | 2-Hydroxyhepta-2,4-diene-1,7-dioate isomerase/5-carboxymethyl-2-oxo- hex-3-ene-1,7-dioate decarboxylase-related protein (D. radiodurans) | gb|AAF11167.1| | 69/168 (41) | 1e-029 |
| 60 | 166 | 59915-60415c | Membrane transporter of cationic drugs (Halobacterium sp. strain NRC-1) | |NP444228.1| | 38/105 (36) | 7 e-017/PICK> |
| 61 | 116 | 60466-60816c | Multidrug resistance efflux protein (S. lividans) | gb|AAK95484.1| | 30/58 (51) | 6e-011 |
| 62 | 308 | 60844-61770c | Bifunctional protein (methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase) (S. coelicolor) | emb|CAB97427.1| | 174/286 (60) | 1e-093 |
| 63 | 824 | 61867-64341c | Glycine cleavage system T protein, putative (uncultured proteobacterium), | gb|AAL76414.1| | 345/810 (42) | 0.0 |
| related to sarcosine dehydrogenase (M. loti) | dbj|BAB48695.1| | 335/812 (41) | e-170 | |||
| 64 | 287 | 64397-65260c | Probable formyltetrahydrofolate deformylase protein (R. solanacearum) | emb|CAD15575.1| | 152/281 (54) | 2e-080 |
| 65 | 96 | 65268-65555c | Manual prediction; no homology | |||
| 66 | 481 | 66082-67527c | Amino acid permease (C. acetobutylicum) | gb|AAK78828.1| | 105/472 (22) | 3e-023 |
| 67 | 470 | 68009-69421 | Hypothetical protein (P. aeruginosa), related to putative regulatory | gb|AAG05303.1| | 51/111 (45) | 4e-022 |
| protein (S. coelicolor) | emb|CAA22503.1| | 32/112 (28) | 1e-004 | |||
| 68 | 85 | 70398-70655c | Putative transposase (fragment) (C. glutamicum) | |NP_600737.1| | 27/75 (36) | 3e-007 |
| 69 | 147 | 70884-71327c | ORF147 (A. nicotinovorans) | gb|AAK64262.1| | 147/147 (100) | 0.0 |
| 70 | 223 | 71475-72146 | ORF223 (A. nicotinovorans) | gb|AAK64269.1| | 223/223 (100) | 0.0 |
| 71 | 70 | 72227-72439c | ORF70 (A. nicotinovorans) | gb|AAK64270.1| | 70/70 (100) | 3e-038 |
| 72 | 204 | 72530-73144c | ORF204 (A. nicotinovorans), related to mobA (X. axonopodis) | gb|AAK64261.1| | 204/204 (100) | e-102 |
| tn|AE011839| | 40/142 (28) | 0.002 | ||||
| 73 | 117 | 73137-73490c | ORF117 (A. nicotinovorans) | gb|AAK64268.1| | 117/117 (100) | 3e-065 |
| 74 | 377 | 73517-74650c | ORF377 (A. nicotinovorans), related to CoxF protein (P. carboxydovorans) | gb|AAK64260.1| | 377/377 (100) | 0.0 |
| tr|X82447| | 78/280 (27) | 6e-14 | ||||
| 75 | 363 | 74643-75734c | ORF363 (A. nicotinovorans), related to CoxE protein (O. carboxidovorans) | gb|AAK64259.1| | 363/363 (100) | 0.0 |
| emb|CAB76247.1| | 112/377 (29) | 1e-034 | ||||
| 76 | 297 | 75752-76645c | ORF297 (A. nicotinovorans), related to CoxD protein (O. carboxidovorans) | gb|AAK64258.1| | 297/297 (100) | e-164 |
| emb|CAA57830.1| | 131/277 (47) | 6e-065 | ||||
| 77 | 294 | 76848-77732c | Hypothetical nitrile amino hydrolase (A. nicotinovorans) | gb|AAK64257.1| | 294/294 (100) | e-167 |
| 78 | 235 | 77729-78436c | ORF235 (A. nicotinovorans), related to CoxG protein (P. carboxydovorans) | gb|AAK64256.1| | 235/235 (100) | e-104 |
| tr|X82447| | 32/141 (22) | 1e-06 | ||||
| 79 | 397 | 78452-79645c | Dihydroxypyridine hydroxylase (A. nicotinovorans) | gb|AAK64255.1| | 397/397 (100) | 0.0 |
| 80 | 310 | 79642-80574c | Hypothetical polyketide cyclase (A. nicotinovorans) | gb|AAK64254.1| | 310/310 (100) | 0.0 |
| 81 | 106 | 80574-80894c | ORF106 (A. nicotinovorans), related to hypothetical protein (C. perfringens) | gb|AAK64267.1| | 106/106 (100) | 6e-058 |
| tr|AP003188| | 32/102 (31) | 4e-05 | ||||
| 82 | 794 | 80977-83361c | Ketone dehydrogenase, large subunit (A. nicotinovorans) | gb|AAK64253.1| | 794/794 (100) | 0.0 |
| 83 | 367 | 83529-84632 | Hypothetical endopeptidase (A. nicotinovorans) | gb|AAK64252.1| | 367/367 (100) | 0.0 |
| 84 | 116 | 84625-84975 | ORF116 (A. nicotinovorans), related to ethanolamine utilization protein | gb|AAK64251.1| | 116/116 (100) | 6e-064 |
| (C. perfringens) | tr|AP003188| | 25/73 (34) | 8e-05 | |||
| 85 | 407 | 85010-86233c | Hypothetical transcriptional regulator (A. nicotinovorans) | gb|AAK64250.1| | 407/407 (100) | 0.0 |
| 86 | 394 | 86297-87481 | Hypothetical transcriptional regulator (A. nicotinovorans) | gb|AAK64249.1| | 388/394 (98) | 0.0 |
| 87 | 296 | 87630-88520 | Ketone dehydrogenase medium subunit (A. nicotinovorans) | emb|CAC37486.2| | 296/296 (100) | 0.0 |
| 88 | 160 | 88520-89002 | Ketone dehydrogenase, small subunit (A. nicotinovorans) | emb|CAA04551.1| | 160/160 (100) | 0.0 |
| 89 | 103 | 89719-90030 | ORF103 (A. nicotinovorans) | gb|AAK64266.1| | 103/103 (100) | 1e-046 |
| 90 | 64 | 90030-90224 | ORF64 (A. nicotinovorans) | gb|AAK64265.1| | 64/64 (100) | 6e-032 |
| 91 | 78 | 90370-90606c | ORF78 (A. nicotinovorans) | gb|AAK64264.1| | 78/78 (100) | 1e-036 |
