Abstract
In this study, we have evaluated the cytotoxic effect of combining two HDAC inhibitors, SAHA and TSA, with TRAIL in human multiple myeloma cell lines. Low doses of SAHA or TSA enhanced the cytotoxic and apoptotic effects of TRAIL and upregulated the surface expression of TRAIL death receptors (DR4 and/or DR5). SAHA and TSA induced G1 phase cell cycle growth arrest by upregulating p21WAF1 and p27Kip1 expression and by inhibiting E2F transcriptional activity. The enhanced TRAIL effect after pretreatment with HDAC inhibitors was consistent with the upregulation of the proapoptotic Bcl-2 family members (Bim, Bak, Bax, Noxa, and PUMA), the downregulation of the antiapoptotic members of the Bcl-2 family (Bcl-2 and Bcl-XL), and IAPs. SAHA and TSA dissipated the mitochondrial membrane potential and enhanced the release of Omi/HtrA2 and AIF from the mitochondria to the cytosol. The cytotoxic effect of both SAHA and TSA was caspase- and calpain-independent. Inhibition of NFκB activation by the proteasome inhibitor, MG132, enhanced the apoptotic effect of TSA. Our study demonstrated the enhancing effects of HDAC inhibitors on apoptosis when combined with TRAIL and, for the first time, emphasized the role of AIF in mediating the cytotoxic effects of HDAC inhibitors.
Keywords: TRAIL/Apo-2L, apoptosis, histone deacetylase inhibitor, multiple myeloma, death receptors
Introduction
Multiple myeloma, an almost incurable disease, is the second most common blood cancer. It is characterized by the presence of malignant plasma cells that are predominantly located in the bone marrow. The treatment of myeloma is mainly directed at the management of complications and reduction of malignant cell masses. Initial chemotherapeutic treatment could be successful; however, resistance development urges the use of higher toxic doses accompanied by hematopoietic stem cell transplantation [1]. Histone deacetylase (HDAC) inhibitors and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) were able to induce apoptosis in human multiple myeloma cell lines [2]; they showed a high degree of selectivity and were nontoxic. Furthermore, TRAIL induced apoptosis in dexamethasone-resistant and cytotoxic drug-resistant multiple myeloma cell [3].
TRAIL crosslinks with two death receptors, TRAIL-R1 (DR4) and TRAIL-R2 (DR5) [4]. The crosslinking of TRAIL to DR4/DR5 results in receptor trimerization and the recruitment of the adapter protein FADD, through the 80-amino acid intracellular death domain of the TRAIL receptor. FADD facilitates the binding and autocatalytic activation of procaspase-8/procaspase-10 and the formation of the death-inducing signaling complex (DISC). DISC formation initiates a cascade of proteolytic activation of downstream caspases and an irreversible commitment of the cells to undergo apoptosis [5]. TRAIL-R3, TRAIL-R4, and osteoprotegerin are other three TRAIL receptors. TRAIL-R3 and TRAIL-R4 have extracellular domains similar to DR4 and DR5, but lack a functional cytoplasmic death domain and may serve as “decoy” receptors [6]. The selectivity of TRAIL can be explained by the higher expression of the decoy receptors (DcR1 and DcR2) in normal cells than in tumor cells [7]. This differential expression will result in higher DcR/DR ratio in normal cells and, consequently, lower response to TRAIL-induced apoptosis as decoy receptors do not transduce death signals.
Histone acetyltransferases (HATs) and HDACs determine the acetylation status of histones and, consequently, the chromatin structure and gene expression [8]. HDAC inhibitors inhibit HDACs, resulting in histone hyperacetylation and easier access of transcription factors with consequent increase in gene expression. Surprisingly, gene profiling studies showed that HDAC inhibitors modulated only 2% of the genes with a similar proportion of gene activation and repression apoptosis [9]. Thus, HDAC inhibitors regulate only a subset of gene expression in regulating growth, differentiation, and apoptosis.
We have previously reported that the combination of TRAIL with other chemotherapeutic agents (vincristine, vinblastine, paclitaxel, etoposide, camptothecin, and doxorubicin) could induce more apoptosis in both TRAIL-sensitive and TRAIL-resistant cell lines [10]. In this study, we investigated the effect of combining two HDAC inhibitors, suberoylanilide hydroxamic acid (SAHA) and trichostatin A (TSA), with TRAIL in human multiple myeloma cell lines. Our hypothesis is that SAHA or TSA will augment the apoptotic effect of TRAIL by regulating apoptosis- and cell cycle-related genes. The purpose of this study is to investigate the molecular mechanisms by which HDAC inhibitors enhance the apoptosis-inducing potential of TRAIL in ARP-1 multiple myeloma cells. We observed the upregulation of the TRAIL DR4/DR5 receptors, the pro-apoptotic Bcl-2 family members (Bim, Bak, Bax, Noxa, and PUMA), the enhanced release of the mitochondrial proteins apoptosis-inducing factor (AIF) and Omi/HTRA2 to the cytosol, and the upregulation of the negative modulators of the G1 phase of the cell cycle, p21WAF1 and p27Kip1, after treatment with SAHA or TSA. Apoptosis proteins (IAPs; XIAP, cIAP1, and cIAP2) and the anti-apoptotic members of the Bcl-2 family were downregulated after treatment with SAHA or TSA. Moreover, the cytotoxic effects of SAHA and TSA were caspase- and calpain-independent. Finally, this study provides an insight into the mechanisms by which HDAC inhibitors augment the cytotoxic effects of TRAIL and induce cell cycle growth arrest, and emphasizes the role of AIF in HDAC inhibitor-mediated apoptosis.
Materials and Methods
Reagents
The antibodies used were as follows: goat polyclonal antibodies for acetylated histones H3 and H4, pRb, Noxa, and PUMA β/δ (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal antibodies for DR4 and DR5 receptors (R&D Systems, Minneapolis, MN); p21WAF1, p27Kip1, Bax, and AIF (Santa Cruz Biotechnology); Omi/HtrA2 (Calbiochem, La Jolla, CA); rabbit polyclonal antibodies for XIAP (IMGENEX, San Diego, CA); PARP (Biosource International, Camarillo, CA); and cIAP1, cIAP2, Bcl-XL, Bcl-2, Bak, and Bim (Santa Cruz Biotechnology). TRAIL and SAHA were purchased from BIOMOL (Plymouth Meeting, PA), TSA from Alexis Biochemicals (Carlsbad, CA) and all other chemicals used were of analytical grade from Fisher Scientific (Suwanee, GA) or Sigma (St. Louis, MO).
Cell Culture
ARP-1, 8226, and MM1S cells were kindly provided by Dr. R. Fenton (School of Medicine, University of Maryland, Baltimore, MD). Cells were cultured in RPMI 1640 medium (GIBCO, Grand Island, NY) supplemented with 10% FBS (GIBCO) in a humidified incubator supplied with 5% carbon dioxide.
Extraction of Cellular Histones
Histones were extracted according to the method of Yoshida et al. [11]. ARP-1 cells (about 2 x 106 cells), treated with SAHA or TSA for different time points, were collected by centrifugation at 1000 rpm for 5 minutes and washed twice with 1x phosphate-buffered saline (PBS). Cells were suspended in 1 ml of ice-cold lysis buffer (10 mM Tris-HCl, 50 mM sodium bisulfite, 1% Triton X-100, 10 mM MgCl2, 8.6% sucrose, pH 6.5), Dounce-homogenized, and centrifuged at 1000g for 10 minutes to collect the nuclei, followed by washing three times with the lysis buffer and once with a buffer consisting of 10 mM Tris-HCl, 13 mM EDTA, pH 7.4, successively. The pellet was suspended in 0.1 ml of ice-cold H2O, acidified with concentrated H2SO4 to give a final concentration of 0.4 N, and incubated at 4°C for at least 1 hour. The suspension was centrifuged for 5 minutes at 15,000 rpm and the supernatant was incubated overnight with 1 ml of acetone at -20°C. The coagulated histones were collected by centrifugation, air-dried, and dissolved in H2O, and their concentrations were estimated by a protein assay kit (Bio-Rad, Hercules, CA).
