Abstract
Biofilms are structured communities of cells that are encased in a self-produced polymeric matrix and are adherent to a surface. Many biofilms have a significant impact in medical and industrial settings. The model gram-positive bacterium Bacillus subtilis has recently been shown to form biofilms. To gain insight into the genes involved in biofilm formation by this bacterium, we used DNA microarrays representing >99% of the annotated B. subtilis open reading frames to follow the temporal changes in gene expression that occurred as cells transitioned from a planktonic to a biofilm state. We identified 519 genes that were differentially expressed at one or more time points as cells transitioned to a biofilm. Approximately 6% of the genes of B. subtilis were differentially expressed at a time when 98% of the cells in the population were in a biofilm. These genes were involved in motility, phage-related functions, and metabolism. By comparing the genes differentially expressed during biofilm formation with those identified in other genomewide transcriptional-profiling studies, we were able to identify several transcription factors whose activities appeared to be altered during the transition from a planktonic state to a biofilm. Two of these transcription factors were Spo0A and sigma-H, which had previously been shown to affect biofilm formation by B. subtilis. A third signal that appeared to be affecting gene expression during biofilm formation was glucose depletion. Through quantitative biofilm assays and confocal scanning laser microscopy, we observed that glucose inhibited biofilm formation through the catabolite control protein CcpA.
Many bacteria exhibit two distinct modes of growth, a free-floating planktonic mode and a sessile biofilm mode. Biofilms are structured communities of cells that are adherent to a surface, an interface, or each other and encased in a self-produced polymeric matrix (7, 8). They are thought to be the predominant growth state of bacteria in many natural environments. Biofilms also have a significant impact in medical and industrial settings, due in part to the increased antimicrobial resistance of bacteria in biofilms (18). Despite this, the genes and regulatory signals that determine whether a planktonic cell will transition to a biofilm are still poorly understood.
Bacillus subtilis has been a model organism for the study of gram-positive bacterial physiology. It was recently demonstrated that both laboratory and wild isolates of B. subtilis form biofilms in a process that is dependent on the transcription factor Spo0A (3, 12). Spo0A also acts to integrate intracellular and extracellular signals to direct the development of environmentally resistant spores (11). However, sporulation is not required for biofilm formation, and the requirement for Spo0A in biofilm formation is bypassed by mutations in abrB (12). In addition to Spo0A, the starvation-activated transcription factor sigma-H is required for the complex biofilm structures formed by the wild isolates of B. subtilis (3). These data indicate that biofilm formation is a genetically programmed event that involves a change in gene expression.
Here, we present a functional genomics approach to studying the regulation of biofilm formation by B. subtilis. We used DNA microarrays, comprised of 4,074 of the 4,100 open reading frames of B. subtilis, to follow the changes in gene expression that occurred during the transition from a planktonic state to a biofilm state. By comparing the genes differentially expressed during biofilm formation with the genes previously identified as differentially expressed due to the presence of a transcription factor or environmental signal, we were able to identify several transcription factors and environmental signals that appeared to affect the expression of the genes under biofilm formation conditions (BFC). Two of the identified transcription factors were Spo0A and sigma-H. The remaining transcription factors, LytS, ResE, sigma-W, YbdK, YcbA, and YfiJ, and the environmental signals, catabolite repression and oxygen depletion, had not previously been shown to affect biofilm formation by B. subtilis. By quantifying biofilm formation at different glucose concentrations, we were able to show that biofilm formation by B. subtilis is catabolite repressed through the transcription factor CcpA.
MATERIALS AND METHODS
Plasmid construction.
Plasmid pBL159, used for disrupting ccpA, was constructed by PCR amplifying the cat gene from pGEM-cat (36) with Pfu (Promega) using primers CM1 (5′-AAGCATGCGTTACCCTTATTATCAAGA-3′) and CM2 (5′-AAGCATGGGGAGCTGTAATATAAAAAC-3′). The cat gene was subsequently cloned into an end-filled SapI site in pUC19 to generate pBL132. A derivative of pBL132 was constructed by amplifying a 318-bp internal fragment of ccpA, starting at base 127 relative to the A of the ATG start codon, by PCR using primers BL196 (5′-GAACGTCTCGGTTACCGTCC-3′) and BL197 (5′-CAGAAGCGGCAAGTACAATCG-3′). The PCR product was blunt cloned into the SmaI site in pBL132 to generate pBL159.