| 92 | 196 | 90622-1212c | ORF124 (A. nicotinovorans) | gb|AAK64246.1| | 79/100 (79) | 2e-036 |
| 93 | 425 | 91550-92827c | 6-Hydroxy-l-nicotine oxidase (A. nicotinovorans) | gb|AAK64245.1| | 425/425 (100) | 0.0 |
| 94 | 816 | 92895-95345c | NdhL (A. nicotinovorans) | gb|AAK64263.1| | 816/816 (100) | 0.0 |
| 95 | 165 | 95338-95835c | NdhS (A. nicotinovorans) | gb|AAK64244.1| | 165/165 (100) | 6e-091 |
| 96 | 283 | 95832-96683c | NdhM (A. nicotinovorans) | gb|AAK64243.1| | 283/283 (100) | e-157 |
| 97 | 236 | 97125-97835c | Transposase (A. nicotinovorans) | emb|CAA65743.1| | 236/236 (100) | e-143 |
| 98 | 87 | 97835-98095c | Transposase (A. nicotinovorans) | gb|AAK64271.1| | 87/87 (100) | 2e-045 |
| 99 | 88 | 98374-98640c | MoaD2 (M. tuberculosis H37Rv) | emb|CAA17674.1| | 35/88 (39) | 5e-007 |
| 100 | 371 | 98647-99762c | Molybdopterin cofactor synthesis protein MoaA2 (A. nicotinovorans) | emb|CAA55583.1| | 368/372 (99) | 0.0 |
| 101 | 429 | 100053-101342 | Molybdopterin cofactor synthesis protein MoeA (A. nicotinovorans) | emb|CAA71780.2| | 397/415 (96) | 0.0 |
| 102 | 168 | 101339-101845 | Molybdopterin co-factor synthesis protein MoaC (A. nicotinovorans) | emb|CAA71781.1| | 165/169 (98) | 1e-074 |
| 103 | 155 | 101958-102425 | Molybdopterin synthase, large subunit MoaE (A. nicotinovorans) | emb|CAA71782.1| | 154/155 (99) | 8e-082 |
| 104 | 262 | 102436-103224 | Molybdate-binding periplasmic protein ModA (A. nicotinovorans) | emb|CAA71776.1| | 211/244 (86) | e-106 |
| 105 | 239 | 103343-104062 | Molybdenum transport transmembrane protein ModB (A. nicotinovorans) | emb|CAA71777.1| | 239/239 (100) | 0.0 |
| 106 | 349 | 104059-105108 | Molybdenum transport ATPase ModC (A. nicotinovorans) | emb|CAA71778.1| | 347/349 (99) | 0.0 |
| 107 | 583 | 106666-108417c | Beta-glucosidase (E. chrysanthemi) | gb|AAA80156.1| | 232/602 (38) | e-104 |
| 108 | 424 | 108429-109703c | Membrane protein, putative (C. crescentus CB15) | gb|AAK24286.1| | 94/367 (25) | 3e-029 |
| 109 | 367 | 109837-110940 | Putative LacI family transcriptional regulatory protein (S. coelicolor) | emb|CAC32300.1| | 108/346 (31) | 2e-033 |
| 110 | 101 | 111227-111532c | Conserved hypothetical protein (D. radiodurans) | gb|AAF09855.1| | 26/100 (26) | 4e-004 |
| 111 | 497 | 111798-113291c | Amino acid permease (C. acetobutylicum) | gb|AAK78828.1| | 101/453 (22) | 2e-021 |
| 112 | 459 | 113841-115220 | 6-HDNO (A. oxydans) | emb|CAA29416.1| | 458/458 (100) | 0.0 |
| 113 | 465 | 115728-117125 | Amino acid transporter (M. loti) | dbj|BAB53436.1| | 107/485 (22) | 3e-017 |
| 114 | 194 | 117206-117790c | Transcriptional regulator (A. pretiosum subsp. auranticum) | gb|AAM54107.1| | 49/196 (25) | 3e-007 |
| 115 | 67 | 118378-118581c | GENEMARK prediction; no homology | |||
| 116 | 311 | 118800-119735c | GLIMMER, GENEMARK prediction; no homology | |||
| 117 | 192 | 120002-120580 | GLIMMER prediction; no homology | |||
| 118 | 68 | 121016-121222 | Manual prediction; no homology | |||
| 119 | 347 | 121239-122282 | GLIMMER, GENEMARK prediction; no homology | |||
| 120 | 110 | 122242-122577c | Manual prediction; no homology | |||
| 121 | 93 | 122574-122855c | Manual prediction; no homology | |||
| 122 | 350 | 122852-123904c | Similar to E. coli hypothetical protein o375 (bacteriophage WO) | dbj|BAA89622.1| | 132/327 (40) | 3e-068 |
| 123 | 146 | 124013-124453c | Conserved hypothetical protein (S. coelicolor) | emb|CAB41070.1| | 35/91 (38) | 4e-012 |
| 124 | 594 | 124450-126234c | MC40 (Micrococcus sp. strain 28), related to conjugative transfer gene | gb|AAK62514.1| | 176/450 (39) | 6e-087 |
| complex protein-like protein (R. equi) | gb|AAG21737.1| | 149/482 (30) | 7e-054 | |||
| 125 | 530 | 126231-127823c | Putative ATP-binding protein (Micrococcus sp. strain 28) | gb|AAK62515.1| | 190/458 (41) | 4e-085 |
| 126 | 502 | 127823-129331c | Putative integral membrane protein (Micrococcus sp. strain 28) | gb|AAK62516.1| | 154/437 (35) | 3e-072 |
| 127 | 436 | 129318-130628c | Transfer gene complex protein-like protein (R. equi) | gb|AAG21743.1| | 39/107 (36) | 8e-011 |
| 128 | 246 | 130630-131370c | GENEMARK prediction, related to hypothetical protein (R. equi) | dbj|BAB16652.1| | 47/177 (26) | 0.001 |
| 129 | 89 | 131399-131668c | GENEMARK prediction; no homology | |||
| 130 | 201 | 131682-132287c | MC47 (Micrococcus sp. strain 28) | gb|AAK62521.1| | 52/118 (44) | 9e-017 |
| 131 | 480 | 132326-133768c | Conserved hypothetical protein (M. tuberculosis CDC1551) | gb|AAK44775.1| | 81/286 (28) | 5e-015 |
