Abstract
Phosphoglyceride-linked enterobacterial common antigen (ECAPG) is a cell surface glycolipid that is synthesized by all gram-negative enteric bacteria. The carbohydrate portion of ECAPG consists of linear heteropolysaccharide chains comprised of the trisaccharide repeat unit Fuc4NAc-ManNAcA-GlcNAc, where Fuc4NAc is 4-acetamido-4,6-dideoxy-d-galactose, ManNAcA is N-acetyl-d-mannosaminuronic acid, and GlcNAc is N-acetyl-d-glucosamine. The potential reducing terminal GlcNAc residue of each polysaccharide chain is linked via phosphodiester linkage to a phosphoglyceride aglycone. We demonstrate here the occurrence of a water-soluble cyclic form of enterobacterial common antigen, ECACYC, purified from Escherichia coli strains B and K-12 with solution nuclear magnetic resonance (NMR) spectroscopy, electrospray ionization mass spectrometry (ESI-MS), and additional biochemical methods. The ECACYC molecules lacked an aglycone and contained four trisaccharide repeat units that were nonstoichiometrically substituted with up to four O-acetyl groups. ECACYC was not detected in mutant strains that possessed null mutations in the wecA, wecF, and wecG genes of the wec gene cluster. These observations corroborate the structural data obtained by NMR and ESI-MS analyses and show for the first time that the trisaccharide repeat units of ECACYC and ECAPG are assembled by a common biosynthetic pathway.
Lipopolysaccharide (LPS) is the major cell surface glycolipid of gram-negative bacteria. However, the cell surface of all gram-negative enteric bacteria contains an additional glycolipid, the phosphoglyceride-linked enterobacterial common antigen (ECAPG) (16, 21, 24, 32). The carbohydrate portion of ECA is a linear heteropolysaccharide comprised of the trisaccharide repeat unit →3)-α-d-Fuc4NAc-(1→4)-β-d-ManNAcA-(1→4)-α-d-GlcNAc-(1→, where Fuc4NAc, ManNAcA, and GlcNAc denote 4-acetamido-4,6-dideoxy-d-galactose, N-acetyl-d-mannosaminuronic acid, and N-acetyl-d-glucosamine, respectively (Fig. 1A) (19, 22, 32). Individual ECA polysaccharide chains are covalently linked to diacylglycerolphosphate via a glycosidic linkage between the potential reducing terminal GlcNAc residue and the phosphate residue of the aglycone (16, 17, 30); the phosphoglyceride aglycone anchors the ECA chains to the outer membrane (2, 33). Accordingly, phosphoglyceride-linked ECA chains are referred to as ECAPG. While the function of ECAPG remains to be established, recent studies suggest that it may be involved in the resistance of Shiga toxin-producing Escherichia coli O157:H7 to organic acids (6).
FIG. 1.
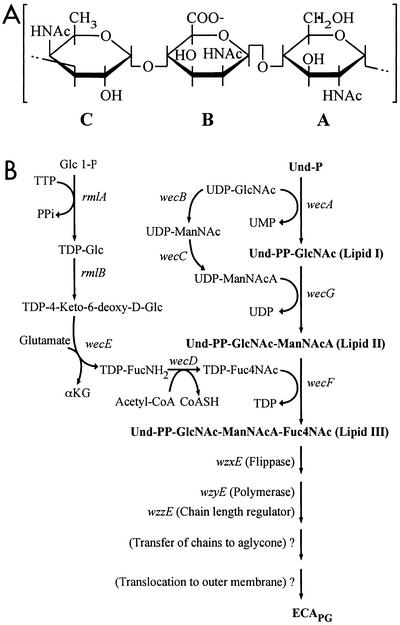
Biosynthetic pathway for the assembly of ECA. (A) Structure of trisaccharide repeat unit of ECA. Amino sugars A, B, and C are N-acetyl-α-d-glucosamine (GlcNAc), N-acetyl-β-d-mannosaminuronic acid (ManNAcA), and 4-acetamido-4,6-dideoxy-α-d-galactose (Fuc4NAc), respectively. (B) Enzymatic reactions and genetic loci involved in the biosynthesis of ECA. The structural genes of the enzymes that catalyze individual reactions are indicated in italics. Abbreviations: Und-P, undecaprenyl-monophosphate; Und-PP, undecaprenylpyrophosphate; TTP, thymidine triphosphate; PPi, inorganic pyrophosphate; α-KG, α-ketoglutaric acid; acetyl-CoA, acetyl-coenzyme A; CoASH, coenzyme A; TDP, dTDP. The individual amino sugars are abbreviated as described above.
Many of the genes involved in the assembly of ECA polysaccharide chains in E. coli are located in the wec gene cluster located at 85.4 min on the E. coli K-12 chromosome (8), and the functions of these genes have been determined in considerable detail (3, 29, 32). The ECA trisaccharide repeat unit is assembled as an undecaprenylpyrophosphate-linked intermediate (lipid III) (Fig. 1B) (4, 5, 31, 32). Accordingly, synthesis of the repeat unit is initiated by the transfer of GlcNAc 1-P from UDP-GlcNAc to undecaprenylphosphate to yield undecaprenylpyrophosphate-GlcNAc (lipid I) catalyzed by WecA (5, 31, 32). Subsequent reactions involve the successive transfer of ManNAcA and Fuc4NAc from the donors UDP-ManNAcA and TDP-Fuc4NAc, catalyzed by WecG and WecF, respectively.
Although synthesis of lipid III occurs on the cytosolic face of the cytoplasmic membrane, currently available information suggests that Wzy-catalyzed polymerization of repeat units to form linear polysaccharide chains occurs on the periplasmic face of the membrane. This requires the transbilayer movement of lipid III to the periplasmic face of the membrane, and it has been suggested that this step is mediated by a “flippase” encoded by the wzxE gene (o416) (20). Finally, polymerization is followed by the transfer of polysaccharide chains from the lipid carrier to an as yet unidentified acceptor to yield phosphoglyceride-linked chains, and the completed ECAPG molecules are then translocated to the outer membrane. However, essentially nothing is known regarding the genes and mechanisms involved in the latter two steps.
ECAPG is regarded as the major form of ECA, and it is present in all gram-negative enteric bacteria (16). ECAPG accounts for approximately 0.2% of the cellular dry weight of E. coli K-12 (18, 22). Two related forms, ECALPS and ECACYC, have also been identified in certain organisms. ECALPS molecules possess the same linear ECA polysaccharide chains found in ECAPG, but in the case of ECALPS these chains are covalently linked to the core region of LPS (15, 16) instead of a phosphoglyceride aglycone. In contrast, ECACYC is a water-soluble polymer that contains only ECA trisaccharide repeat units (16). In addition, the degree of polymerization of ECACYC molecules is quite different from that observed for linear ECA polysaccharide chains. For example, the polysaccharide chains of ECAPG synthesized by E. coli K-12 exhibit a population that ranges from 1 to 14 repeat units in length, with a modal value of 5 to 7 repeat units (3). In contrast, ECACYC molecules isolated from Shigella sonnei contain only four to six trisaccharide repeat units (11).