XTT Colorimetric Assay
XTT cell proliferation assay is a colorimetric assay for the nonradioactive quantification of cell proliferation and viability, where XTT (tetrazolium salt) is metabolized by viable cells only to a water-soluble formazan dye that can be measured spectrophotometrically [12]. Briefly, 100 µl of cell suspension (1 x 106 cells/ml) was added to a 96-well plate, 100 µl of different concentrations of the drugs (TSA, SAHA, or TRAIL dissolved in RPMI) was added to the wells, and cells were incubated for various time points in a humidified incubator at 37°C and 5% CO2. A mixture of 50 µl of XTT (0.2 mg/ml) and PMS (2.5 µM) dissolved in RPMI was added to each well, and the cells were further incubated for 4 hours at 37°C. Finally, the absorbance of the dye was measured spectrophotometrically at 450 and 690 nm as a reference wavelength. To detect the role of different caspases, serine proteases or calpains in the cytotoxic effect of HDAC inhibitors, ARP-1 cells were pretreated with 30 µM of the pan-caspase inhibitor Z-VAD-FMK (Calbiochem), 50 µM TPCK (Calbiochem), or 30 µM Calpeptin (Calbiochem) for 2 hours prior to SAHA or TSA treatment, respectively.
DR4/DR5 Staining for Flow Cytometry
ARP-1 cells (1 x 106) were fixed with 2% Paraformaldehyde for 10 minutes at room temperature, washed twice with PBS, incubated with DR4 or DR5 mouse primary antibody (R&D Systems) for 1 hour, washed twice with PBS, and finally incubated with FITC-conjugated secondary antibody (Molecular Probes, Eugene, OR) for 30 minutes in the dark. Fluorescence was analyzed using Becton Dickinson FACScan flow cytometer.
Propidium Iodide (PI) Staining for Cell Cycle Analysis
PI is a fluorescent nucleic acid binding dye that binds preferentially to double-stranded nucleic acids, allowing fluorescent intensity to be used as an indicator of the cellular DNA content [13]. Synchronized ARP-1 and MM1S cells were treated with SAHA (500 nM) or TSA (50 nM) for 24 hours and then fixed in 70% ethanol for 30 minutes at 4°C. Cells were washed with PBS, pretreated with RNase, DNase-free (1 unit), for 30 minutes at 37°C, chilled on ice to 4°C, and stained with 1 µg/ml PI in PBS. PI stained cells were analyzed using a Becton Dickinson FACScan flow cytometer.
Assessment of Apoptosis by Annexin V or DAPI Staining
Annexin V FITC staining was performed as per the manufacturer's instructions (Oncogene Research, San Diego, CA). Briefly, 1 x 106 ARP-1 cells were incubated with Annexin V FITC for 15 minutes at room temperature in the dark, and cells were centrifuged and resuspended in 1x binding buffer and PI for analysis by flow cytometry. DAPI staining was performed by fixing of the cells with 2% paraformaldehyde for 30 minutes at room temperature, incubation with DAPI (0.5 µg/ml) for 20 minutes at room temperature, washing with PBS, and finally counting of the apoptotic nuclei using fluorescence microscopy.
RNase Protection Assay (RPA)
Total RNA was extracted using Trizol reagent (Life Technologies, Inc., Gaithersburg, MD). The RPA was performed as per the manufacturer's instructions (PharMingen, San Diego, CA). Briefly, hAPO-3d multiprobe template was used for T7 RNA polymerase-directed synthesis of [32P]UTP-labeled antisense RNA probes. Eight micrograms of RNA was incubated with α[32P]UTP-labeled single-stranded RNA probes overnight at 56°C, and treated with RNase for 45 minutes at 30°C. The RNA-RNA complexes were resolved by electrophoresis in 6% denaturing polyacrylamide gels and analyzed by autoradiography.
Immunoblotting
Cells were lysed in a buffer containing 10 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.5 mM EDTA, 1 mM EGTA, 1% SDS, 1 mM sodium orthovanadate, and a mixture of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 µg/ml pepstatin A, and 2 µg/ml aprotinin). Equal amounts of lysate protein were run on 12% SDS-PAGE gels and transferred to PVD membrane (Perkin-Elmer, Boston, MA). PVDF blots were blocked with 5% nonfat dry milk in TBS buffer (20 mM Tris-HCl, pH 7.4, 500 mM NaCl), and incubated with primary antibody (diluted in TBS containing 1% BSA) overnight at 4°C. Immunoblots were washed three times (15, 5, and 5 minutes each) with TBST (TBS and 0.01% Tween 20). Immunoreactivity was detected by sequential incubation with horseradish peroxidase-conjugated secondary antibody and ECL detection.
Subcellular Fractionation
Extraction of the cytosolic fraction was performed as previously described [14]. Briefly, cells were harvested by centrifugation at 1000g for 10 minutes at 4°C. The cell pellets were washed once with ice-cold PBS and resuspended with 5 vol of ice-cold buffer (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 17 µg/ml PMSF, 8 µg/ml aprotinin, and 2 µg/ml leupeptin, pH 7.4). Cells were homogenized with a 22-gauge needle, and the nuclei were pelleted by centrifugation at 750g for 10 minutes at 4°C. The supernatant was centrifuged at 10,000g for 25 minutes. The supernatant (cytosolic fraction) was saved for further analysis and checked for purity by Western blot analysis using anti-cytochrome oxidase 2 antibody. The protein concentrations were determined by the Bradford method (Bio-Rad).
Measurement of the Mitochondrial Membrane Potential ΔΨm
The mitochondrial membrane potential was determined by the retention of JC-1 dye as previously described [15]. JC-1 exhibits potential-dependent accumulation in the mitochondria indicated by a fluorescence emission shift from green (∼529 nm) to red (∼590 nm). Consequently, mitochondrial depolarization is indicated by a decrease in the red/green fluorescence intensity ratio. ARP-1 cells (5 x 105) cells were loaded with JC-1 dye (1 µg/ml) during the last 30 minutes of incubation with SAHA or TSA. Cells were washed in PBS twice. Fluorescence was monitored in a fluorimeter by measuring both the monomer (527-nm emission; green) and J-aggregate (590-nm emission; red) forms of JC-1 following 488-nm excitation. ΔΨm was calculated as a ratio of the fluorescence of J-aggregate and monomer forms of JC-1.
Immunofluorescence Staining
ARP-1 cells were incubated with SAHA or TSA for 12 hours, fixed with 2% paraformaldehyde, and incubated for 1 hour at room temperature in 0.1% Triton X-100/1% BSA in PBS to enhance permeability and to block nonspecific binding. Fixed cells were incubated for 1 hour at room temperature with AIF mouse monoclonal antibody (1:100), washed three times with PBS, and then resuspended in a mixture of goat anti-mouse IgG (1:2000) FITC conjugate (Molecular Probes) and 1 µg/ml DAPI (Molecular Probes) for 45 minutes at room temperature. Cells were washed three times in PBS and then mounted for fluorescence detection using a Nikon Eclipse E-800 fluorescence microscope.
Electrophoretic Mobility Shift Assay (EMSA)
Double-stranded oligonucleotides containing the consensus binding site for NF-κB (Promega, Madison, WI) and E2F-1 (Santa Cruz Biotechnology) were 5′ end-labeled using polynucleotide kinase and [γ-32P]dATP. Nuclear extracts (3.0 µg) from ARP-1 cells treated with 50 nM TSA were incubated with 1 µl of labeled oligonucleotide (20,000 cpm) and 6 µl of incubation buffer (10 mM Tris-HCl, 40 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 2% glycerol, and 2 µg of poly(dI-dC) for 20 minutes at room temperature). The specificity of NF-κB and E2F-1 DNA-binding activity was confirmed by the competition with excess cold (unlabeled) wild type. DNA-protein complexes were resolved by electrophoresis in 6% nondenaturing polyacrylamide gels and analyzed by autoradiography.
Luciferase Reporter Assay
ARP-1 cells were cotransfected with E2F1 luciferase reporter plasmid (a generous gift from A. W. Hamburger, University of Maryland) and a cytomegalovirus 4 promoter-driven β-gal expression plasmid using Lipofectamine (Invitrogen, Carlsbad, CA). Luciferase activity was measured and normalized by β-gal as per the manufacturer's instructions (Promega).
Statistical Analyses
Results are shown as mean ± SD. Student's t test was used to determine significant differences. In all cases, P < .05 was considered statistically significant.