| 132 | 170 | 133808-134320c | GLIMMER, GENEMARK prediction; no homology | |||
| 133 | 357 | 134317-135390c | Putative secreted protein (S. coelicolor) | emb|CAC36669.1| | 56/139 (40) | 7e-020 |
| 134 | 105 | 135513-135827 | Predicted transcriptional regulator (C. glutamicum ATCC 13032) | ebj|BAB97654.1| | 25/76 (32) | 3e-004 |
| 135 | 155 | 135902-136369c | Conserved hypothetical protein (M. leprae) | emb|CAC29577.1| | 40/130 (30) | 4e-009 |
| 136 | 388 | 136370-137536c | Putative secreted protein (S. coelicolor) | emb|CAB59459.1| | 104/314 (33) | 1e-037 |
| 137 | 327 | 137521-138504c | Manual prediction; no homology | |||
| 138 | 216 | 139080-139730 | GLIMMER prediction; no homology | |||
| 139 | 653 | 139845-141806 | GLIMMER prediction; no homology | |||
| 140 | 241 | 141796-142521c | Putative transposase (C. michiganesis subsp. insidiosus) | gb|AAC29483.1| | 135/231 (58) | 5e-069 |
| 141 | 82 | 142522-142769c | Putative transposase (C. michiganesis subsp. sepedonicus) | gb|AAC29482.1| | 22/35 (62) | 9e-004 |
| 142 | 231 | 143016-143711c | GLIMMER prediction; no homology | |||
| 143 | 314 | 143708-144652c | Soj family protein (M. tuberculosis CDC1551) | gb|AAK46019.1| | 59/193 (30) | 8e-015 |
| 144 | 208 | 144897-145523c | GLIMMER prediction; no homology | |||
| 145 | 192 | 147471-148049 | GLIMMER, GENEMARK prediction; no homology | |||
| 146 | 78 | 148142-148378c | GENEMARK prediction; no homology | |||
| 147 | 145 | 148567-149004c | GENEMARK prediction; no homology | |||
| 148 | 119 | 149103-149459c | GENEMARK prediction; no homology | |||
| 149 | 182 | 149661-150209c | Hypothetical protein (R. solanacearum) | emb|CAD17238.1| | 87/172 (50) | 1e-043 |
| 150 | 192 | 150470-151048 | Recombinase XFa0019 (imported) (X. fastidiosa) | gb|AAF85588.1| | 90/185 (48) | 3e-039 |
| 151 | 487 | 151064-152527 | Probable transposase transposon Tn552 (S. aureus) | emb|CAA36949.1| | 174/405 (42) | 1e-095 |
| 152 | 218 | 152770-153426 | Probable ATP-binding protein Tn552 (S. aureus transposon) | emb|CAA36948.1| | 99/254 (38) | 3e-040 |
| 153 | 167 | 153519-154022 | DNA recombinase homolog Pin (C. glutamicum) | gb|AAD25057.1| | 115/162 (70) | 3e-062 |
| 154 | 323 | 154559-155530 | GLIMMER prediction; no homology | |||
| 155 | 190 | 155660-156232c | GENEMARK prediction; no homology | |||
| 156 | 232 | 156229-156927c | Hypothetical protein (C. crescentus CB15), related to adenylate cyclase | gb|AAK24668.1| | 87/225 (38) | 6e-037 |
| (Synechocystis sp. strain PCC 6803) | dbj|BAA17880.1| | 51/192 (26) | 7e-006 | |||
| 157 | 232 | 156962-157660c | Hypothetical protein (C. crescentus CB15), related to adenylate cyclase 1 | gb|AAK24668.1| | 114/216 (52) | 4e-059 |
| protein (S. meliloti) | emb|CAC41696.1| | 31/155 (20) | 2e-004 | |||
| 158 | 96 | 157802-158092 | Recombinase XFa0019 (imported) (X. fastidiosa) | gb|AAF85588.1| | 25/40 (62) | 5e-008 |
| 159 | 479 | 158578-160017 | Unknown (P. multocida) | gb|AAK03624.1| | 104/331 (31) | 1e-037 |
| 160 | 133 | 160142-160543c | Manual prediction; no homology | |||
| 161 | 201 | 160569-161174c | Probable DNA ligase (LigC) (M. tuberculosis H37Rv) | emb|CAA18053.1| | 71/193 (36) | 1e-027 |
| 162 | 302 | 161797-162705 | Smf family protein (M. tuberculosis CDC1551) | gb|AAK47290.1| | 107/239 (44) | 3e-041 |
| 163 | 107 | 162772-163095 | GENEMARK prediction; no homology | |||
| 164 | 339 | 163680-164699 | Hypothetical protein (M. tuberculosis) | gb|AAK47101.1| | 81/361 (22) | 4e-005 |
| 165 | 48 | 164889-165035 | GENEMARK prediction; no homology | |||
| 1 | 165032-165137c | Continues as 1-296c |
CDS, coding sequence; c, complementary strand.
An E value of >0.4 indicates no homology.
Continues from position 165137 to 165032.
ORF analysis.
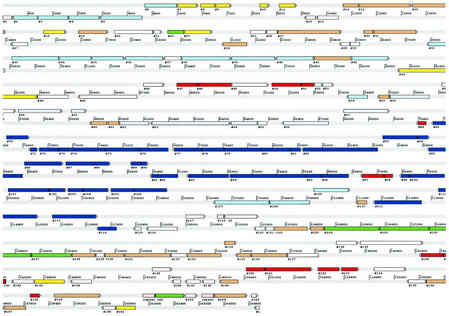
A graphical representation of the 165 known or predicted ORFs is depicted in Fig. 1, and their relationships to representative homologs in databases are detailed in Table 1. Based on gene similarity search results, functions associated with ORFs may be classified into the following categories: utilization of nutrients, transposition, conjugation, and replication (Fig. 1).
FIG. 1.
Schematic representation of the ORFs on the two strands of the pAO1 DNA. Dark blue, ORFs related to nicotine utilization; light blue, ORFs related to carbohydrate; violet, ORFs related to amino acid and sarcosine; red, ORFs related to IS elements and composite transposons; yellow, ORFs related to replication; green, ORFs related to conjugation; brown, conserved hypothetical ORFs; white, ORFs with no known function.
Utilization of nutrients.