Although the structure of ECACYC has been characterized, nothing is known about its function, and there is no information available regarding the genetics and biosynthesis of this novel molecule. This is due, in large part, to the general belief that the occurrence of ECACYC within members of the Enterobacteriaceae is rather restricted, since it has only been found in cell extracts of Shigella sonnei phase I (11, 19), Yersinia pestis (39), and Plesiomonas shigelloides (7, 37); the last organism has now been included in the Enterobacteriaceae.
The results presented in this communication describe the occurrence and characterization of ECACYC in E. coli strain B as determined by a variety of methods, including nuclear magnetic resonance (NMR) spectroscopy and electrospray ionization mass spectrometry (ESI-MS). ECACYC was initially found to copurify with the C-terminal PAS (Per-Arnt-Sim) domain of the human hypoxia-inducible factor 2 (HIFd) (38) following its overexpression as a recombinant protein in E. coli B. However, the detection of ECACYC in these preparations was fortuitous because it was not found to be associated with HIFd and its synthesis was independent of the overexpression of this protein. ECACYC was also found in cell extracts of E. coli K-12, and similar to the results obtained with E. coli B, its synthesis was independent of the overexpression of HIFd. Finally, the results of genetic and biochemical analyses show for the first time that the trisaccharide repeat units of ECACYC and ECAPG are assembled by a common biosynthetic pathway.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
E. coli strains used in this study are listed in Table 1. Transductions were carried out with phage P1 vir according to Silhavy et al. (36). Introduction of plasmid pKG31 into recipient strains was carried out either by transformation or electroporation with standard procedures. Cultures for the routine propagation of bacteria were grown at 37°C in Luria-Bertani (LB) broth or on LB agar containing 0.2% glucose (28). Where indicated, 15N-labeled protein and 13C-labeled ECA were isolated from cells grown in M9 minimal medium (28) containing 0.1% 15NH4Cl and 0.3% glucose (either natural abundance 13C or uniformly labeled [99%] with 13C [Cambridge Isotope Laboratories]). Tetracycline, ampicillin, and chloramphenicol were added to media when appropriate to give final concentrations of 10 μg/ml, 50 μg/ml, and 30 μg/ml, respectively.
TABLE 1.
Bacterial strains
| Strain | Relevant genotype and information | Reference or source |
|---|---|---|
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm met (DE3) | Novagen |
| BL21(DE3)/pLysS | F−ompT hsdSB (rB− mB−) gal dcm met (DE3) pLysS (Cmr) | Novagen |
| HMS174(DE3) | F−recA1 hsdR(rK12− mK12+) Rifr (DE3) | Novagen |
| AB1133 | thr-1 leuB6 Δ(gpt-proA)66 hisG4 argE3 thi-1 rfbD1 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdgK51 supE44 | Laboratory Collection, received as CGSC 1133a |
| MC4100 | F−araD139 Δ(arg-lac)169 λ− e14−flhD5301 fruA25 relA1 rpsL150 rbsR22 deoC1 | Laboratory Collection, received as CGSC 6152a |
| 21548 | As AB1133 but wecA::Tn10 | 36 |
| 21568 | As AB1133 but wecG::Tn10 | 36 |
| PND788 | As MC4100 but ompR::Tn10(Tet) wecF::Tn10(Cam) 8RS88[degP-lacZ] | 36 |
| PR4185 | BL2(DE3)/pLysS/pKG31 | This study |
| PR4186 | BL21(DE3)/pKG31 | This study |
| PR4153 | As PR4153 but wecA::Tn10 [P1(21548) × BL2(DE3)/pLysS, then pKG31 (transformation)] | This study |
| PR4164 | As PR4164 but wecG::Tn10 [P1(21568) × BL21(DE3), then pKG31 (transformation)] | This study |
| PR4161 | As PR4161 but wecF::Tn10(Cam) [P1(PND788) × BL21(DE3), then pKG31 (transformation)] | This study |
E. coli Genetic Stock Center; M. Berlyn, Biology Department, Yale University, New Haven, CT 06520.
Plasmid pKG31 was constructed based on sequence alignments and secondary-structure predictions that identified a minimal PAS domain within human HIF2 from amino acids 240 to 350. The DNA sequence for this domain was amplified by PCR and inserted in a modified form of the pHis-parallel 1 expression vector (35) where the His6 tag was replaced by the β1 domain of streptococcal protein G (GB1). The resulting plasmid contains GB1 and HIFd separated by a 13-amino-acid linker that contains a tobacco etch virus protease cleavage site, allowing facile removal of the fusion and linker from HIFd during purification.
Purification of cyclic ECA.
ECACYC was found to copurify with GB1-HIFd following its overexpression in E. coli strains PR4185 and PR4186 (Table 1). GB1-HIFd fusion protein expression was induced by adding 0.5 mM isopropyl-β-d-galactopyranoside in 1 liter of either LB or M9 minimal medium containing 15NH4Cl and [13C]glucose (either natural abundance or 99% enriched and uniformly labeled), and expression was allowed to proceed overnight at 20°C. The cells were harvested by centrifugation and handled at 4°C for all remaining purification steps. The pellet was resuspended in 25 ml of 50 mM sodium phosphate buffer (pH 7.6)-15 mM NaCl-5 mM dithiothreitol, lysed by high-pressure extrusion, centrifuged, and filtered (0.22 μm), and the supernatant was purified with a Source 15Q anion-exchange column (Amersham Biosciences) preequilibrated with the above buffer. GB1-HIFd eluted from the column during the course of washing the column with 2 volumes of the same buffer. Protein-containing fractions were pooled and concentrated in an Amicon pressure-driven ultrafiltration cell with YM10 10-kDa filters.
The concentrated GB1-HIFd was digested with tobacco etch virus protease (13), followed by removal of the cleaved GB1 fragment by passage of the digest through an immunoglobulin G-Sepharose affinity column (Amersham Biosciences). HIFd, which eluted in the flowthrough volume of this column, was concentrated in an Amicon ultrafiltration system with a YM3 3-kDa filter and then loaded onto a HiLoad 26/60 Superdex 75 column (Amersham Biosciences) equilibrated in 50 mM sodium phosphate buffer (pH 7.2)-15 mM NaCl-5 mM dithiothreitol. The chromatographic mobility of HIFd was consistent with an apparent molecular mass of 14.3 kDa, which agreed with the predicted monomeric molecular mass to within 8%. Chromatograms were obtained by monitoring the UV absorbance at 280 nm (Ε280 of HIFd = 16,170 M−1 cm−1), and protein-containing fractions were analyzed for HIFd by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In addition, the identity of purified HIFd samples was verified by ESI-MS.
ECACYC was isolated from 30 mg of 15N-labeled protein (purified as described above) by ethanol extraction (70% ethanol, 5 min at 80°C). Denatured protein was removed by centrifugation at 10,000 × g at 4°C. Ethanol was removed from the supernatant solution by rotary evaporation at 50°C, and the aqueous phase was then lyophilized. The polysaccharide was further purified by reverse-phase high-pressure liquid chromatography (HPLC) (Vydac C18 column, 0.5 by 25 cm) with a linear gradient of 0 to 5% acetonitrile in H2O. The chromatogram was obtained by monitoring the absorbance from 190 to 600 nm, and the carbohydrate-containing fractions eluting from this column were identified by their absorbance at 206 nm. The presence of ECA in these samples was confirmed by one-dimensional 1H-NMR spectroscopy.