The qcrA- and hutP-lacZ fusion constructs were generated through the PCR amplification of the promoter region with primers containing specific restriction enzyme sites (EcoRI and BamHI, underlined in the primer sequences). Primers BL203 (5′-GCGAATTCAGTTAAAGGGACGG-3′) and BL204 (5′-GCGGATCCACAAATTGGCGAAAG-3′) were used to amplify the qcrA promoter region, and primers BL208 (5′-GCGAATTCTAACTGCTTTGTCAGG-3′) and BL209 (5′-GCGGATCCCCGAAGCAGCGATCCCAG-3′) were used for the hutP promoter region. These primers amplified from −429 to +300 for qcrA and from −361 to +303 for hutP, with +1 being the A of the ATG start codon of the respective genes. The regions amplified were chosen to include any necessary promoter region motifs (33, 37). The PCR products were ligated to the EcoRI and BamHI sites of the pKS2 vector (22) to generate plasmids pBL163 and pBL164 for qcrA and hut, respectively.
Plasmid pBL165 carries gfpmut2 (6), which encodes a variant of the green fluorescent protein (GFP) that fluoresces more intensely, and cat flanked by the 5′ and 3′ ends of amyE. This plasmid expresses GFP from a strong promoter, Pspac-hy (30). To construct pBL165, gfpmut2 was PCR amplified from pKL147 (20) with primers BL21 (5′-GGCCAAGCTTAAGGAGGTGATCATTAAAAATGAGTAAAGGAGAAGAAC-3′) and BL22 (5′-GGCCGGATCCTGCAGGTCTGGACATT-3′). The underlined sequences are HinDIII and BamHI restriction enzyme sites, and the italic sequence is a ribosome-binding site 7 bp upstream of the ATG start codon of gfpmut2 (indicated in boldface). After cleavage of this PCR product with HinDIII and BamHI, it was ligated to the HinDIII-to-BamHI vector fragment of pPL82 (21). This resulted in replacement of the gene encoding the Pspac-hy repressor, lacI, with gfpmut2, allowing constitutive expression of gfpmut2.
Strain construction.
The B. subtilis strains used in this study were constructed by transformation with chromosomal DNA or plasmids using standard protocols (13). All of the strains are derivatives of the parental strain BAL218 (JH642) and contain trpC2 and pheA1 mutations (28). The ccpA mutant strain BAL795 (ccpA::pBL159 cat) was isolated by transforming BAL218 with pBL159 and selecting for chloramphenicol-resistant transformants. Plasmid pBL159 cannot replicate in B. subtilis, and chloramphenicol-resistant transformants have pBL159 integrated into the chromosome via homologous recombination at ccpA.
Strains BAL812 [amyE::(qcrA′-lacZ neo)] and BAL829 [amyE::(hutP′-lacZ neo)] were constructed by transforming BAL218 with pBL163 and pBL164, respectively, and selecting for the neomycin resistance associated with the lacZ fusion. Neomycin-resistant transformants that had amyE replaced with the lacZ fusion were screened for based on an amylase-negative phenotype on 1% starch agar plates (13).
Wild-type and ccpA mutant strains expressing GFP were used for confocal scanning laser microscopy (CSLM) analysis of biofilm formation. BAL835 [amyE::(Pspac-hy-gfpmut2 cat)] was generated by transformation of BAL218 with plasmid pBL165, selecting for chloramphenicol resistance associated with Pspac-hy-gfpmut2. Transformants that had amyE replaced with gfp were selected based on an amylase-negative phenotype. As ccpA and Pspac-hy-gfpmut2 were both linked to cat, the cat associated with Pspac-hy-gfpmut2 was disrupted with the gene encoding spectinomycin resistance to generate BAL836 [amyE::(Pspac-hy-gfpmut2 cat::spc)]. This was accomplished by transforming BAL835 [amyE::(Pspac-hy-gfpmut2 cat)] with pJL62 (19), selecting for spectinomycin-resistant colonies and screening for chloramphenicol sensitivity. BAL837 [ccpA::pBL159 (cat) amyE::(Pspac-hy-gfpmut2 cat::spc)] was generated by transformation of BAL795 (ccpA::pBL159 cat) with chromosomal DNA from BAL836 and selecting for spectinomycin resistance.