The predicted metabolic abilities linked to pAO1 center on carbohydrate, nicotine, and amino acid and sarcosine utilization (Fig. 1).
ORF62 to ORF67, which may confer to pAO1-carrying bacteria the ability to utilize sarcosine, with ORF63 being related to mitochondrial monomeric sarcosine dehydrogenases (5), may be viewed as part of the pathway of nicotine utilization. Removal of the pyrrolidine side chain of nicotine by an as-yet-uncharacterized enzyme results in the formation of γ-methylaminobutyrate (14). One subsequent β-oxidation step would give rise to sarcosine. The arrangement of the ORFs resembles that of a chromosomally located operon of Arthrobacter globiformis (34).
ORF69 to ORF96 encompass the ORFs of known and hypothetical genes encoding nicotine-degrading enzymes and proteins believed to take part in the assembly of these enzymes and their interaction with the plasma membrane (3). ORF83, similar to endopeptidases, was hypothesized to cleave the peptide bond of the 2,3,6-trihydroxypyridine ring in its amide resonance form (3). The nitrilase of ORF77 would then remove the amino group with the formation of the citric acid cycle intermediate α-ketoglutarate. Alternatively, ring opening of 2,3,6-trihydroxypyridine could take place between C-2 and C-3 with the formation of N-formyl-maleamic acid (HOOC-CH=CH-CO-NH-CHO) (19). Deformylation followed by deamination would give rise to maleic acid (HOOC-CH=CH-COOH). The formation of the citric acid cycle intermediate fumarate from maleate would require the activity of a cis-trans isomerase (23). The pAO1sequence revealed no ORF with obvious similarity to this type of enzyme. Which of these alternatives takes place in the bacteria remains to be determined.
Separated by the IS1473 element (33) are genes of enzymes required for the biosynthesis of the molybdenum cofactor and genes of a molybdenum ABC-type transporter (32). Reanalysis of this DNA region revealed an additional ORF99, which is related to MoaD enzymes of molybdopterin biosynthesis (39) and which may be translationally coupled to MoaA.
The gene encoding 6HDNO was identified on pAO1 at a distance of 17 kb from the nic gene cluster, in close vicinity to two ORFs with similarity to amino acid permeases and to a transcriptional regulator. They may form a transcriptional unit. The possible involvement of the amino acid permease-like hypothetical proteins in nicotine uptake will be addressed below.
Drug resistance.
ORF19, which is related to vancomycin resistance, and two ORFs, ORF60 and ORF61, which are related to the two subunits of multidrug efflux pumps, are present on pAO1. However, both the strains carrying and not carrying pAO1 showed the same vancomycin resistance, and the predicted resistances to tertiary ammonium salts, ethidium bromide, and heavy metals were also indistinguishable between the two strains (not shown).
Insertion elements and transposons.
Besides IS1473 (ORF97 and ORF98), pAO1 carries a second hypothetical insertion sequence (IS) element with significant similarity (E value of 2e-37) to the transposase of IS1473. As in the case of IS1473, ORF140 and ORF141 may form the InsB and InsA peptides of the transposase, fused by a programmed −1 translational frameshift (33), characteristic of members of the IS1 family.
There are ORFs of two possible composite transposons on pAO1, both of which are related to transposons of the gram-positive bacterium Staphylococcus aureus. ORF48 shows similarity to transposase A (TpnA) of transposon Tn554 (36). Tn544 consists of the genes tpnA, tpnB, and tpnC, encoding functions required for transposition, followed by erythromycin and spectinomycin resistance determinants. On pAO1, ORF50 and ORF51, which resemble transposase B (TpnB) and transposase C (TpnC) of transposon Tn544, respectively, are separated from TpnA by ORF49, which is similar to a hypothetical unknown protein of Mycobacterium gordonae. It may represent an as-yet-unknown resistance factor. The ORF that is similar to TpnA is preceded by ORF47, which is related to the phage integrase family, a possible indication of an episome-related transposition event.
The second putative composite transposon on pAO1 is related to the S. aureus Tn552 (42). Tn552 may transpose replicatively by cointegrate formation and resolution, a process requiring both a transposase and a resolvase. In addition, it mediates resistance to β-lactam antibiotics. As with Tn552, the hypothetical transposon on pAO1 is formed from ORF152, which is related to the ATP-binding DNA transposition protein of Tn552; ORF151, which is related to the Tn552 transposase; and ORF153, which is related to recombinases, including that of Tn552. However, no ORFs similar to β-lactamases are present. Positioned 5′to the Tn552 ORFs is ORF150, which is also related to recombinases. Remarkably, ORF158 represents a truncated version of this same recombinase, with no start codon and ribosomal binding site. ORF156 and ORF157, situated between the putative Tn552 and the remnant of the recombinase, could have been generated by a duplication, both showing similarity to the same hypothetical adenylate cyclase-related protein of Caulobacter crescentus. Apparently, they are evidence of a recombination event having taken place at this site, possibly mediated by Tn552, and including an imprecise excision of one transposon, leaving the truncated recombinase (42).
Replication.
Clusters of ORFs encoding hypothetical proteins that could be involved in replication are distributed along the pAO1 DNA. The first cluster starts with ORF7, ORF8, and ORF9, which show similarity to ParB chromosome-partitioning proteins, contains ORF11 (similar to single-stranded DNA-binding proteins), and ends with ORF18 (similar to KfrA, a transcriptional regulator of partitioning genes [48]) present on many plasmids. Plasmid-encoded partitioning functions consist of two proteins, ParA and ParB, which form one operon autoregulated by the Par proteins. The ParA protein is an ATPase and assembles with ParB subunits into a nucleoprotein complex that binds to a cis-acting centromere-like site (18). Unusual on pAO1 is the presence of an apparently fragmented ParB consisting of three distinct ORFs (ORF7 to ORF9) and ORF143, related to ParA (soj locus in Bacillus subtilis), situated on pAO1 at a great distance (∼28 kb) from the ParB ORFs in a set of ORFs (ORF142 to ORF149) with no known homology. As predicted for ParA proteins, ORF143 exhibits at its N terminus an ATP/GTP-binding consensus sequence (P loop). Homologs of soj/parA map in bacteria to the chromosomal and plasmid origin regions of replication (28, 37). The origin of replication of many plasmids contains tandem direct repeats. No such particular DNA sequences could be clearly identified 5′or 3′ of the Par ORFs. The repeats serve as binding sites for the replication protein DnaA (43), but no DnaA homolog could be identified on pAO1. The family of RepABC low-copy-number plasmids do not contain the repeated sequences (iterons) common in other plasmids (40). They depend on the activity of the Rep proteins for replication. Homologs to these proteins were not found on pAO1. Therefore, at present the site of the origin of replication of pAO1 remains undetermined.