Results
SAHA and TSA Induce the Acetylation of H3 and H4 Histones
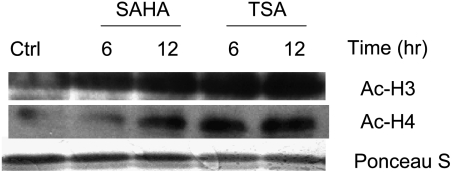
To verify whether the effects of SAHA or TSA were mediated through the hyperacetylation of histones, the effect of SAHA (500 nM) or TSA (50 nM) on histone acetylation in ARP-1 cells was studied. ARP-1 cells treated with SAHA or TSA for 6 or 12 hours exhibited a significant increase in acetylation over the control (Figure 1). Equal loading of histones was confirmed by Ponceau S staining.
Figure 1.
SAHA and TSA induce the accumulation of acetylated histones in ARP-1 cells. ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 6 and 12 hours followed by extraction of cellular histones and Western blot analysis to detect the levels of expression of acetylated H3 (Ac-H3) and acetylated H4 (Ac-H4). Ponceau S staining was used to confirm the proper loading of histones. Ctrl represents the control lane.
Low Concentration of SAHA or TSA Augments TRAIL-Induced Apoptosis and Cytotoxicity in Multiple Myeloma
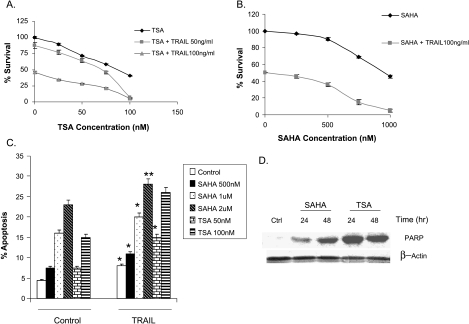
HDAC inhibitors are known apoptosis inducers [16,17]. Cytotoxicity and apoptosis induced by pretreatment of ARP-1 multiple myeloma cells with SAHA or TSA followed by TRAIL were studied using XTT cell proliferation assay and Annexin V PI staining, respectively. Pretreatment of ARP-1 cells with different concentrations of TSA for 24 hours followed by TRAIL (50 ng/ml or 100 ng/ml) treatment for another 24 hours significantly augmented the cytotoxic effect of TRAIL (Figure 2A). The maximum cytotoxic effect was achieved by combining 100 nM TSA and TRAIL (50 or 100 ng/ml). Also, the pretreatment of ARP-1 cells with different doses of SAHA (0–1 µM) for 24 hours followed by TRAIL (100 ng/ml) augmented TRAIL cytotoxicity, and the combination of 1 µM SAHA and 100 ng/ml TRAIL attained the maximum cytotoxic effect (Figure 2B).
Figure 2.
SAHA and TSA augment TRAIL cytotoxic and apoptotic effects. (A) ARP-1 cells were pretreated with different TSA concentrations for 24 hours followed by treatment with different TRAIL concentrations (50 and 100 ng/ml) for another 24 hours. XTT cell proliferation assay was used to estimate cell survival. Percentage of survival was estimated as a percentage of the value of the untreated control. Data represent the mean for four replicates ± SD. (B) ARP-1 cells were treated with different concentrations of SAHA for 24 hours followed by treatment with 100 ng/ml TRAIL for another 24 hours and the percentage of survival was determined by XTT assay as described above. Data represent the mean for four replicates ± SD. (C) ARP-1 cells were treated with different concentrations of SAHA and TSA, alone or in combination with 50 ng/ml TRAIL, for 24 hours and the percentage of apoptotic cells was determined by Annexin V staining. Data represent the mean for four replicates ± SD. *Significant difference from the respective control at P < .05. (D) ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 24 and 48 hours and the ∼85 kDa cleavage product of PARP was detected by Western blot analysis. β-Actin was used as a loading control. Ctrl represents the control lane.
Annexin PI staining showed that SAHA or TSA cotreatment with TRAIL for 24 hours augmented the apoptotic effect of TRAIL in ARP-1 cells (Figure 2C), indicating that pretreatment or cotreatment of multiple myeloma cells with SAHA or TSA and TRAIL augments both the cytotoxic and apoptotic activities of TRAIL. SAHA- or TSA-induced apoptosis was confirmed by PARP cleavage (Figure 2D). The IC50 values for TRAIL, SAHA, and TSA in different multiple myeloma cell lines showed that the MM1S cell line was relatively resistant to the cytotoxic effect of TRAIL in comparison to other cell lines. However, the cytotoxic effect of SAHA and TSA was comparable in all cell lines (Table 1).
Table 1.
IC50 Determination for TRAIL, SAHA, and TSA in Different Multiple Myeloma Cell Lines.
| Cell Line | IC50 | ||
| SAHA (nM) | TSA (nM) | TRAIL (ng/ml) | |
| ARP-1 | 950 | 80 | 83 |
| MM1S | 1125 | 78 | 285 |
| 8226 | 1200 | 70 | 80 |
Cells were incubated with different concentrations of TRAIL, SAHA, or TSA for 48 hours followed by determination of the percentage of survival using XTT assay. The experiment was repeated four times for each concentration. The IC50 was estimated from the percentage of survival curve.
Upregulation of the Surface Expression of DR4 and DR5 and Induction of G1 Phase Cell Cycle Growth Arrest by HDAC Inhibitors
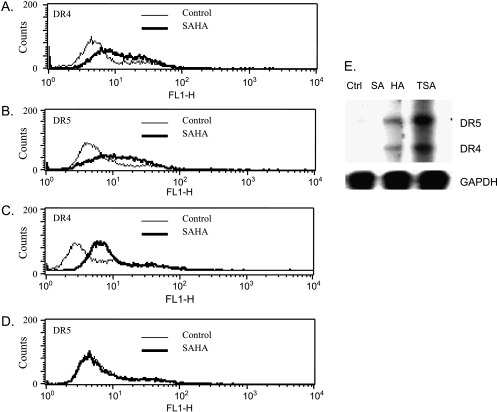
The membrane expression of the TRAIL receptors, DR4/DR5, plays an important role in TRAIL-induced apoptosis [10]. We speculated that SAHA and TSA could upregulate the surface expression of both receptors. Treatment of ARP-1 cells with SAHA (500 nM) or TSA (50 nM) for 24 or 48 hours resulted in differential upregulation of the membrane expression of the DR4/DR5 receptors. SAHA treatment for 24 hours significantly upregulated the expression of both DR4 and DR5 receptors (data not shown), with a maximum upregulation of DR4 and DR5 after 48 hours of treatment (Figure 3, A and B, respectively). TSA treatment for 24 hours exhibited a differential modulation by upregulating the expression of DR4 receptor but not DR5 (Figure 3, C and D, respectively); further treatment for up to 48 hours did not show any further change in the expression of DR4 or DR5 (data not shown). Upregulation of DR4/DR5 receptors could be explained by either an increase in the stability of the DR4/DR5 proteins and/or an increase in the transcription of their genes. The latter possibility was confirmed by the increase in the mRNA transcripts of both DR4 and DR5 after SAHA or TSA treatment for 12 hours (Figure 3E). The induction of DR5 mRNA transcription by TSA without showing an increase in the surface expression could be attributed to the trafficking of the receptor from the cytosol to the membrane, or to the absence of a direct correlation between the level of DR5 mRNA transcript and its translated protein.
Figure 3.
SAHA and TSA upregulate the surface expression and transcription of DR4/DR5 in ARP-1 cells. ARP-1 cells were treated with SAHA (500 nM) for 48 hours and the surface expression of DR4 (A) and DR5 (B) was measured as described under the Materials and Methods section. ARP-1 cells were treated with TSA (50 nM) for 48 hours, and the surface expression of DR4 (C) and DR5 (D) was measured. (E) ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 12 hours; total RNA was extracted to perform RPA (hAPO-3d template set) as described under the Materials and Methods section. GAPDH was used as a loading control. Ctrl represents the control lane.