NMR spectroscopy.
All NMR experiments were recorded on Varian Inova 500- and 600-MHz spectrometers, generally at 27°C. NMR spectra of the ECACYC that copurified with HIFd were recorded in samples containing 0.8 mM protein in 50 mM sodium phosphate (pH 7.2)-15 mM NaCl-5 mM dithiothreitol-10% D2O. NMR spectroscopic analysis of HPLC-purified ECACYC was performed on a sample that contained the polysaccharide at a final concentration of approximately 100 μM. ECA samples were dissolved in 550 μl of either 99.96% D2O or H2O:D2O (90:10, by volume) mixtures. One-dimensional 1H-NMR spectra were recorded with presaturation of the water resonance during the 2-s relaxation delay. Two-dimensional total correlation spectroscopy spectra were recorded with an MLEV-17 spin-lock pulse sequence (7 and 100 ms), and two-dimensional nuclear Overhauser enhancement spectroscopy spectra were recorded with a mixing time of 300 and 500 ms. Carbon and nitrogen chemical shift assignments were based on 1H-13C and 1H-15N heteronuclear single-quantum coherence (HSQC) spectra, respectively. Additional chemical shift data were obtained from standard three-dimensional NMR experiments generally used to assign protein backbone and side chain atoms, including HNCACB, CBCA(CO)NH, HNCO (34), and (HBCBCA)COCAHA (14). Chemical shifts were referenced to the methyl-1H signals of sodium 2,2-dimethyl-2-silapentane 5-sulfonate, with direct referencing for all 1H shifts and indirect referencing for 15N and 13C shifts (23).
Crude cell lysate NMR samples were prepared from 200-ml cultures of BL21(DE3) or HMS174(DE3) cells transformed with pKG31 that were allowed to grow overnight at 20°C in M9 minimal medium containing 15NH4Cl, with and without induction of GB1-HIFd fusion protein expression. Cells were harvested, lysed, centrifuged, and filtered as described before. The supernatant was concentrated to 0.5 ml and used to record standard 1H-15N HSQC spectra (12).
Mass spectrometry.
A Micromass Quattro II Triple quadrupole mass spectrometer (Micromass) equipped with the manufacturer's electrospray source was used for ESI-MS experiments. Samples were dissolved in 48% methanol and 4% ammonium hydroxide for ESI-negative ion analysis. Samples were continuously introduced into the source at a rate of 5 μl/min by an infusion pump (Harvard Apparatus), and mass spectra were acquired over an m/z range of 300 to 1,700 per 5 s.
FACE analyses.
ECA samples were hydrolyzed at 100°C with 0.1, 0.25, 0.5, and 1.0 N HCl for 30 min and then dried under reduced pressure with a Speed Vac apparatus (Savant Instruments). The hydrolyzed samples were analyzed by fluorophore-assisted carbohydrate electrophoresis (FACE) with a FACE apparatus, oligosaccharide profiling kit, and reagents according to the directions of the manufacturer (Glyko). Accordingly, reducing termini were labeled with the neutral fluorophore 2-aminoacridone and then resolved on an oligosaccharide profiling gel. Labeled oligosaccharides were detected on the gel, and electronic images of the gel were generated with a Bio-Rad Fluor-S Multi-Imager equipped with a 530DF60 filter.
RESULTS
Copurification of ECA with HIFd.
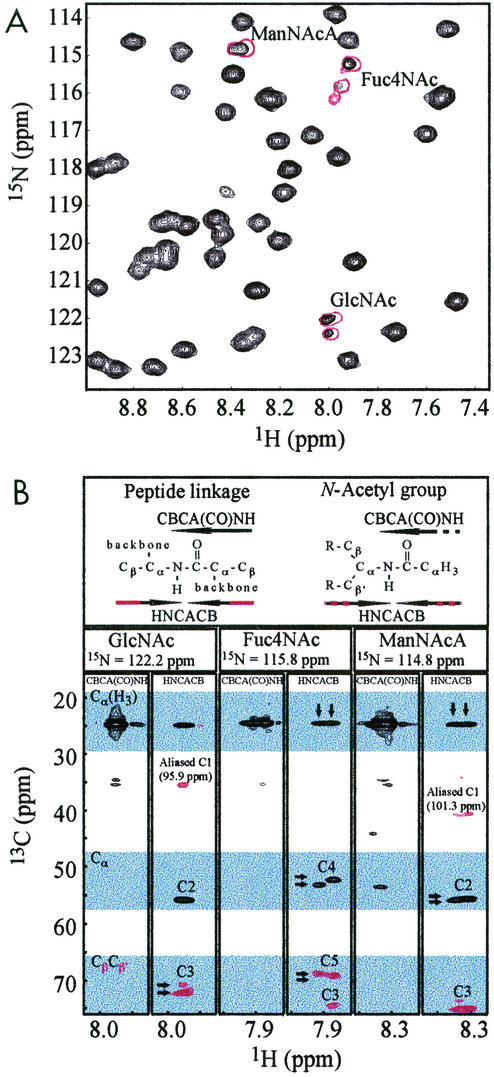
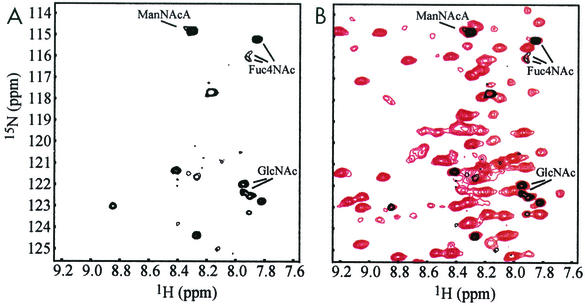
HIFd is believed to play an integral role in the function of human hypoxia-inducible factor 2, a eukaryotic transcription factor that responds to reduced intracellular oxygen levels (38). To conduct structural studies of this domain, we expressed the 13.2-kDa HIFd protein fragment in E. coli PR4186 (Table 1), a derivative of E. coli B. Initial 1H-15N HSQC spectra of HIFd displayed a well-dispersed resonance pattern indicative of a well-folded protein. Interestingly, three sets of 15N/1H correlations in these spectra exhibited particularly narrow line widths and peak doubling (Fig. 2A). These resonances gave unusual correlations in standard triple-resonance experiments commonly used for protein backbone chemical shift assignment, including HNCACB and CBCA(CO)NH spectra.
FIG. 2.