A second cluster possibly related to replication contains ORF43, the largest ORF identified on pAO1, which is similar to DNA helicases, and ORF44, ORF45, and ORF46, with no homology to known proteins. Dam methylases play important roles in the initiation of replication and the interaction of the replicated oriC segments with the cell membrane (30). The protein encoded by ORF23, which is similar to S-adenosylmethionine-dependent methyltransferases, may serve this function in the replication of pAO1.
Conjugation.
Conjugation in bacteria is a complex process involving many gene products in aggregate stabilization, pilus biogenesis, surface exclusion, and conjugal DNA metabolism (29). There are few ORFs on pAO1 with similarity to known proteins related to these functions. ORF124 may encode a TrsK-like membrane protein, a component of the conjugative transfer gene complex. ORF127 may be associated with conjugation, similar to topoisomerases of the TrbL transfer gene complex protein-like proteins; ORF126 may encode a putative membrane protein; and ORF125 may encode a putative ATP-binding protein. ORF132 shows low similarity (E value of 0.58) to outer membrane fimbrial usher proteins involved in fimbrial export and assembly of the fimbrial subunits. ORF22 is similar to sigma factors of the extracytoplasmic function family. They control the expression of genes involved in outer membrane protein folding, including proper pilus assembly (16, 38). Some of the ORFs from ORF115 to ORF139, with no homology or with homology to conserved proteins of unknown function, may be involved in conjugation mechanisms, since, as shown below, pAO1 is a conjugative plasmid.
Natural transformation in bacteria involves several steps connected to competence induction, DNA binding, and uptake and reconstitution of plasmid DNA. DprA proteins (Sfm in E. coli) play an essential role in natural competence in Haemophilus influenzae, Helicobacter pylori, and Streptococcus pneumoniae (2, 4, 25, 46), but at which step is not known. Inactivation of dprA in S. pneumoniae did not affect the internalization of DNA and may be required at a later stage of transformation (4). The presence of a DprA-like ORF162 seems to be the first reported instance of a dprA homolog from a plasmid. The similarity of ORF162 to DprA proteins extends over their entire length, including the central conserved part of these proteins (2). Remarkably, a 376-bp sequence 5′to the DprA-like ORF is exactly duplicated as a direct repeat at the 5′ end of ORF164 (Fig. 1). The presence of two large direct repeats on both sites of the DprA ORF may be suggestive of DprA being involved in recombination processes following plasmid DNA uptake during natural transformation.
pAO1 is a conjugative plasmid.
We took advantage of a recently described plasmid, pKGT452β, which carries a transposon with a chloramphenicol resistance gene (17) to examine the conjugative potential of pAO1. The plasmid can be transformed into Arthrobacter species by electroporation, where it does not replicate but by transposition gives rise to chloramphenicol-resistant bacteria. An A. nicotinovorans strain not carrying pAO1 was transformed with pKGT452β, and Cmr colonies appeared on selection plates with frequencies of 10 to 100 colonies per μg of pKGT452β DNA. Bacteria not carrying pAO1 are unable to degrade nicotine and do not turn dark, in contrast to pAO1-carrying bacteria, when grown on citrate plates with nicotine (14, 21). The pAO1-negative A. nicotinovorans Nic− Cmr strain was mated on filters on LB plates with A. nicotinovorans/pAO1 Nic+ Cm−, and exconjugants were selected on citrate plates with chloramphenicol and nicotine. Cmr Nic+ colonies appeared at a frequency of 10−3 to 10−2 of Cmr Nic− colonies.
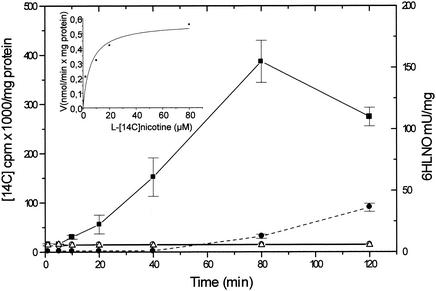
l-[14C]nicotine uptake by A. nicotinovorans carrying pAO1, by the A. nicotinovorans strain not carrying pAO1, and by E. coli.
l-Nicotine uptake by A. nicotinovorans and its relation to the pAO1 state of the bacteria have not been described before. When A. nicotinovorans carrying pAO1, the A. nicotinovorans strain not carrying pAO1, and E. coli XL1-Blue bacteria were incubated with l-[14C]nicotine and the time-dependent nicotine uptake was measured, only those bacteria harboring the pAO1 plasmid showed l-nicotine accumulation (Fig. 2). The addition of l-[14C]nicotine did not result, during the time of the measurements, in an increase in the turbidity of the bacterial cultures or in the protein concentration of the bacterial extracts. A similar background of unspecifically bound radioactivity was observed for the bacteria of all three cultures at 1 and 5 min of incubation with l-[14C]nicotine (Fig. 2). Nicotine uptake started after a lag of approximately 10 min after the addition of the alkaloid to the A. nicotinovorans/pAO1 cultures, an indication that it had to be induced. 6HLNO activity, taken as a marker for the induction of the nicotine-degrading enzymes, became detectable after 80 min, and at 120 min the cultures started to turn light blue, which coincided with a decrease in the radiolabeled material recovered from the bacteria (Fig. 2). The activity of the second enzyme of the pathway was measured because the 6HLNO assay is very sensitive, and NDH and 6HLNO are coinduced and their genes appear to form an operon (22). At up to 80 min, the radioactivity measured in the bacterial lysates reflects the accumulation of l-[14C]nicotine by the bacteria. The appearance of enzyme activity and of nicotine blue showed that the catabolic pathway was fully induced. One possible explanation for the decrease in the radiolabeled material recovered in the lysate of the bacteria at 120 min may be that the [14C]CH3 of nicotine had entered the C-1 pool and was released in part as [14C]CO2. For the l-[14C]nicotine uptake, a Km of 5.6 ± 2.2 μM and a Vmax of 0.6 ± 0.06 nmol/min · mg of protein were determined (Fig. 2, inset), which are similar to those of some high-affinity, low-capacity amino acid transporters (10).