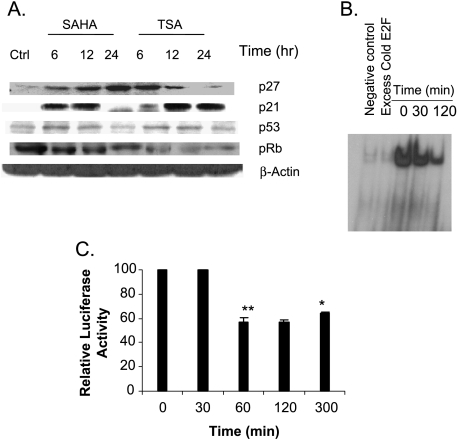
HDAC inhibitors are known to induce growth arrest, differentiation, and/or apoptosis [18]. To gain insight into their mechanism of growth arrest, cell cycle analysis was performed to determine the phase at which SAHA or TSA induces growth arrest. Table 2 shows a significant increase in the G1 phase population of both ARP-1 and MM1S cells after 24 hours of SAHA (500 nM) or TSA (50 nM) treatment, indicating a G1 phase growth arrest. To understand the mechanism by which SAHA and TSA induce G1 phase growth arrest, we investigated their effect on the expression of the negative modulators of the G1 phase of the cell cycle, p21WAF1 and p27Kip1. The expression levels of p21WAF1, p27Kip1, and p53 were upregulated after 6 to 12 hours of treatment with SAHA or TSA (Figure 4A). To further investigate the role of p21WAF1 in the observed G1 growth arrest, the expression of the retinoblastoma tumor suppressor protein (pRb) was monitored after treatment with SAHA or TSA. pRb, in its hypophosphorylated form, can directly bind and inactivate the promoter-bound transcription factor E2F, which is required in the progression of the cell cycle from the G1 to S phase by activating several genes involved in DNA replication such as DNA polymerase α, proliferating cell nuclear antigen (PCNA), Orc1, Cdc6, and the Mcm proteins [19,20]. Both drugs significantly decreased the expression of the hyperphosphorylated form of pRb (Figure 4A), which is consistent with the early upregulation of the cdk inhibitor p21WAF1 and the observed G1 phase growth arrest. Moreover, TSA was able to decrease the binding and transcriptional activity of E2F1 after 30 minutes of treatment (Figure 4, B and C, respectively), which is consistent with the observed decrease of the hyperphosphorylated form of pRb.
Table 2.
SAHA and TSA Induce G1 Phase Cell Cycle Growth Arrest in ARP-1 and MM1S Cells.
| Control | SAHA | TSA | |
| ARP1 | |||
| G1 | 31.8 ± 0.4 | 54 ± 0.7* | 42.6 ± 0.3 |
| S | 52.6 ± 0.2 | 39 ± 0.8 | 46.4 ± 0.6 |
| G2/M | 15.6 ± 0.3 | 7 ± 0.2 | 11 ± 0.4 |
| MM1S | |||
| G1 | 39.1 ± 0.4 | 52.9 ± 0.2* | 75.6 ± 0.7* |
| S | 41.3 ± 0.1 | 33.4 ± 0.2 | 24 ± 0.3 |
| G2/M | 19.6 ± 0.8 | 13.7 ± 0.5 | 0.4 ± 0.01 |
Synchronized ARP-1 and MM1S cells treated for 24 hours with SAHA (500 nM) or TSA (50 nM) were stained with PI and the percentage of cells in each phase of the cell cycle was determined by flow cytometry. Data represent the mean ± SD for three experiments.
Significant difference from the respective control at P < .05.
Figure 4.
HDAC inhibitors induce G1 phase growth arrest and decrease the binding and transcriptional activity of E2F-1. (A) Synchronized ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 6, 12, and 24 hours. The expressions of p21WAF1, p27Kip1, p53, and phosphorylated Rb were determined at each time point by immunoblotting. β-Actin was used as a loading control. Ctrl represents the control lane. (B) Nuclear proteins were extracted from ARP-1 cells after treatment with TSA (50 nM) for 30 and 120 minutes, and EMSA was performed as described under the Materials and Methods section. Cold E2F lane represents excess unlabelled E2F1 consensus sequence. (C) ARP-1 cells were transfected with E2F1 luciferase reporter plasmid and CMV 4 promoter-drivenβ -gal expression plasmid to normalize the transfection efficiency. Cells were treated with TSA (50 nM) for different time intervals (30, 60, 120, and 300 min) and the relative luciferase activity was determined according to the manufacturer's instructions. Data represent the mean for three replicates ± SD. *Significant difference from the control at P < .05.
Involvement of the Mitochondria in HDAC Inhibitor-Induced Apoptosis
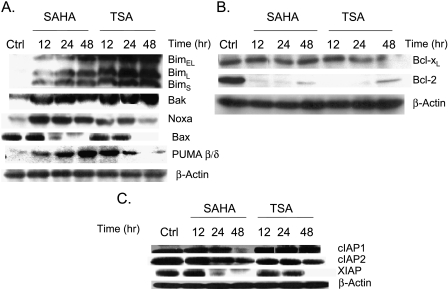
The pro-apoptotic members of the Bcl-2 protein family play a major role in the disruption of the mitochondrial membrane potential ΔΨm and the release of different mitochondrial pro-apoptotic proteins [21]. ARP-1 cell treatment with SAHA (500 nM) or TSA (50 nM) for different time intervals differentially upregulated the expression of the pro-apoptotic-members of the Bcl-2 family: the three isoforms of Bim, Bak, Noxa, PUMA β/δ, and Bax (Figure 5A). On the contrary, the level of expression of the anti-apoptotic Bcl-2 protein was almost abolished by SAHA or TSA treatment for different time intervals (Figure 5B). Bcl-XL showed only a slight decrease in expression after 24-hour treatment with SAHA and a significant decrease after 24 and 48 hours of TSA treatment.
Figure 5.
Differential modulation of the expression of the pro-apoptotic and anti-apoptotic members of the Bcl-2 family and IAPs by SAHA and TSA. (A) ARP1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 12, 24, and 48 hours and the protein expression of Bim, Bak, Noxa, Bax, and PUMA β/δ was determined by immunoblotting at each time point. (B) ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 12, 24, and 48 hours and the protein expression of Bcl-XL and Bcl-2 was determined by immunoblotting. β-Actin was used as a loading control. Ctrl represents the control lane. (C) ARP-1 cells were treated with SAHA 500 nM or TSA 50 nM for 12, 24, and 48 hours, and the protein expression of cIAP1, cIAP2, and XIAP was determined by immunoblotting. β-Actin was used as a loading control. Ctrl represents the control lane.
IAPs are well-known inhibitors of apoptosis by binding to caspases and inhibiting their activity [22]; consequently, the degradation of IAPs will favor any apoptotic stimuli. Smac/Diablo and Omi/HtrA2 are able to inactivate IAPs by disrupting the IAP-caspase complexes and/or inducing auto-ubiquitination and degradation of IAPs [23]. ARP-1 cell treatment with low concentrations of SAHA (500 nM) or TSA (50 nM) for different time intervals (12, 24, and 48 hours) resulted in different effects on the expression level of three different IAP proteins. SAHA treatment downregulated the expression of cIAP1 after 48 hours only, whereas cIAP2 and XIAP expressions were downregulated by both SAHA and TSA in a time-dependent manner (Figure 5C).
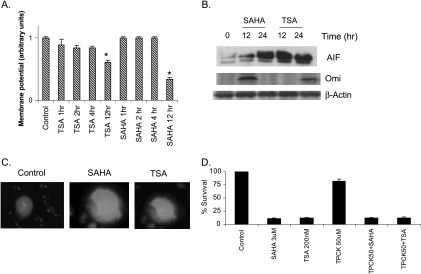
The disruption of the ΔΨm is accompanied by the release of crucial mitochondrial proteins, such as Omi/HtrA2, Smac/Diablo, and AIF, in the cytoplasm to induce apoptosis [24]. ARP-1 cell treatment with SAHA or TSA significantly lowered ΔΨm after 12 hours of treatment (Figure 6A).
Figure 6.
Involvement of the mitochondria in SAHA- and TSA-induced apoptosis. (A) ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 1, 2, 4, and 12 hours followed by staining with JC-1 dye and measurement of the mitochondrial membrane potential (ΔΨm) as described under the Materials and Methods section. The untreated cells (control) were assigned an arbitrary value and the value of the other treatments was estimated relative to the control. Data represent the mean for four replicates ± SD. *Significant difference from the control at P < .05. (B) ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 12 and 24 hours, the cytosolic fraction was isolated, and the expression of AIF and Omi/HtrA2 was determined at each time point by immunoblotting. β-Actin was used as a loading control. (C) ARP-1 cells were treated with SAHA (500 nM) or TSA (50 nM) for 12 hours followed by double immunofluorescence staining of the nucleus (blue color) and AIF (green) as described under the Materials and Methods section. (D) ARP-1 cells were either pretreated with TPCK (50 µM) for 2 hours or left untreated, followed by treatment with SAHA (3 µM) or TSA (200 nM) for 24 hours. The percentage of survival was determined by XTT assay as described previously.