Identification of nonprotein amide resonances. (A) Expansion of the 1H-15N HSQC spectrum of HIFd with copurified ECA (black) and HPLC purified ECA (red). Signals from the amides of the N-acetyl groups of ECA are characterized by notably narrow line widths and signal doubling compared to protein signals. (B) CBCA(CO)NH and HNCACB strips for each N-acetyl group of ECA. Black and red indicate positive and negative cross peaks, respectively. These spectra correlate carbon resonances with the amide 15N and 1H shifts. In particular, the CBCA(CO)NH experiment gives a positive cross peak between the Cα-like site to the carbonyl (Cα[H3]). The HNCACB experiment shows positive cross peaks for the same Cα-like site (Cα[H3]) and the Cα-like site with respect to the HN (Cα) with the same amide. In addition, negative cross peaks are observed in the HNCACB experiment for the Cβ-like signals (see header). Note that the Cβ-like anomeric signals (≈100 ppm) occur outside of the chemical shift range of the 13C dimension (15 to 75 ppm) and are aliased into this range near 40 ppm. Arrows indicate the characteristic signal doubling of ECACYC signals, the origin of which may be sample heterogeneity or slow time scale dynamics (see Discussion).
When used on uniformly 15N/13C protein samples, these methods link 15N/1H chemical shifts to the 13Cα and 13Cβ shifts of carbons on either side of the amide linkage. However, the signals in these spectra generated by the three 15N/1H pairs clearly indicated that they were from amides linked to groups not normally found within protein samples. This is demonstrated in segments of CBCA(CO)NH and HNCACB spectra corresponding to these amides, which show that each 15N/1H pair has only a single carbon at approximately 25 ppm present on the distal side of the amide linkage rather than the two expected Cα and Cβ signals (Fig. 2B). 1H-13C HSQC spectra show that this carbon is directly attached to protons at approximately 2 ppm, establishing that it is an N-acetyl group. The triple resonance spectra (Fig. 2B) also indicated that each amide had three signals to the proximal side, one from a Cα-like site (≈55 ppm) and two from Cβ-like sites (≈70 and 100 ppm). All of these data are clearly inconsistent with standard amino acid structure, strongly suggesting that this sample contained nonprotein material.
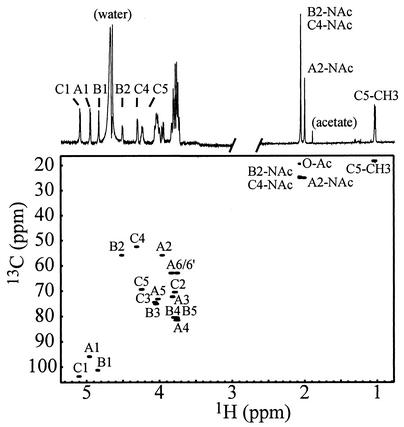
To identify the source of these signals, we purified this material by ethanol extraction and reverse-phase HPLC to obtain a protein-free sample. A one-dimensional 1H-NMR spectrum of this material showed a typical carbohydrate pattern, including very intense signals at approximately 2 ppm corresponding to various N-acetyl groups (Fig. 3). Three characteristic signals corresponding to anomeric protons indicated that this compound contained three monosaccharides. These signals appeared with equal intensities at 4.86, 4.97, and 5.12 ppm, indicating that the monosaccharides were present in equal amounts. The identities of these were provided by preliminary monosaccharide composition analyses conducted on this NMR sample. These revealed that the carbohydrate contained three N-acetylated amino sugars in equal amounts: N-acetylglucosamine, 4-acetamido-4,6-dideoxyhexosamine, and N-acetylhexosamine uronic acid. Glucose was also detected in lesser amounts (data not shown).
FIG. 3.
One-dimensional 1H and two-dimensional 1H-13C HSQC spectra of protein-free ECACYC at 35°C in D2O. The letter code used for the assignments refers to the corresponding amino sugar labels in Fig. 1A. The region of the one-dimensional 1H spectrum between 0.8 and 2.5 ppm is shown with threefold decreased intensity. It is important to note that the anomeric signal of Fuc4NAc (C1) has the same intensity as A1 and B1, indicating that the chemical environment of the repeating units is identical to that found in ECACYC.
To more completely assign the NMR chemical shifts of this carbohydrate, we used a combination of two-dimensional homonuclear and heteronuclear NMR methods. The majority of the signals in the 1H-NMR spectrum were easily assigned with total correlation spectroscopy data, starting at the anomeric proton resonances. Carbon resonances were assigned from 1H-13C HSQC spectra, and nitrogen resonances were assigned by 1H-15N HSQC of 15N-labeled ECA. These assignments are provided in Table 2. The program SUGABASE (http://boc.chem.uu.nl/sugabase/sugabase.html) was used to search for carbohydrate structures that correlated with these NMR data and the monosaccharide composition. Our 1H and 13C chemical shift assignments generally agree with those previously reported for ECACYC from Plesiomonas shigelloides (37) and Yersina pestis (39) and also with the 13C assignments reported for Plesiomonas shigelloides ECAPG (7) (Table 2). However, a comparison of these chemical shift assignments of ECACYC revealed small variations that are likely due to heterogeneity occurring from natural modifications, possible degradation during purification, and variations in experimental conditions.
TABLE 2.
Comparison of 1H- and 13C-NMR chemical shifts (δ in ppm) for ECAa
| Atom | δ (ppm)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GlcNAc
|
ManNAcA
|
Fuc4NAc
|
|||||||||||||
| A | B | C | D | E | A | B | C | D | E | A | B | C | D | E | |
| H1 | 4.97 | 4.95 | 4.96 | — | 4.97 | 4.86 | 4.83 | 4.90 | — | 4.86 | 5.12 | 5.09 | 5.13 | — | 5.12 |
| H2 | 3.98 | — | 3.98 | — | 3.98 | 4.53 | — | 5.57 | — | 4.53 | 3.81 | — | 3.82 | — | 3.80 |
| H3 | 3.84 | — | 3.82 | — | 3.84 | 4.06 | — | 4.08 | — | 4.06 | 4.07 | — | 4.06 | — | 4.07 |
| H4 | 3.77 | — | 3.76 | — | 3.78 | 3.78 | — | 3.84 | — | 3.77 | 4.33 | — | 4.34 | — | 4.33 |
| H5 | 4.04 | — | 3.76 | — | 4.03 | 3.82 | — | 4.07 | — | 3.82 | 4.26 | — | 4.21 | — | 4.26 |
| H6 | 3.84, 3.79 | — | — | — | 3.86, 3.78 | ||||||||||
| CH3 | 1.05 | — | 1.04 | — | 1.05 | ||||||||||
| NAcH | 2.02 | — | — | — | — | 2.08 | — | — | — | — | 2.08 | — | — | — | — |
| OAcH | 2.08 | ||||||||||||||
| C1 | 94.1 | 94.3 | 94.2 | 95.3 | 95.0 | 99.5 | 99.9 | 99.9! | 100.0 | 100.6 | 101.9 | 102.0 | 102.4! | 100.1 | 102.9 |
| C2 | 54.1 | — | 54.2 | 54.0 | 55.0 | 54.0 (58.2) | — | 53.8 | 54.0 | 54.9 | 68.6 | — | 68.5 | 68.0 | 69.3 |
| C3 | 70.4 (68.8) | — | 70.6 | 70.3 | 71.1 | 73.3 | — | 72.7? | 71.3 | 73.8 | 72.6 | — | 73.2? | 75.9? | 73.8 |
| C4 | 79.7 | — | 79.0 | 79.8 | 80.1 | 78.8 | — | 79.6 | 79.8 | 79.9 | 50.7 (51.6) | — | 50.7 | 51.1 | 51.6 |
| C5 | 71.3 | — | 71.4 | 73.0 | 72.2 | 78.7 | — | 77.0 | 75.6? | 79.3 | 67.6 (67.2) | — | 67.9 | 66.8 | 68.1 |