FIG. 2.
l-[14C]nicotine uptake by A. nicotinovorans carrying pAO1 and 6HLNO activity. l-[14C]nicotine (1.5 μCi) was added to bacterial cultures, and nicotine uptake and 6HLNO activity (dashed line), were determined at the indicated time points as described in Materials and Methods. Diamonds, A. nicotinovorans carrying pAO1; triangles, A. nicotinovorans not carrying pAO1; squares, E. coli XL1-Blue. Results are means ± standard deviations calculated from at least three independent experiments. The inset shows the l-[14C]nicotine concentration-dependent uptake determined as outlined in Materials and Methods.
There is no ORF with similarity to a transporter within the nic gene cluster. Putative transporters for sugar, sarcosine, and molybdenum seem to be responsible for the uptake of substrates of enzymes encoded by the corresponding gene cluster. Surprisingly, the gene for 6HDNO (ORF112) was found situated between ORF111 and ORF113, both of which have significant similarity to amino acid permeases. In view of the close proximity of these putative amino acid transporters to a gene connected with nicotine metabolism, it seemed of interest to test whether amino acids could interfere with nicotine uptake.
Nicotine blue formation by A. nicotinovorans/pAO1 cultures in the presence of amino acids.
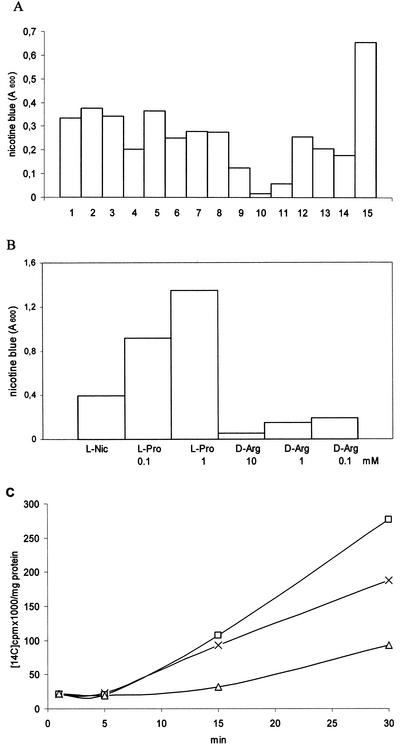
The generation of nicotine blue by A. nicotinovorans/pAO1 cultures can be monitored by the increase in absorption at 600 nm and is an indication that nicotine is taken up by the bacteria and metabolized to the aerobic end product of the nicotine catabolic pathway. The effect of l- or d-amino acids on the development of nicotine blue by A. nicotinovorans/pAO1 cultures is shown in Fig. 3A. Since nicotine is a base, the l and d isomers of arginine, lysine, and histidine were tested. In addition, proline (which is structurally related to the pyrrolidine ring of nicotine), the polar amino acids glutamine and serine, and the hydrophobic aromatic amino acid phenylalanine (in this case as ld-phenylalanine) were employed. The d-stereoisomers of Lys, His, and Arg, and to a lesser extend l-arginine, inhibited the formation of nicotine blue, whereas l-proline, and to a lesser extend d-proline, stimulated its formation (Fig. 3A). The effects of d-Arg and l-Pro were concentration dependent (Fig. 3B).
FIG. 3.
Formation of nicotine blue and l-[14C]nicotine uptake in the presence of l- and d-amino acids. (A) To A. nicotinovorans/pAO1 stationary-phase bacteria, a 100 μM concentration of each of the following amino acids was added: bar 2, l-Glu; bar 3, l-Ser; bar 4, l-Arg; bar 5, l-Lys; bar 6, l-Ala; bar 7, l-His; bar 8, d-Pro; bar 9, d-Lys; bar 10, d-Arg; bar 11, d-His; bar 12, d-Ala; bar 13, d-Ser; bar 14, D, l-Phe; and bar 15, l-Pro. The cultures were induced with 50 μM l-nicotine, and nicotine blue formation was monitored after 3 h. Bar 1, nicotine blue formation in the presence of nicotine only. (B) Concentration-dependent effects of l-Pro and d-Arg on the formation of nicotine blue. (C) l-[14C]nicotine (1.5 μCi) was added to the A. nicotinovorans cultures in the absence of amino acids (crosses) or in the presence of 1 mM l-Pro (squares) or 1 mM d-Arg (triangles), and the time-dependent accumulation of radioactivity by the bacteria was determined as described in Materials and Methods.
l-[14C]nicotine uptake in the presence of d-Arg and l-Pro.
In order to establish whether the different amounts of nicotine blue formed in the presence of d-Arg and l-Pro indeed reflected differences in nicotine uptake, l-[14C]nicotine uptake by A. nicotinovorans carrying pAO1 was determined in the presence of these amino acids. The results presented in Fig. 3C demonstrate that this was the case.
From these experiments, nicotine uptake could be regarded as a cotransport with l-Pro. On the other hand, l-[14C]nicotine import by bacteria not carrying pAO1 and by E. coli XL1-Blue could not be induced by the presence of l-Pro. Thus, l-Pro could not replace l-nicotine as an inducer of the nicotine uptake system. Proline uptake itself was unrelated to pAO1, since E. coli as well as A. nicotinovorans, irrespective of the presence of pAO1, imported the amino acid very efficiently (not shown).
DISCUSSION
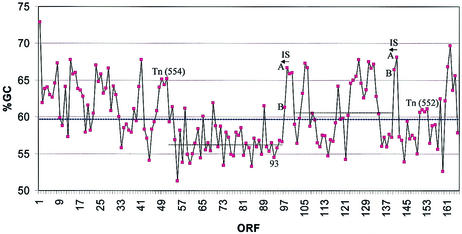
An analysis of the G+C content of the ORFs clearly indicates that, as in the case of other large plasmids (35, 48), the pAO1 DNA shows a modular composition and that transposition events participated in the generation of its present state (Fig. 4). The DNA segment between the putative Tn554 and IS1473, encompassing ORF52 to ORF96 and including the nic gene cluster (3), exhibits a lower overall G+C content of approximately 56%, compared to the rest of the plasmid (over 60%). It may be mentioned in this context that the 6HLNO gene (ORF93 [Fig. 4]) was suggested to have been acquired by pAO1 through horizontal gene transfer from a eucaryotic source, based on the lower G+C content of the 6HLNO gene and on the amino acid similarity of 6HLNO to amine oxidases of eucaryotic origin (45). However, the 6HLNO ORF is itself part of this low-G+C DNA segment of pAO1 (Fig. 4). In addition, recent data bank searches have revealed that 6HLNO is most closely related to a bacterial protein (tyramine oxidase of Micrococcus luteus [accession number AB010716]).