The release of the mitochondrial proteins such as cytochrome c, Smac/Diablo, AIF, and Omi/HtrA2 in the cytosol stimulates a cascade of events ending in apoptosis. Omi/HtrA2 is known to play a dual role in the induction of apoptosis by binding to IAPs and through its serine protease activity [25]. AIF can shuttle directly to the nucleus and induce peripheral chromatin condensation and large-scale fragmentation of DNA; none of these effects is prevented by the pan-caspase inhibitor, ZVAD-FMK [26,27]. SAHA or TSA treatment significantly upregulated the expression of both AIF and Omi/HtrA2 in the cytosolic fraction of ARP-1 cells (Figure 6B). The enhanced release of Omi/HtrA2 was consistent with the observed decrease in the expression levels of the IAPs. The shuttling of AIF to the nucleus was confirmed by immunofluorescence staining after 12 hours of treatment with SAHA or TSA (Figure 6C). To determine the role of serine proteases in SAHA- and TSA-mediated cytotoxicity, the chymotrypsin-like serine protease inhibitor, TPCK, was used for the pretreatment of the ARP-1 cells before SAHA or TSA treatment. TPCK was unable to minimize or inhibit the cytotoxic effect of SAHA or TSA (Figure 6D), indicating that their cytotoxic effect is Omi/HtrA2-independent.
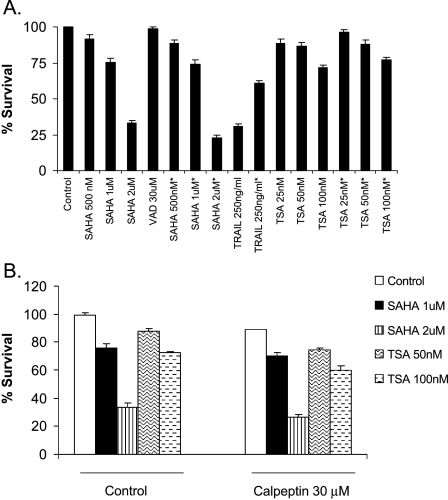
SAHA-Induced and TSA-Induced Cytotoxicities are Caspase-Independent and Calpain-Independent
Despite the central role of caspases in the execution of cell death, other proteins such as AIF and Omi/HtrA2 are able to induce cell death in a caspase-independent manner [28]. To investigate the role of different caspases in SAHA- or TSA-mediated cytotoxic effect, ARP-1 cells were pretreated with the pan-caspase inhibitor, Z-VAD-FMK, followed by SAHA or TSA treatment. There was no significant change in the percentage of survival in the Z-VAD-FMK-treated or -untreated cells, whereas TRAIL, which is known to be caspase-dependent, showed a significant decrease in the percentage of survival as predicted (Figure 7A). The proteolytic enzyme, calpain, is also known to be activated by the loss of mitochondrial potential [29] and can be inhibited by calpeptin. Calpeptin pretreatment of ARP-1 cells was unable to reduce the cytotoxicity of SAHA or TSA (Figure 7B), and even a higher dose of calpeptin (50 µM), which was a bit toxic to the cells by itself, did not inhibit SAHA or TSA cytotoxic effects (data not shown).
Figure 7.
SAHA and TSA cytotoxic effects are caspase- and calpain-independent. (A) ARP-1 cells were either pretreated with 30 µM Z-VAD-FMK (VAD) for 2 hours or left untreated, then different doses of SAHA, TSA, or TRAIL (positive control) were added to both the treated and untreated cells for 36 hours. XTT cell proliferation assay was used to estimate cell survival and percentage of survival was estimated as a percentage of the value of the untreated control. Pretreatment with Z-VAD-FMK. Data represent the mean for four replicates ± SD. There was no significant difference at P < .05 between the Z-VAD-FMK-pretreated and -untreated cells after TSA or SAHA treatment. (B) ARP-1 cells were either pretreated with 30 µM calpeptin for 2 hours or left untreated, then different doses of SAHA or TSA were added to both the treated and untreated cells for 36 hours. XTT cell proliferation assay was used to assess cell survival. Data represent the mean for four replicates ± SD. No significant difference was observed between the control and calpeptin-treated cells at P < .05.
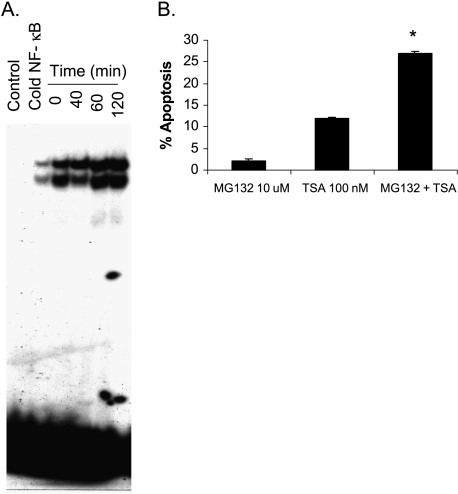
TSA Enhances the DNA Binding Activity of NF-κB in Myeloma Cells
Enhancing the DNA binding and transcriptional activation of the anti-apoptotic transcription factor, NF-κB, was associated with the ineffectiveness of HDAC inhibitors to induce apoptosis in lung cancer [30]. TSA was not able to induce much apoptosis in myeloma cells as shown in Figure 2C, which could be attributed to NF-κB activation. Figure 8A shows a significant enhancement of NF-κB binding after 60 and 120 minutes of TSA (50 nM) treatment and the specificity of binding was confirmed by the reduction in binding after using excess unlabeled NF-κB. The inhibition of NF-κB activity by pretreatment of ARP-1 cells with 10 µM MG132 (Calbiochem) for 1 hour significantly enhanced TSA-induced apoptosis (Figure 8B).
Figure 8.
TSA increases the binding activity of NF-κB in ARP-1 myeloma cells. (A) ARP-1 cells were treated with 50 nM TSA for 0, 40, 60, and 120 minutes followed by extraction of the nuclear proteins and EMSA. The control lane represents the negative control (no nuclear proteins) and the cold NF-κB lane contains excess unlabeled NF-κB (x 100) to compete with the labeled NF-κB for binding to nuclear proteins. (B) ARP-1 cells were treated with MG132 (10 µM) or TSA (100 nM) alone for 24 hours or pretreated with MG132 (10 µM) for 2 hours followed by TSA treatment (100 nM) for 24 hours. DAPI staining was used to assess apoptosis and the percentage of apoptosis was estimated as a percentage of the value of the untreated control. Data represent the mean for four replicates ± SD. *Significant difference from the respective controls at P < .05.
Discussion
We have evaluated the effect of two HDAC inhibitors derived from hydroxamic acid in human multiple myeloma cell lines. SAHA and TSA induced growth arrest and apoptosis, and augmented the apoptotic and cytotoxic effects of TRAIL. SAHA and TSA induced the transcription and surface expression of TRAIL DR4/DR5 death receptors and the expression of the pro-apoptotic members of the Bcl-2 family. However, the level of expression of the IAPs and the anti-apoptotic members of the Bcl-2 family were downregulated. Despite the central role of caspases in apoptosis induction, SAHA and TSA cytotoxic effects were shown to be caspase-independent and were mediated through the release of the mitochondrial protein, AIF.
HDAC inhibitors are a new class of chemotherapeutic drugs that are able to induce tumor cell apoptosis and/or cell cycle arrest; however, the molecular mechanisms supporting their antitumor effects are not clearly understood. SAHA was developed by modifying the HDAC-inhibitory compound, TSA, with a hydroxamic acid moiety as the key structural element. Both compounds have shown efficacy in the treatment of cancer in vivo and in vitro [9] and, because of SAHA low toxicity, they are currently evaluated in clinical trials [31]. Previous reports have shown that the effect of HDAC inhibitors on gene expression was not global and only modulated the expression of few genes [32,33]. Our working hypothesis is that SAHA or TSA could upregulate the expression of TRAIL death receptors DR4/DR5 and pro-apoptotic Bcl-2 family proteins, and downregulate the expression of other anti-apoptotic proteins (Bcl-2 and IAPs); consequently, the pretreatment or cotreatment with SAHA or TSA could augment the cytotoxic and apoptotic effect of TRAIL. Similarly, several recent studies have shown that HDAC inhibitors can enhance the apoptosis-inducing potential of TRAIL in leukemia [34–39], breast cancer [40], melanoma [41,42], adenocarcinoma [43], and malignant mesothelioma [44].