| C6 | 61.1 | — | 61.2 | 60.8 | 61.7 | ||||||||||
| CH3 | 16.6 | — | 16.6 | 16.6 | 17.0 | ||||||||||
| NHAc | 23.2 | — | — | 23.0 | — | 23.1 | — | — | 23.0 | — | 23.0 | — | — | 22.8 | — |
| OHAc | 17.7 | ||||||||||||||
| H(N)* | 7.99 | 8.30 (8.36) | 7.87 (7.91, 7.98) | ||||||||||||
| 15N(H)* | 122 (122.4) | 114.8 | 115.2 (115.8, 116.0) | ||||||||||||
| 13C(O)** | 175.0 | 176.4 | 175.2 (175.6, 174.8) | ||||||||||||
A, assignments of ECACYC from E. coli at 27°C (1H reference DSS = 0.007 ppm; 13C reference DSS = −1.84 ppm). B, partial assignment of ECACYC from P. shigelloides at 25°C (37) (1H reference TMS = 0.00 ppm; 13C reference dioxane = 67.40 ppm). C, Assignments of ECACYC from Y. pestis at 70°C (19) (1H reference acetone = 2.23 ppm; 13C reference acetone = 31.45 ppm). D, 13C assignments of ECAPG from P. shigelloides at 70°C (39) (13C reference TMS = −1.31 ppm). E, assignments of linear and lipid-free ECA from E. coli at 25°C (9) (1H reference TMS = 0.00 ppm; 13C reference dioxane = 67.40 ppm). !, assignment interchanged with respect to original paper (19); ?, assignment may be reversed according to original paper (19, 39); ∗, measured in a 15N-labeled sample in H2O; ∗∗, measured in a 13C-labeled sample in the presence of HIFd in H2O at 30°C. Bold type indicates data obtained in the present study. Chemical shift values in parentheses are from minor species, as discussed in the text. —, chemical shift not reported.
To confirm the glycosidic linkages for this polysaccharide, we acquired several nuclear Overhauser enhancement spectroscopy spectra. Strong interresidue nuclear Overhauser enhancement contacts (particularly those for Fuc4NAc H1-ManNAcA H4, ManNAcA H1-GlcNAc H3, and GlcNAc H1-Fuc4NAc H3) were observed in agreement with linkages identified in ECA (Fig. 1A). Interestingly, the nuclear Overhauser enhancement spectroscopy cross peaks had the same sign as the diagonal peaks, indicating a global rotational correlation time corresponding to a molecular mass above ≈1.5 kDa. From these observations, we conclude that the carbohydrate material present in purified preparations of HIFd is an ECA polysaccharide.
Identification of cyclic ECA.
As discussed earlier, ECAPG is a component of the cell surface of all gram-negative enteric bacteria (16, 21, 24, 32). In addition to ECAPG, the occurrence of ECALPS has also been reported in certain members of the Enterobacteriaceae, including E. coli K-12 (15, 16). However, it is important to emphasize that the occurrence of ECACYC has been demonstrated in only a few organisms (11, 16, 19, 37, 39), and it has not been identified in E. coli. Therefore, we conducted experiments to determine if indeed the water-soluble ECA that was present in HIFd preparations obtained from E. coli B was ECACYC.
Analyses of the HIFd-copurifying ECA by ESI-MS provided strong evidence that this material was ECACYC. Accordingly, mass spectra of ECA present in HIFd samples prior to ethanol extraction and HPLC purification revealed molecular ions of mass 2,430 ± 1 Da, 2,472 ± 1 Da, 2,514 ± 1 Da, 2,556 ± 1 Da, and 2,598 ± 1 Da, in a ratio of approximately 1:2:6:3:1, respectively. The molecular ion of 2,430 ± 1 Da is in agreement with that calculated for ECACYC containing four trisaccharide repeat units (2,429 Da). Partial O-acetylation at the C-6 of GlcNAc has been previously described as a common modification of ECA (11, 19) and would result in a mass increase of 42 Da for a single acetylation event. Thus, the observed molecular ions correspond to cyclic ECA molecules containing four trisaccharide repeat units substituted with one, two, three, and four O-acetyl groups, respectively. Molecular ions for five or six repeat units, which have been reported for ECACYC from Shigella sonnei (37), were not found in the mass spectra of ECACYC from E. coli. In this regard, it should be noted that the ECACYC obtained from Plesiomonas shigelloides was also found to contain only four repeat units (37). However, the basis for the apparent organism-dependent variation in the degree of polymerization is not understood.
Additionally, NMR analysis of the ECA isolated by copurification with HIFd confirmed that this material is indeed ECACYC. Aside from ECACYC, all other forms of ECA (including ECAPG, ECALPS, and various biosynthetic intermediates) are extremely hydrophobic due to the chemical nature of the lipid molecules to which they are linked. These forms of ECA are poorly soluble, and they form large micelles in aqueous solution that would dramatically increase the line width and decrease the signal-to-noise ratio of NMR signals. As evidence of this, Basu et al. used high temperature (70°C) and large sample tubes (10 mm diameter) to record a one-dimensional 13C spectrum of ECAPG (7). In contrast, HIFd-associated ECA gave highly resolved NMR spectra at 27°C in a standard 5-mm NMR tube (Fig. 3A). Moreover, the 1H one-dimensional and 1H-13C HSQC spectra of the ECA material did not reveal 1H and 13C signals of aliphatic groups, which are typically found between 1 and 2 ppm for 1H and around 30 ppm for 13C, that would be indicative of the lipid components of ECAPG and ECALPS (7). In addition, 31P-NMR analysis of HIFd-associated ECA did not indicate the presence of phosphodiesters, which would be expected to occur in both ECAPG and ECALPS, or phosphomonesters, which would be expected to occur in ECALPS (data not shown).
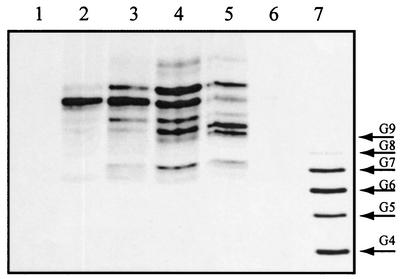
Finally, it is important to note that ECAPG, ECALPS, and ECACYC are characterized by the absence of a free terminal reducing sugar. Indeed, no signals for terminal reducing sugar residues were detected in the 1H- and 13C-NMR spectra obtained from the ECA associated with purified preparations of HIFd. The lack of a free reducing terminus was confirmed by FACE analyses. In this method, saccharides and oligosaccharides are labeled at the reducing terminus with the appropriate fluorescent probe to yield a derivative with a net negative charge, and the fluorescent derivative is then analyzed by gel electrophoresis. The electrophoretic mobility of the derivative is dependent on its charge-mass ratio as well as its hydrodynamic volume. Fluorescent labeling of HIFd-associated ECA with 2-aminoacridone proved to be unsuccessful (Fig. 4, lane 6), consistent with the lack of a free reducing terminus. In contrast, mild acid treatment of HIFd-associated ECA generated several oligosaccharide fragments with free reducing termini, as indicated by the 2-aminoacridone-labeled products shown in Fig. 4, lanes 2 to 5.