FIG. 4.
G+C contents of the ORFs of pAO1. Solid line, mean G+C content of pAO1 DNA (59.7%); broken line from Tn554 to IS1473, average G+C content of 56.6%; broken line between IS elements, average G+C content of 60.6%. The average G+C content of remaining ORFs is 62%. ORF93, 6HLNO. A and B indicate the two ORFs of the transposases.
Catabolic transposons are frequently located on transmissible plasmids, and, as is characteristic for composite transposons, are flanked by related, but not identical, IS elements. They may be very large (over 50 kb), and the best known are involved in the degradation of aromatic compounds (47). The DNA segment flanked by IS1473 and its related IS element, which represent direct repeats, may form such a transposon (Fig. 4). The nic gene cluster and the cluster of ORFs on the hypothetical catabolic transposon (including those of MoCo biosynthesis and the putative nicotine transporters with 6HDNO) may have been acquired by transposition-related events by an ancestral plasmid, carrying genes for carbohydrate utilization (ORF1 to ORF46), extending its metabolic capabilities to the exploitation of nicotine.
ORFs connected to uptake and utilization of nutrients represent 47.7% of the pAO1 sequence. Besides carbohydrate utilization (22.5%), the most prominent feature of the plasmid is the large cluster (25.2%) of ORFs related to nicotine utilization. The identification of a MoaD ORF99 completes the list of ORFs of proteins essential for the biosynthesis of the molybdenum dinucleotide cofactor of NDH and 6-hydroxy-pseudooxynicotine dehydrogenase (ketone dehydrogenase) (3). MoaE and MoaD form the large and small subunits, respectively, of the molybdopterin synthase involved in the insertion of the sulfur atoms into precursor Z (39). ORF78 shows some similarity (38.6% identity in a stretch of 44 amino acids out of 78) to MetB of the second step of methionine synthesis, which belongs to the trans-sulfuration enzyme family. Its gene product may functionally replace MoeB, which is implicated in sulfur transfer to the molybdopterin synthase and is the only enzyme of molybdenum cofactor synthesis with no equivalent ORF on pAO1. The presence of genes of a complex biosynthetic pathway for a cofactor is a unique feature of pAO1.
Since the tobacco plant produces only l-nicotine, the presence of a 6-hydroxy-d-nicotine-specific oxidase may be surprising (21). The need for an enzyme with d-nicotine specificity could be warranted if a d-nicotine derivative would arise as a natural product of nicotine metabolism. It has been reported that strains of tobacco plants produce the l and d isomers of nornicotine (26). Nornicotine may be a metabolite of nicotine (22), and nornicotine, in contrast to nicotine, appears to be racemized (26). The first enzyme of pAO1-dependent nicotine degradation, nicotine dehydrogenase, hydroxylates C-6 of the pyridine rings of the l and d isomers of nicotine and nornicotine nonstereospecifically (14). However, the next enzymatic step, catalyzed by 6HLNO, proceeds only on the l isomer. Therefore, for d-nornicotine degradation, a 6-hydroxy-d-nornicotine-specific oxidase would be required. The finding that the gene of 6HDNO is separated by several thousand nucleotides from the genes of the l-nicotine-specific enzymes and in close proximity to ORFs with similarity to amino acid permeases may be an indication that 6HDNO fulfills yet another function.
This work presents for the first time the identification of an l-nicotine uptake system. It may have evolved from an amino acid permease. The two amino acid permease-like ORFs belong to different classes. One belongs to permeases of positively charged amino acids, which do not require a periplasmic amino acid-binding protein (10, 12, 20). The second of the 12 transmembrane domains of ORF113 is related to the consensus sequence of these permeases (PROSITE PS00218). The second amino acid-related permeases (ORF111) shows an amino acid sequence signature characteristic for Na+ solute symporters (PS50283). Uptake of proline in E. coli (and other bacteria) has been shown to be via an Na+ proline symporter (11). The increase in nicotine uptake in the presence of proline may point to a nicotine-proline symport. A conclusion as to whether these amino acid permease-related ORFs do indeed represent the l-nicotine transporter(s) of pAO1 has to await the functional characterization of the corresponding genes and gene products.
Acknowledgments
We acknowledge the excellent technical assistance of E. Schiefermayr and I. Deuchler, and we thank Carmen Brizio, University of Bari, Bari, Italy, for her help with kinetic data and computer software.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to R.B.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baitsch, D., C. Sandu, R. Brandsch, and G. L. Igloi. 2001. Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J. Bacteriol. 183:5262-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge, M., M. Moscoso, M. Prudhomme, B. Martin, and J.-P. Claverys. 2002. Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45:411-421. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron, F., A. Otto, P. Brache, R. Day, L. Denoroy, R. Brandsch, and D. Bataile. 1998. Molecular cloning and tissue distribution of rat sarcosine dehydrogenase. Eur. J. Biochem. 257:556-561. [DOI] [PubMed] [Google Scholar]
- 6.Bonfield, J. K., K. F. Smith, and R. Staden. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17:123-133. [Google Scholar]
- 8.Brandsch, R., and K. Decker. 1984. Isolation and partial characterization of plasmid DNA from Arthrobacter oxydans. Arch. Microbiol. 138:15-17. [DOI] [PubMed] [Google Scholar]
- 9.Brühmüller, M., H. Möhler, and K. Decker. 1972. Covalently bound flavin in d-6-hydroxynicotine oxidase from Arthrobacter oxidans. Purification and properties of d-6-hydroxynicotine oxydase. Eur. J. Biochem. 29:143-151. [DOI] [PubMed] [Google Scholar]
- 10.Burkovski, A., and R. Krämer. 2002. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biotechnol. 58:265-274. [DOI] [PubMed] [Google Scholar]
- 11.Cairney, J., C. F. Higgins, and I. R. Booth. 1984. Proline uptake through the major transport system of Salmonella typhimurium is coupled to sodium ions. J. Bacteriol. 160:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celis, R. T. F. 1981. Chain-termination mutants affecting a periplasmic binding protein involved in the active transport of arginine and ornithine in Escherichia coli. J. Biol. Chem. 256:773-779. [PubMed] [Google Scholar]