A previous study has shown that the simultaneous administration of high concentrations of SAHA (2 µM) and TRAIL significantly increased the apoptotic activity and mitochondrial damage of TRAIL in human leukemia cells [45] without any significant change in the expression of TRAIL receptors. Other HDAC inhibitors such as LAQ824 upregulated the transcription and protein expression of DR4/DR5 in human leukemia cells [35]. Also SAHA and TSA were shown to upregulate DR5 expression in various human malignant tumor cells [38]. In this study, we have also shown that pretreatment of multiple myeloma cells with SAHA or TSA significantly increased the apoptotic and cytotoxic effects of TRAIL. However, this effect was accompanied by a significant increase in the membrane receptor expression of DR4/DR5 and their mRNA transcript, indicating that the effect of SAHA on DR4/DR5 expression is specific for each cell type and that different HDAC inhibitors do not exhibit the same effect on DR4/DR5 expression, although they act by the same mechanism of action.
To further characterize the mechanisms of the TRAIL-augmenting effect of SAHA or TSA, the effect of treatment with low concentrations of SAHA or TSA on the expression of the anti-apoptotic IAPs was investigated. IAPs are well-known inhibitors of caspases; their downregulation significantly increased apoptosis in glioma cells [46], neuroblastoma [47], and human leukemia [48], and their upregulation caused tumor cell resistance to drug-induced apoptosis [49]. In this study, we have shown that cIAP-2 and XIAP expression was downregulated by SAHA and TSA in a time-dependent manner, whereas cIAP-1 level of expression was downregulated only after 48 hours of treatment with SAHA. The effect on the expression of XIAP and cIAP2 is in agreement with the effect of SAHA on the MM1S multiple myeloma cells [50] despite the fact that we used a 10x less concentration of SAHA. Valproic acid and SAHA were also shown to modulate survivin expression in human melanoma cells [41]. Our data are consistent with the observed increase in TRAIL-induced apoptosis when pretreated with SAHA or TSA and also with the increase in Omi/HtrA2 release from the mitochondria because IAPs are known substrates for this serine protease [23].
Previous reports have shown that the cytotoxic effect of SAHA is caspase-dependent in breast cancer cells [32] and leukemia cells [51]. However, SAHA cytotoxicity was shown to be caspase-independent and calpain-dependent in MM1S human multiple myeloma cells [50]. In this study, we observed that the pretreatment with the pan-caspase inhibitor, Z-VAD-FMK, or with the calpain inhibitor, calpeptin, did not reduce the cytotoxicity of SAHA or TSA, indicating that SAHA or TSA cytotoxicity is caspase- and calpain-independent in ARP-1 multiple myeloma cells. The discrepancy in the role of calpain dependency in SAHA-mediated cytotoxicity could be explained by the fact that we used a different cell line and the effect could be cell type-specific.
Preclinical studies provided evidence that proteasome inhibitors may be a potential treatment approach for hematopoietic malignancies and it was suggested that downregulation of NFκB pathway is the mechanism of action of proteasome inhibitors [30]. In this study, the proteasome inhibitor, MG-132, significantly increased the apoptotic potential of TSA in myeloma cells. We have previously shown that NFκB regulates the expression of both pro-apoptotic (e.g., DR4, DR5, and caspase-8) and anti-apoptotic proteins (e.g., IAPs, FLIP, and Bcl-XL) [52]. These factors may determine the sensitivity of tumor cells to TRAIL- and/or HDAC inhibitor-induced apoptosis. The observed increase in NFκB DNA binding activity after TSA treatment is consistent with the observed effect of SAHA on NFκB in human leukemia cells [53].
Apoptosis can be induced by both mitochondrial-dependent (intrinsic) and mitochondrial-independent (extrinsic) pathways [6]. Mitochondrial membrane depolarization and the release of different mitochondrial proteins such as cytochrome c [54], Smac/DIABLO [14], Omi/HtrA2 [55], endonuclease G [28], and AIF [56] appear to be the early event in the intrinsic pathway. To investigate the mitochondrial role in the HDAC inhibitors' TRAIL-augmenting effect, we observed a significant drop in the mitochondrial membrane potential after 12 hours of SAHA or TSA treatment, which is in agreement with other previous reports [57,58]. Based on the abovementioned results showing SAHA and TSA cytotoxic effects as caspase-independent, we hypothesized that other mitochondrial proteins such as AIF and/or Omi/HtrA2 could be released from the mitochondria and executed the process of cell death [22,56,59]. The increase in the cytosolic release of AIF and Omi/HtrA2 after SAHA or TSA treatment supports our hypothesis; however, the inability of TPCK to inhibit the cytotoxicity of SAHA or TSA and the shuttling of AIF to the nucleus as demonstrated by immunofluorescence supports a major role of AIF and not Omi/HtrA2 in the cytotoxic effect induced by SAHA or TSA. The enhanced cytosolic release of AIF and Omi/HtrA2 is in agreement with the effect of different HDAC inhibitors, such as MS-275 [60] and LAQ824 [35], in human leukemia cells.
The Bcl-2 family of proteins comprises both pro-apoptotic and anti-apoptotic members; the anti-apoptotic members such as Bcl-2 and Bcl-XL inhibit the release of the mitochondrial apoptogenic factors, whereas the pro-apoptotic members trigger the release of such factors [24]. The proapoptotic members include the BH3-only proteins (Noxa, PUMA, Bid, Bad, and Bim) and other multiple domain proteins such as Bak and Bax. In this study, we have shown that the treatment of multiple myeloma cells with low doses of SAHA or TSA for different time intervals selectively upregulated the expression of the pro-apoptotic members of the Bcl-2 family and downregulated the expression of the anti-apoptotic members. Other HDAC inhibitors such as FKK228 showed an increase in the level of the BH3-only protein Bim in human gastric MKN45 and colorectal DLD1 adenocarcinoma cells [61]. SAHA and TSA also modulated the expression of Bcl-2 and Bcl-XL in human melanoma cells [41]. Our data are also in agreement with the downregulation of the expression of the anti-apoptotic members (Bcl-2 and Bcl-XL) in pancreatic adenocarcinoma cells [62] by TSA and in multiple myeloma cell lines [63] using the HDAC inhibitor, depsipeptide (FR901228).
Induction of growth arrest is a main trait of HDAC inhibitors; however, the growth arrest phase differs among the different HDAC inhibitors. Our data support a G1 growth phase arrest in both ARP-1 and MM1S cells, mediated through the upregulation of the G1 phase-negative modulators p21WAF1 and p27Kip1 and transcriptional inhibition of E2F. The observed upregulation of p21WAF1 in other multiple myeloma cell lines by the HDAC inhibitor, LAQ824 [2], and SAHA [50], albeit at a higher concentration, is in agreement with our data. However, the observed p27Kip1 upregulation is not in agreement with the unaltered level of expression observed before by another group [50], which again could be attributed to the different cell line used. The upregulation of both p21WAF1 and p27Kip1 in breast cancer cell lines by SAHA is also in agreement with our data [32].
Conclusion
We have evaluated the cytotoxic and apoptotic effects of two HDAC inhibitors derived from hydroxamic acid in human multiple myeloma cells. SAHA and TSA enhanced TRAIL-induced apoptosis through: 1) upregulation of DR4 and/or DR5 death receptors; 2) upregulation of the pro-apoptotic proteins of the Bcl-2 family; 3) downregulation of the anti-apoptotic members of the Bcl-2 family; 4) downregulation of the IAPs; and 5) enhancement of AIF release from the mitochondria. HDAC inhibitors induced G1 phase growth arrest by: 1) upregulation of the expression of p21WAF1 and p27Kip1; and 2) pRb hypophosphorylation and E2F transcriptional inhibition. This study should arouse the attention to the role of AIF in HDAC inhibitor-induced cytotoxicity during the course of clinical evaluation of these drugs and demonstrated that combining HDAC inhibitors and TRAIL could be a useful strategy in the treatment of multiple myeloma.