FIG. 4.
FACE analysis of acid hydrolyzed ECA. Lane 1, 2-aminoacridone control; lanes 2, 3, 4, and 5, 2-aminoacridone-derivatized products resulting from the treatment of HPLC-purified ECA with 1 N, 0.5 N, 0.25 N, and 0.1 N HCl for 30 min at 100°C, respectively; lane 6, unhydrolyzed HPLC-purified ECA control that was incubated with 2-aminoacridone under derivatizing conditions; lane 7, oligosaccharide standards containing four to nine glucose residues labeled with 8-aminonaphthalene-1,3,6-trisulfonic acid (three negative charges).
We note that the FACE results here could possibly be produced by a new form of linear ECA which has its reducing end blocked by a novel modification. However, such modification would lead to unique NMR signals from the terminal Fuc4NAc, particularly for the anomeric proton. No such signals were observed (Fig. 3). Furthermore, the molecular mass of such a molecule would be significantly above 2,447 Da (molecular ion of unmodified linear polymer), which is not supported by our ESI-MS data.
In summary, these data establish that ECA purified in this manner does not consist of a linear ECA polysaccharide that was generated by some uncharacterized degradative process. Rather, we conclude that the HIFd-associated polysaccharide consists of ECACYC molecules, each of which contains four trisaccharide repeat units and an average of approximately two O-acetyl groups.
Synthesis of ECACYC in E. coli B and K-12 strains is independent of HIFd overexpression.
ECACYC has not been previously identified in E. coli. Accordingly, the data presented thus far raise questions as to whether ECACYC biosynthesis is limited to E. coli B strains or is induced by HIFd overexpression. To address these questions, the soluble fraction of cell lysates of E. coli B strain BL21(DE3) and E. coli K-12 strain HMS174(DE3) cultures transformed with pKG31 were analyzed for ECACYC, with and without induction of HIFd overexpression. NMR analysis of 15N-labeled cell lysates showed intense amide signals characteristic of soluble ECACYC in 15N-1H HSQC spectra of HMS174(DE3) lysates independent of HIFd expression (Fig. 5). Similar results were obtained for BL21(DE3) lysates (data not shown). The chemical shift values of the amide 15N and 1H resonances of ECACYC overlaid very closely and were unaffected by HIFd (Fig. 2A and 5), suggesting that there is no interaction between ECACYC and HIFd. In these spectra, additional signals from other nitrogen-containing metabolites or small proteins can be observed. These data demonstrate that HIFd overexpression has no effect on ECACYC biosynthesis and that ECACYC is present in both E. coli B and K-12 strains.
FIG. 5.
NMR signals of ECACYC in crude cell lysate. (A) Expansion of the 1H-15N HSQC spectrum of 15N-labeled crude cell lysate of HMS174(DE3) cells without induction of GB1-HIFd expression. (B) Overlay of 1H-15N HSQC spectra of 15N-labeled crude cell lysate of HMS174(DE3) cells with (red) and without (black) induction of GB1-HIFd expression. The characteristic strong amide signals of soluble ECA are indicated. The crude cell lysate spectrum of noninduced E. coli cells showed only a few 1H-15N resonances, indicating that ECACYC is present along with only a few other nitrogen-containing metabolites or small proteins present at high concentrations. In contrast, overexpression of a small protein like GB1-HIFd gives rise to many additional signals readily observed in this experiment.
Genetic loci involved in synthesis of ECACYC.
A considerable amount is known about the genes involved in synthesis of the linear ECA chains of ECAPG and ECALPS (3, 8, 29, 32). In contrast, essentially nothing is known about the genetic determinants of ECACYC. The wecA, wecF, and wecG genes of E. coli K-12 encode the GlcNAc 1-P, Fuc4NAc, and ManNAcA transferases, respectively, that are involved in the assembly of the ECA trisaccharide repeat unit of linear ECA polysaccharide chains (Fig. 1B) (8, 26, 27). Accordingly, null mutations in these genes completely abolish the synthesis of ECAPG and ECALPS (26, 27, 29), as determined by a variety of assays, including colony immunoblot assay (25), passive hemagglutination assay (31), and SDS-PAGE and fluorography analyses of cell envelopes prepared from cells grown in the presence of radiolabeled GlcNAc (31). However, it is not known whether these genes also function for the synthesis of ECACYC.
In this regard, it should be noted that the relatively small size of ECACYC and the lack of a hydrophobic aglycone component of ECACYC preclude its detection by many of the methods used to study the biosynthesis of the other ECA forms. In addition, it is not yet known whether the antibodies used for the colony immunoblot and passive hemagglutination assays recognize ECACYC. Accordingly, HIFd was overexpressed in strains PR4153 (wecA::Tn10/pKG31), PR4164 (wecG::Tn10/pKG31), and PR4161 (wecF::Tn10/pKG31) grown in the presence of 15NH4Cl, and attempts were made to detect the presence of 15N-ECACYC in cell extracts of each of these strains as revealed in 1H-15N HSQC spectra. No N-acetyl signals attributable to ECA were detected in any of these preparations (data not shown). These observations corroborate the conclusion stemming from NMR studies described earlier that the carbohydrate material present in purified preparations of HIFd is indeed an ECA polysaccharide. Furthermore, these findings provide the first evidence that the trisaccharide repeat unit of both ECACYC and linear ECA polysaccharide chains is assembled by a common pathway.
DISCUSSION
ECAPG is a cell surface component of all gram-negative enteric bacteria (16), and it accounts for approximately 0.2% of the cellular dry weight of E. coli K-12 (18, 22). In contrast, ECACYC has thus far been found to occur in only a few members of the Enterobacteriaceae (11, 16, 19, 39). The data presented in this study now demonstrate that ECACYC is also synthesized by E. coli, and it is entirely possible that future studies will reveal that ECACYC is a constituent of many other gram-negative enteric bacteria. The apparent limited occurrence of ECACYC among members of the Enterobacteriaceae may simply reflect the fact that only a few organisms have been examined for the presence of ECACYC because of the lack of readily available assays for its detection.
ECACYC was initially found to copurify with the C-terminal PAS domain of the human hypoxia-inducible factor 2 (HIFd) (38) following its overexpression as a recombinant protein in E. coli B. However, detailed analysis of the NMR spectra of HIFd, including three-dimensional 15N- and 15N, 13C-edited nuclear Overhauser enhancement spectroscopy spectra, did not reveal interactions between ECACYC and HIFd. Furthermore, NMR analyses revealed strong signals for ECACYC in the soluble fraction obtained from crude cell lysates of both E. coli strains B and K-12 in the absence of HIFd synthesis. These data indicate that the synthesis of ECACYC is independent of the overexpression of HIFd, and they also suggest that its occurrence in E. coli is not strain specific.