- 13.Decker, K., H. Eberwein, F. A. Gries, and M. Brühmüller. 1961. Über den Abbau des Nicotins durch Bakterienenzyme. IV. l-6-Hydroxy-Nicotin als erstes Zwischenprodukt. Biochem. Z. 334:227-244. [PubMed] [Google Scholar]
- 14.Decker, K., and H. Bleeg. 1965. Induction and purification of stereospecific nicotine oxidizing enzymes from Arthrobacter oxydans. Biochim. Biophys. Acta 105:313-334. [DOI] [PubMed] [Google Scholar]
- 15.Eberwein, H., F. A. Gries, and K. Decker. 1961. Über den Abbau des Nikotins durch Bakterienenzyme II, Isolierung und Charakterisierung eines nikotinabbauenden Bodenbakteriums. Hoppe-Seyler's Z. Physiol. Chem. 323:236-248. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich, A., C. Prust, T. Hartsch, A. Henne, and B. Averhoff. 2002. Molecular analysis of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartemann, K. H., and R. Eichenlaub. 2001. Isolation and characterization of IS1409, an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1, and development of a system for transposon mutagenesis. J. Bacteriol. 183:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdes, K., J. Moller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 19.Gherna, R. L., S. H. Richardson, and S. C. Rittenberg. 1965. The bacterial oxidation of nicotine. VI. The metabolism of 2,6-dihydroxypseudooxynicotine. J. Biol. Chem. 240:3669-3674. [PubMed] [Google Scholar]
- 20.Glansdorf, N. 1996. Biosynthesis of arginine and polyamines, p. 408-433. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 21.Gloger, M., and Decker, K. 1969. Zum Mechanismus der Induktion nicotinabbauender Enzyme in Arthrobacter oxidans. Z. Naturforsch. 246:1016-1025. [PubMed] [Google Scholar]
- 22.Grether-Beck, S., G. L. Igloi, S. Pust, E. Schiltz, K. Decker, and R. Brandsch. 1994. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol. Microbiol. 13:929-936. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama, K., Y. Asai, Y. Uchida, M. Kobayashi, M. Terasawa, and H. Yukawa. 1997. Gene cloning and characterization of maleate cis-trans isomerase from Alcaligenes faecalis. Biochem. Biophys. Res. Commun. 239:74-79. [DOI] [PubMed] [Google Scholar]
- 24.Jones, D., and R. M. Keddie. 1992. The genus Arthrobacter, p. 1283-1299. In A. Balows, A., H. G. Truper, M. Dwarkin, W. Harder, and K. H. Schleifer (ed.), The Prokaryotes, vol. 2. Springer Verlag, Heidelberg, Germany.
- 25.Karudapuram, S., X. Zhao, and G. J. Barcak. 1995. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J. Bacteriol. 177:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kisaki, T., and E. Tamaki. 1961. Phytochemical studies on tobacco alkaloids. I. Optical rotatory power of nornicotine. Arch. Biochem. Biophys. 92:351-355. [DOI] [PubMed] [Google Scholar]
- 27.Kodama, Y., H. Yamamoto, N. Amano, and T. Amchi. 1992. Reclassification of two strains of Arthrobacter oxydans and proposal of Arthrobacter nicotinovorans sp. nov. Int. J. Syst. Bacteriol. 42:234-239. [DOI] [PubMed] [Google Scholar]
- 28.Lin, D. C. H., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 29.Llosa, M., F. X. Gomis-Rüth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two step mechanism from DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Marinus, M. G. 1996. Methylation of DNA, p. 782-791. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 31.Martinez, B., J. Tomkins, L. W. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menéndez, C., A. Otto, G. L. Igloi, P. Nick, R. J. Brandsch, B. Schubach, B. Böttcher, and R. K. Brandsch. 1997. Molybdate-uptake genes and molybdopterin-biosynthesis genes on a bacterial plasmid. Eur. J. Biochem. 250:524-531. [DOI] [PubMed] [Google Scholar]
- 33.Menéndez, C., G. L. Igloi, and R. Brandsch. 1997. IS 1473, a putative insertion sequence identified in the plasmid pAO1 from Arthrobacter nicotinovorans: isolation, characterization and distribution among Arthrobacter species. Plasmid. 37:35-41. [DOI] [PubMed] [Google Scholar]
- 34.Meskys, R., R. J. Harries, V. Casaite, J. Basran, and N. S. Scrutton. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur J. Biochem. 268:3390-3398. [DOI] [PubMed] [Google Scholar]
- 35.Murata, T., M. Ohnishi, T. Ara, J. Kaneko, C.-G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsuba, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implication for evolution of large plasmid genomes. J. Bacteriol. 184:3194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, E., L. Huwyler, and M. do Carmo de Freire Bastos. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 4:3357-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picardeau, M., J. R. Lobry, and B. J. Hinnebusch. 2000. Analysing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plastids. Genome Res. 10:1594-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 39.Rajagopalan, K. V. 1997. Biosynthesis and processing of the molybdenum cofactors. Biochem. Soc. Trans. 25:757-761. [DOI] [PubMed] [Google Scholar]
- 40.Ramirez-Romero, M. A., N. Soberon, A. Perez-Oseguera, J. Tellez-Sos, and M. A. Cevallos. 2000. Structural elements required for replication and incompatibility of the Rhizobium etli symbiotic plasmid. J. Bacteriol. 182:3117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowland, S. J., and K. G. Dyke. 1990. Tn552 a novel transposable element from Staphylococcus aureus. Mol. Microbiol. 4:961-975. [DOI] [PubMed] [Google Scholar]
- 43.Schaper, C., and W. Messer. 1995. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270:17622-17626. [DOI] [PubMed] [Google Scholar]
- 44.Schenk, S., A. Hoelz, B. Kraus, and K. Decker. 1998. Gene structure and properties of enzymes of the plasmid-encoded nicotine catabolism of Arthrobacter nicotinovorans. J. Mol. Biol. 284:1323-1339. [DOI] [PubMed] [Google Scholar]
- 45.Schenk, S., and K. Decker. 1999. Horizontal gene transfer involved in the convergent evolution of the plasmid-encoded enantioselective 6-hydroxynicotine oxidases. J. Mol. Evol. 48:178-186. [DOI] [PubMed] [Google Scholar]
- 46.Smeets, L. C., J. J. Bijlsma, E. J. Kuipers, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 27:99-102. [DOI] [PubMed] [Google Scholar]
- 47.Tan, H.-M. 1999. Bacterial catabolic transposons. Appl. Microbiol. Biotechnol. 51:1-12. [DOI] [PubMed] [Google Scholar]
- 48.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]