Abbreviations
- DR
death receptor
- DcR
decoy receptor
- HDAC
histone deacetylase
- IAP
inhibitor of apoptosis protein
- PARP
poly(ADP-ribose) polymerase
- SAHA
suberoylanilide hydroxamic acid
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- TSA
trichostatin A
- z-VAD-fmk
z-Val-Ala-Asp-fluoromethylketone
- AIF
apoptosis-inducing factor
References
- 1.Gupta D, Hideshima T, Anderson KC. Novel biologically based therapeutic strategies in myeloma. Rev Clin Exp Hematol. 2002;6:301–324. doi: 10.1046/j.1468-0734.2002.00082.x. [DOI] [PubMed] [Google Scholar]
- 2.Catley L, Weisberg E, Tai YT, Atadja P, Remiszewski S, Hideshima T, Mitsiades N, Shringarpure R, LeBlanc R, Chauhan D, et al. NVP-LAQ824 is a potent novel histone deacetylase inhibitor with significant activity against multiple myeloma. Blood. 2003;102:2615–2622. doi: 10.1182/blood-2003-01-0233. [DOI] [PubMed] [Google Scholar]
- 3.Mitsiades N, Mitsiades CS, Poulaki V, Anderson KC, Treon SP. Intracellular regulation of tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human multiple myeloma cells. Blood. 2002;99:2162–2171. doi: 10.1182/blood.v99.6.2162. [DOI] [PubMed] [Google Scholar]
- 4.Suliman A, Lam A, Datta R, Srivastava RK. Intracellular mechanisms of TRAIL: apoptosis through mitochondrial-dependent and -independent pathways. Oncogene. 2001;20:2122–2133. doi: 10.1038/sj.onc.1204282. [DOI] [PubMed] [Google Scholar]
- 5.Kandasamy K, Srivastava RK. Role of the phosphatidylinositol 3′-kinase/PTEN/Akt kinase pathway in tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in non-small cell lung cancer cells. Cancer Res. 2002;62:4929–4937. [PubMed] [Google Scholar]
- 6.Srivastava RK. Intracellular mechanisms of TRAIL and its role in cancer therapy. Mol Cell Biol Res Commun. 2000;4:67–75. doi: 10.1006/mcbr.2001.0265. [DOI] [PubMed] [Google Scholar]
- 7.Pollack IF, Erff M, Ashkenazi A. Direct stimulation of apoptotic signaling by soluble apo2l/tumor necrosis factor-related apoptosis-inducing ligand leads to selective killing of glioma cells. Clin Cancer Res. 2001;7:1362–1369. [PubMed] [Google Scholar]
- 8.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 9.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 10.Singh TR, Shankar S, Chen X, Asim M, Srivastava RK. Synergistic interactions of chemotherapeutic drugs and tumor necrosis factor-related apoptosis-inducing ligand/Apo-2 ligand on apoptosis and on regression of breast carcinoma in vivo. Cancer Res. 2003;63:5390–5400. [PubMed] [Google Scholar]
- 11.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 12.Roehm NW, Rodgers GH, Hatfield SM, Glasebrook AL. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J Immunol Methods. 1991;142:257–265. doi: 10.1016/0022-1759(91)90114-u. [DOI] [PubMed] [Google Scholar]
- 13.McCarthy RC, Fetterhoff TJ. Issues for quality assurance in clinical flow cytometry. Arch Pathol Lab Med. 1989;113:658–666. [PubMed] [Google Scholar]
- 14.Kandasamy K, Srinivasula SM, Alnemri ES, Thompson CB, Korsmeyer SJ, Bryant JL, Srivastava RK. Involvement of proapoptotic molecules Bax and Bak in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced mitochondrial disruption and apoptosis: differential regulation of cytochrome c and Smac/DIABLO release. Cancer Res. 2003;63:1712–1721. [PubMed] [Google Scholar]
- 15.Reers M, Smith TW, Chen LB. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry. 1991;30:4480–4486. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- 16.Finzer P, Kuntzen C, Soto U, zur Hausen H, Rosl F. Inhibitors of histone deacetylase arrest cell cycle and induce apoptosis in cervical carcinoma cells circumventing human papillomavirus oncogene expression. Oncogene. 2001;20:4768–4776. doi: 10.1038/sj.onc.1204652. [DOI] [PubMed] [Google Scholar]
- 17.Lavelle D, Chen YH, Hankewych M, DeSimone J. Histone deacetylase inhibitors increase p21(WAF1) and induce apoptosis of human myeloma cell lines independent of decreased IL-6 receptor expression. Am J Hematol. 2001;68:170–178. doi: 10.1002/ajh.1174. [DOI] [PubMed] [Google Scholar]
- 18.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 19.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava RK. TRAIL/Apo-2L: mechanisms and clinical applications in cancer. Neoplasia. 2001;3:535–546. doi: 10.1038/sj.neo.7900203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L, DuBois G, Lazebnik Y, Zervos AS, Fernandes-Alnemri T, et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J Biol Chem. 2002;277:432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasula SM, Gupta S, Datta P, Zhang Z, Hegde R, Cheong N, Fernandes-Alnemri T, Alnemri ES. Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J Biol Chem. 2003;278:31469–31472. doi: 10.1074/jbc.C300240200. [DOI] [PubMed] [Google Scholar]
- 24.Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 25.van Loo G, van Gurp M, Depuydt B, Srinivasula SM, Rodriguez I, Alnemri ES, Gevaert K, Vandekerckhove J, Declercq W, Vandenabeele P. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ. 2002;9:20–26. doi: 10.1038/sj.cdd.4400970. [DOI] [PubMed] [Google Scholar]
- 26.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 27.Cregan SP, Fortin A, MacLaurin JG, Callaghan SM, Cecconi F, Yu SW, Dawson M, Dawson VL, Park DS, Kroemer G, et al. Apoptosis-inducing factor is involved in the regulation of caspase-independent neuronal cell death. J Cell Biol. 2002;158:507–517. doi: 10.1083/jcb.200202130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saelens X, Festjens N, Walle LV, van Gurp M, van Loo G, Vandenabeele P. Toxic proteins released from mitochondria in cell death. Oncogene. 2004;23:2861–2874. doi: 10.1038/sj.onc.1207523. [DOI] [PubMed] [Google Scholar]
- 29.Pink JJ, Wuerzberger-Davis S, Tagliarino C, Planchon SM, Yang X, Froelich CJ, Boothman A. Activation of a cysteine protease in MCF-7 and T47D breast cancer cells during beta-lapachone-mediated apoptosis. Exp Cell Res. 2000;255:144–155. doi: 10.1006/excr.1999.4790. [DOI] [PubMed] [Google Scholar]
- 30.Mayo MW, Denlinger CE, Broad RM, Yeung F, Reilly ET, Shi Y, Jones DR. Ineffectiveness of histone deacetylase inhibitors to induce apoptosis involves the transcriptional activation of NF-kappa B through the Akt pathway. J Biol Chem. 2003;278:18980–18989. doi: 10.1074/jbc.M211695200. [DOI] [PubMed] [Google Scholar]
- 31.Reddy P, Maeda Y, Hotary K, Liu C, Reznikov LL, Dinarello CA, Ferrara JL. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc Natl Acad Sci USA. 2004;101:3921–3926. doi: 10.1073/pnas.0400380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L, Pardee AB. Suberoylanilide hydroxamic acid as a potential therapeutic agent for human breast cancer treatment. Mol Med. 2000;6:849–866. [PMC free article] [PubMed] [Google Scholar]
- 33.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 34.Boehrer S, Brieger A, Schaaf S, Kukoc-Zivojnov N, Nowak D, Ruthardt M, Hoelzer D, Mitrou PS, Weidmann E, Chow KU. In the erythroleukemic cell line HEL prostate-apoptosis-response-gene-4 (par-4) fails to down-regulate Bcl-2 and to promote apoptosis. Leuk Lymphoma. 2004;45:1445–1451. doi: 10.1080/10428190410001663617. [DOI] [PubMed] [Google Scholar]
- 35.Guo F, Sigua C, Tao J, Bali P, George P, Li Y, Wittmann S, Moscinski L, Atadja P, Bhalla K. Cotreatment with histone deacetylase inhibitor LAQ824 enhances Apo-2L/tumor necrosis factor-related apoptosis inducing ligand-induced death-inducing signaling complex activity and apoptosis of human acute leukemia cells. Cancer Res. 2004;64:2580–2589. doi: 10.1158/0008-5472.can-03-2629. [DOI] [PubMed] [Google Scholar]