Subsequent work has revealed that the methods used here to purify HIFd were chiefly responsible for the fortuitous discovery of ECACYC in E. coli K-12. Accordingly, the initial anion-exchange step for the isolation of the fusion protein, GB1-HIFd, is not very efficient because the protein binds weakly to the Source 15Q anion-exchange resin with the described phosphate buffer. Subsequent refinements of the isolation protocol employed a lower-ionic-strength buffer (50 mM Tris, pH 7.6), which resulted in increased binding of the protein to the resin without concomitant binding of ECACYC, allowing the separation of these molecules from one another (P. Erbel, unpublished results). In contrast, attempts to employ gel filtration chromatography and molecular cutoff filters to separate the highly negatively charged and unusually shaped ECACYC (2.4 kDa to 2.6 kDa) from HIFd (13.2 kDa) were unsuccessful.
Bruix et al. (10) were also unsuccessful in their initial attempts to use size exclusion chromatography to separate ECA from the comparably sized chemotactic protein CheY (14.0 kDa) from E. coli. CheY-associated ECA was ultimately isolated by repeated phenol extraction of the protein (9), generating material that was identified as a linear and lipid-free ECA polysaccharide. It is significant to note that these investigators identified a free reducing terminal Fuc4NAc residue (α-anomer at 5.23 ppm and β-anomer at 4.77 ppm) in their purified preparations. Since GlcNAc is the potential reducing terminal amino sugar of ECA trisaccharides, this observation suggests the possibility that these polysaccharides resulted from degradation of ECACYC that was originally present in the fractions containing CheY. All of these data suggest that ECACYC could be a more general contaminant in samples of other recombinant proteins expressed in E. coli.
ECACYC is readily identified by characteristic signal doubling in a 1H-15N HSQC spectrum (Fig. 2), possibly caused by chemical heterogeneity (e.g., differential O-acetylation) and restrained rotational motion around the C-N bonds of N-acetyl groups. In this context, it is interesting that Staaf and colleagues (7, 37) suggested that ECACYC undergoes slow conformational changes based on NMR and molecular dynamics studies on unlabeled ECACYC isolated from Plesiomonas shigelloides. Accordingly, such slow conformational exchange processes might also account for the signal doubling observed in this study.
Based on the molar extinction coefficient of HIFd at 280 nm, the amount of purified HIFd from 1 g (wet weight) of cells was calculated to be approximately 15 mg. Consequently, the amount of ECACYC can be estimated from the peak volumes of the N-acetyl signals of ECACYC relative to the protein backbone amide signals (ratio is 1:2) in 1H-15N HSQC spectra (Fig. 2A). With this ratio, and taking into account both the average molecular mass of ECACYC and the fourfold redundancy of the N-acetyl signals, it was estimated that approximately 0.4 mg of ECACYC was isolated from 200 mg (dry weight) of E. coli cells. Surprisingly, this suggests that ECACYC and ECAPG are present in similar amounts in E. coli. It is important to stress that nothing is known about possible factors that may affect the amounts of ECACYC and ECAPG synthesized by cells. Thus, a more accurate determination of the cellular quantity of these molecules will require direct assays as well as additional information about the possible regulation of their synthesis.
This investigation revealed that the trisaccharide repeat units of ECACYC and linear ECA polysaccharide chains are assembled as a lipid-linked intermediate (lipid III) by a common biosynthetic pathway that involves enzymes encoded by the wecA, wecG, and wecF genes of the wec gene cluster of E. coli K-12. Although it has previously been assumed that these genes play a role in ECACYC synthesis, there in fact exists no direct evidence to validate this assumption. Accordingly, the data presented here constitute the first information regarding genetic loci involved in the synthesis of this molecule.
It is highly likely that the pathways for the assembly of water-soluble ECACYC and the linear ECA chains of ECAPG and ECALPS diverge following synthesis of lipid III. Indeed, it has been suggested that ECACYC may be a component of the cytoplasm (1, 16). In this event, it seems likely that the assembly of ECACYC would most likely occur on the inner leaflet of the cytoplasmic membrane by a WzyE-independent mechanism, and it would not require WzxE-mediated translocation of lipid III across the cytoplasmic membrane. It also seems reasonable to assume that the enzyme that catalyzes the cyclization reaction is specifically involved in the synthesis of ECACYC. In this regard, the functions of essentially all of the genes in the wec gene cluster have been defined, and none of these genes appear to be specifically involved in the assembly of ECACYC. Therefore, genetic determinants specifically involved in the synthesis of this polymer must be located at sites on the chromosome outside of this gene cluster; however, these genetic loci have not yet been identified.
The functions of ECACYC and ECAPG are not known, and attempts to identify their functions would be greatly facilitated by the availability of mutants specifically defective in the synthesis of either of these molecules. However, as stated above, the identification of genetic determinants specifically involved in the assembly of ECACYC has yet to be accomplished. In addition, the isolation of mutants specifically defective in the synthesis of ECAPG has also proven to be problematic. Accordingly, the wzyE and wzxE genes encode the polymerase and putative flippase involved in the assembly of linear ECA polysaccharide chains, respectively. Although mutations in these genes specifically abolish the synthesis of ECAPG, recent experiments have revealed that the use of such mutants to investigate the functions of ECACYC and ECAPG is not feasible because mutations in these genes are deleterious to the cell; this appears to be due to toxicity resulting from the accumulation of lipid III (P. D. Rick, unpublished results). Furthermore, attempts to isolate mutants specifically defective in the synthesis of ECAPG due to the inability to transfer ECA polysaccharide chains to a diacylglyceride or phosphoglyceride acceptor have not yet been successful.
Despite these obstacles, the discovery that ECACYC is synthesized by E. coli K-12 now affords a tractable experimental system that will greatly facilitate efforts to identify the function of this novel molecule as well as to define the genes and enzymes involved in its assembly.
Acknowledgments
This research was supported by NIGMS grant GM52882 to P.D.R. and grants from the NIH (CA90601 and CA95471), Searle Scholars Program, Robert A. Welch Foundation (I-1424), and UT Southwestern Endowed Scholars Program to K.H.G. N.G. was supported by grants from the NIH (GM38545) and Robert A. Welch Foundation (I-1168) to Mark Lehrman (UT Southwestern Medical Center).
REFERENCES
- 1.Acker, G., D. Bitter-Suermann, U. Meier-Dieter, H. Peters, and H. Mayer. 1986. Immunocytochemical localization of enterobacterial common antigen in Escherichia coli and Yersinia enterocolitica cells. J. Bacteriol. 168:348-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acker, G., W. Knapp, K. Wartenberg, and H. Mayer. 1981. Localization of enterobacterial common antigen in Yersinia enterocolitica by the immunoferritin technique. J. Bacteriol. 147:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, K., J. Klena, and P. D. Rick. 1999. The modality of enterobacterial common antigen polysaccharide chain lengths is regulated by o349 of the wec gene cluster of Escherichia coli K-12. J. Bacteriol. 181:6564-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, K., P. Nunes-Edwards, and P. D. Rick. 1989. In vitro synthesis of a lipid-linked trisaccharide involved in synthesis of enterobacterial common antigen. J. Bacteriol. 171:1326-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, K., and P. D. Rick. 1987. Biosynthesis of enterobacterial common antigen in Escherichia coli. In vitro synthesis of lipid-linked intermediates. J. Biol. Chem. 262:7142-7150. [PubMed] [Google Scholar]
- 6.Barua, S., T. Yamashino, T. Hasegawa, K. Yokoyama, K. Torii, and M. Ohta. 2002. Involvement of surface polysaccharides in the organic acid resistance of Shiga toxin-producing Escherichia coli O157:H7. Mol. Microbiol. 43:629-640. [DOI] [PubMed] [Google Scholar]
- 7.Basu, S., H. M. Kuhn, A. Neszmelyi, K. Himmelspach, and H. Mayer. 1987. Chemical characterization of enterobacterial common antigen isolated from Plesiomonas shigelloides ATCC 14029. Eur. J. Biochem. 162:75-81. [DOI] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 9.Bruix, M., J. Jimenez-Barbero, and P. Cronet. 1995. Determination by NMR spectroscopy of the structure and conformational features of the enterobacterial common antigen isolated from Escherichia coli. Carbohydr. Res. 273:157-170. [DOI] [PubMed] [Google Scholar]
- 10.Bruix, M., J. Pascual, J. Santoro, J. Prieto, L. Serrano, and M. Rico. 1993. 1H- and 15N-NMR assignment and solution structure of the chemotactic Escherichia coli Che Y protein. Eur. J. Biochem. 215:573-585. [DOI] [PubMed] [Google Scholar]
- 11.Dell, A., J. Oates, C. Lugowski, E. Romanowska, L. Kenne, and B. Lindberg. 1984. The enterobacterial common-antigen, a cyclic polysaccharide. Carbohydr. Res. 133:95-104. [DOI] [PubMed] [Google Scholar]
- 12.Gronenborn, A. M., and G. M. Clore. 1996. Rapid screening for structural integrity of expressed proteins by heteronuclear NMR spectroscopy. Protein Sci. 5:174-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapust, R. B., and D. S. Waugh. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8:1668-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kay, L. E. 1993. Pulsed-field gradient-enhanced three-dimensional NMR experiment for correlating 13Cα/β, 13C′, and 1Hα chemical shifts in uniformly carbon-13-labeled proteins dissolved in water. J. Am. Chem. Soc. 1993:2055-2057.
- 15.Kiss, P., J. Rinno, G. Schmidt, and H. Mayer. 1978. Structural studies on the immunogenic form of the enterobacterial common antigen. Eur. J. Biochem. 88:211-218. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn, H. M., U. Meier-Dieter, and H. Mayer. 1988. ECA, the enterobacterial common antigen. FEMS Microbiol. Rev. 4:195-222. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn, H. M., E. Neter, and H. Mayer. 1983. Modification of the lipid moiety of the enterobacterial common antigen by the “Pseudomonas factor”. Infect. Immun. 40:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lugowski, C., and E. Romanowska. 1978. Enterobacterial common antigen: isolation from Shigella sonnei, purification and immunochemical characterization. Eur. J. Biochem. 91:89-97. [DOI] [PubMed] [Google Scholar]
- 19.Lugowski, C., E. Romanowska, L. Kenne, and B. Lindberg. 1983. Identification of a trisaccharide repeating-unit in the enterobacterial common-antigen. Carbohydr. Res. 118:173-181. [DOI] [PubMed] [Google Scholar]
- 20.Macpherson, D. F., P. A. Manning, and R. Morona. 1995. Genetic analysis of the rfbX gene of Shigella flexneri. Gene 155:9-17. [DOI] [PubMed] [Google Scholar]
- 21.Makela, P. H., and H. Mayer. 1976. Enterobacterial common antigen. Bacteriol. Rev. 40:591-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannel, D., and H. Mayer. 1978. Isolation and chemical characterization of the enterobacterial common antigen. Eur. J. Biochem. 86:361-370. [DOI] [PubMed] [Google Scholar]
- 23.Markley, J. L., A. Bax, Y. Arata, C. W. Hilbers, R. Kaptein, B. D. Sykes, P. E. Wright, and K. Wuthrich. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J. Biomol. NMR 12:1-23. [DOI] [PubMed] [Google Scholar]
- 24.Mayer, H., and G. Schmidt. 1979. Chemistry and biology of the enterobacterial common antigen (ECA). Curr. Top. Microbiol. Immunol. 85:99-153. [DOI] [PubMed] [Google Scholar]
- 25.Meier-Dieter, U., G. Acker, and H. Mayer. 1989. Detection of enterobacterial common antigen on bacterial cell surfaces by colony-immunoblotting: effect of its linkage to lipopolysaccharide. FEMS Microbiol. Lett. 50:215-219. [DOI] [PubMed] [Google Scholar]
- 26.Meier-Dieter, U., K. Barr, R. Starman, L. Hatch, and P. D. Rick. 1992. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J. Biol. Chem. 267:746-753. [PubMed] [Google Scholar]
- 27.Meier-Dieter, U., R. Starman, K. Barr, H. Mayer, and P. D. Rick. 1990. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J. Biol. Chem. 265:13490-13497. [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Rahman, A., K. Barr, and P. D. Rick. 2001. Identification of the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in synthesis of enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 183:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rick, P. D., G. L. Hubbard, M. Kitaoka, H. Nagaki, T. Kinoshita, S. Dowd, V. Simplaceanu, and C. Ho. 1998. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology 8:557-567. [DOI] [PubMed] [Google Scholar]
- 31.Rick, P. D., H. Mayer, B. A. Neumeyer, S. Wolski, and D. Bitter-Suermann. 1985. Biosynthesis of enterobacterial common antigen. J. Bacteriol. 162:494-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigen and capsular polysaccharides, p. 104-122. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 33.Rinno, J., J. R. Golecki, and H. Mayer. 1980. Localization of enterobacterial common antigen: immunogenic and nonimmunogenic enterobacterial common antigen-containing Escherichia coli. J. Bacteriol. 141:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sattler, M., J. Schleucher, and C. Griesinger. 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress Nuclear Magnetic Resonance Spectrosc. 34:93-158. [Google Scholar]
- 35.Sheffield, P., S. Garrard, and Z. Derewenda. 1999. Overcoming expression and purification problems of RhoGDI with a family of “parallel” expression vectors. Protein Expr. Purif. 15:34-39. [DOI] [PubMed] [Google Scholar]
- 36.Silhavy, T. J., T. J. Burman, and L. W. Enquist. 1984. Experiments with gene fusions, p. 107. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Staaf, M., C. Hoog, B. Stevensson, A. Maliniak, and G. Widmalm. 2001. Conformational investigation of a cyclic enterobacterial common antigen employing NMR spectroscopy and molecular dynamics simulations. Biochemistry 40:3623-3628. [DOI] [PubMed] [Google Scholar]
- 38.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 39.Vinogradov, E. V., Y. A. Knirel, J. E. Thomas-Oates, A. S. Shashkov, and L. L'Vov. 1994. The structure of the cyclic enterobacterial common antigen (ECA) from Yersinia pestis. Carbohydr. Res. 258:223-232. [DOI] [PubMed] [Google Scholar]