- 36.Insinga A, Monestiroli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, Altucci L, Nervi C, Minucci S, Pelicci PG. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;11:71–76. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- 37.Nebbioso A, Clarke N, Voltz E, Germain E, Ambrosino C, Bontempo P, Alvarez R, Schiavone EM, Ferrara F, Bresciani F, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 38.Nakata S, Yoshida T, Horinaka M, Shiraishi T, Wakada M, Sakai T. Histone deacetylase inhibitors upregulate death receptor 5/TRAIL-R2 and sensitize apoptosis induced by TRAIL/APO2-L in human malignant tumor cells. Oncogene. 2004;23:6261–6271. doi: 10.1038/sj.onc.1207830. [DOI] [PubMed] [Google Scholar]
- 39.Shankar S, Singh TR, Fandy TE, Luetrakul T, Ross DD, Srivastava RK. Interactive effects of histone deacetylase inhibitors and TRAIL on apoptosis in human leukemia cells: involvement of both death receptor and mitochondrial pathways. Int J Mol Med. 2005 (in press) [PubMed] [Google Scholar]
- 40.Chopin V, Slomianny C, Hondermarck H, Le Bourhis X. Synergistic induction of apoptosis in breast cancer cells by cotreatment with butyrate and TNF-alpha, TRAIL, or anti-Fas agonist antibody involves enhancement of death receptors' signaling and requires P21 (waf1) Exp Cell Res. 2004;298:560–573. doi: 10.1016/j.yexcr.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 41.Facchetti F, Previdi S, Ballarini M, Minucci S, Perego P, La Porta CA. Modulation of pro- and anti-apoptotic factors in human melanoma cells exposed to histone deacetylase inhibitors. Apoptosis. 2004;9:573–582. doi: 10.1023/B:APPT.0000038036.31271.50. [DOI] [PubMed] [Google Scholar]
- 42.Zhang XD, Gillespie SK, Borrow JM, Hersey P. The histone deacetylase inhibitor suberic bishydroxamate regulates the expression of multiple apoptotic mediators and induces mitochondria-dependent apoptosis of melanoma cells. Mol Cancer Ther. 2004;3:425–435. [PubMed] [Google Scholar]
- 43.Inoue H, Shiraki K, Ohmori S, Sakai T, Deguchi M, Yamanaka T, Okano H, Nakano T. Histone deacetylase inhibitors sensitize human colonic adenocarcinoma cell lines to TNF-related apoptosis inducing ligand-mediated apoptosis. Int J Mol Med. 2002;9:521–525. [PubMed] [Google Scholar]
- 44.Neuzil J, Swettenham E, Gellert N. Sensitization of mesothelioma to TRAIL apoptosis by inhibition of histone deacetylase: role of Bcl-xL down-regulation. Biochem Biophys Res Commun. 2004;314:186–191. doi: 10.1016/j.bbrc.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 45.Rosato RR, Almenara JA, Dai Y, Grant S. Simultaneous activation of the intrinsic and extrinsic pathways by histone deacetylase (HDAC) inhibitors and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) synergistically induces mitochondrial damage and apoptosis in human leukemia cells. Mol Cancer Ther. 2003;2:1273–1284. [PubMed] [Google Scholar]
- 46.Kim EH, Kim SU, Shin DY, Choi KS. Roscovitine sensitizes glioma cells to TRAIL-mediated apoptosis by downregulation of survivin and XIAP. Oncogene. 2004;23:446–456. doi: 10.1038/sj.onc.1207025. [DOI] [PubMed] [Google Scholar]
- 47.Kim S, Kang J, Qiao J, Thomas RP, Evers BM, Chung DH. Phosphatidylinositol 3-kinase inhibition down-regulates survivin and facilitates TRAIL-mediated apoptosis in neuroblastomas. J Pediatr Surg. 2004;39:516–521. doi: 10.1016/j.jpedsurg.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Kang J, Kisenge RR, Toyoda H, Tanaka S, Bu J, Azuma E, Komada Y. Chemical sensitization and regulation of TRAIL-induced apoptosis in a panel of B-lymphocytic leukaemia cell lines. Br J Haematol. 2003;123:921–932. doi: 10.1046/j.1365-2141.2003.04699.x. [DOI] [PubMed] [Google Scholar]
- 49.Notarbartolo M, Cervello M, Poma P, Dusonchet L, Meli M, D'Alessandro N. Expression of the IAPs in multidrug resistant tumor cells. Oncol Rep. 2004;11:133–136. [PubMed] [Google Scholar]
- 50.Mitsiades N, Mitsiades CS, Richardson PG, McMullan C, Poulaki V, Fanourakis G, Schlossman R, Chauhan D, Munshi NC, Hideshima T, et al. Molecular sequelae of histone deacetylase inhibition in human malignant B cells. Blood. 2003;101:4055–4062. doi: 10.1182/blood-2002-11-3514. [DOI] [PubMed] [Google Scholar]
- 51.Amin HM, Saeed S, Alkan S. Histone deacetylase inhibitors induce caspase-dependent apoptosis and downregulation of daxx in acute promyelocytic leukaemia with t(15;17) Br J Haematol. 2001;115:287–297. doi: 10.1046/j.1365-2141.2001.03123.x. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Kandasamy K, Srivastava RK. Differential roles of RelA (p65) and c-Rel subunits of nuclear factor kappa B in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res. 2003;63:1059–1066. [PubMed] [Google Scholar]
- 53.Gao N, Dai Y, Rahmani M, Dent P, Grant S. Contribution of disruption of the nuclear factor-kappaB pathway to induction of apoptosis in human leukemia cells by histone deacetylase inhibitors and flavopiridol. Mol Pharmacol. 2004;66:956–963. doi: 10.1124/mol.104.002014. [DOI] [PubMed] [Google Scholar]
- 54.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 55.Yang QH, Church-Hajduk R, Ren J, Newton ML, Du C. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 2003;17:1487–1496. doi: 10.1101/gad.1097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene. 2004;23:2785–2796. doi: 10.1038/sj.onc.1207517. [DOI] [PubMed] [Google Scholar]
- 57.Rosato RR, Almenara JA, Cartee L, Betts V, Chellappan SP, Grant S. The cyclin-dependent kinase inhibitor flavopiridol disrupts sodium butyrate-induced p21WAF1/CIP1 expression and maturation while reciprocally potentiating apoptosis in human leukemia cells. Mol Cancer Ther. 2002;1:253–266. [PubMed] [Google Scholar]
- 58.Rahmani M, Dai Y, Grant S. The histone deacetylase inhibitor sodium butyrate interacts synergistically with phorbol myristate acetate (PMA) to induce mitochondrial damage and apoptosis in human myeloid leukemia cells through a tumor necrosis factor-alpha-mediated process. Exp Cell Res. 2002;277:31–47. doi: 10.1006/excr.2002.5548. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 60.Maggio SC, Rosato RR, Kramer LB, Dai Y, Rahmani M, Paik DS, Czarnik AC, Payne SG, Spiegel S, Grant S. The histone deacetylase inhibitor MS-275 interacts synergistically with fludarabine to induce apoptosis in human leukemia cells. Cancer Res. 2004;64:2590–2600. doi: 10.1158/0008-5472.can-03-2631. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Adachi M, Zhao X, Kawamura R, Imai K. Histone deacetylase inhibitors FK228, N-(2-aminophenyl)-4-[N-(pyridin-3-ylmethoxycarbonyl)amino-methyl]benzamide and m-carboxycinnamic acid bis-hydroxamide augment radiation-induced cell death in gastrointestinal adenocarcinoma cells. Int J Cancer. 2004;110:301–308. doi: 10.1002/ijc.20117. [DOI] [PubMed] [Google Scholar]
- 62.Moore PS, Barbi S, Donadelli M, Costanzo C, Bassi C, Palmieri M, Scarpa A. Gene expression profiling after treatment with the histone deacetylase inhibitor trichostatin A reveals altered expression of both pro- and anti-apoptotic genes in pancreatic adenocarcinoma cells. Biochim Biophys Acta. 2004;1693:167–176. doi: 10.1016/j.bbamcr.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 63.Khan SB, Maududi T, Barton K, Ayers J, Alkan S. Analysis of histone deacetylase inhibitor, depsipeptide ( FR901228), effect on multiple myeloma. Br J Haematol. 2004;125:156–161. doi: 10.1111/j.1365-2141.2004.04882.x. [DOI] [PubMed] [Google Scholar]