Microtiter plate assay of B. subtilis biofilm formation.
The microtiter plate assay measures the level of cells adhering to the surfaces of the microtiter plate wells. These assays were performed as described by Hamon and Lazazzera (12) with the following exceptions. The cells were grown in biofilm growth medium (a Luria broth-based medium buffered at pH 7.0 and supplemented with 1 mM MgSO4 and 0.1% glucose [12]), except that the concentration of glucose or other added supplements was varied as appropriate for each experiment. For each assay, the optical densities at 570 nm (OD570) of between 16 and 24 wells were averaged. The standard error of the mean of these wells was <10%. Each assay was repeated on at least three separate occasions, and the averages from all assays were averaged to determine the level of biofilm formation for a strain in the presence of a medium supplement.
Growth of cells for RNA isolation and cell counts.
RNA was isolated from BAL218 cells grown under planktonic formation conditions and BFC. Planktonic conditions were achieved by growing cells in biofilm growth medium (12) at 37°C with shaking at 200 rpm to late-exponential growth (OD600 = 2.5). The cells were then diluted to an OD600 of 0.1 in fresh biofilm medium, and an aliquot was immediately harvested for RNA isolation. This sample was defined to be the planktonic cell population. BFC were achieved by placing 20 ml of the diluted planktonic culture into 250-ml beakers and incubating them at 37°C without shaking. The complete contents of a beaker, which included both planktonic and biofilm cells, was harvested 8, 12, and 24 h after transfer to BFC for RNA isolation. Cells from both planktonic and biofilm growth conditions were harvested by centrifugation and placed immediately at −80°C. RNA was isolated from these cells using an RNeasy Midi Prep kit (Qiagen) according to the manufacturer's instructions. The resulting RNA was checked by gel electrophoresis for DNA contamination and for the presence and integrity of the rRNA bands. The amount of RNA was quantified using a spectrophotometer.
The percentages of cells within the biofilm and planktonic phases were calculated after 8, 12, and 24 h of incubation under BFC. The cells were grown as described above for BFC, and an aliquot of the lower planktonic layer was collected by extraction of 1 ml of medium, at the appropriate time points, with minimal disruption to the cells at the air-surface interface in the forming biofilm. This was defined as the planktonic population sample, and at each time point, the planktonic phase appeared homogeneous with no apparent settling of the cells. The contents of the beaker was then vortexed to generate a homogeneous sample consisting of both biofilm and planktonic cells, and this sample was defined as the total cell population. A separate beaker was used for each time point. Viable-cell counts were performed on the planktonic population sample and the total cell population sample at the appropriate time points. The number of CFU per milliliter of the total cell population minus the number of CFU per milliliter in the planktonic population equals the number of biofilm CFU per milliliter. The number of biofilm CFU per milliliter divided by the number of colony CFU per milliliter of the total cell population provided the percentage of cells in the biofilm.
Microarray experiments and data analysis.
Construction of the DNA microarray slides, probe labeling, and the hybridization conditions used were as described previously (4). The biofilm and planktonic growth condition RNA samples were fluorescently labeled with Cy5 and Cy3 (Perkin-Elmer Life Sciences), respectively, through the generation of cDNA (4). The DNA microarrays were simultaneously hybridized with the planktonic cDNA and a biofilm cDNA to determine the ratio of gene expression under BFC to that under planktonic growth conditions. A ratio of >1.0 represents a higher level of expression under BFC, and a ratio of <1.0 represents a higher level of expression under planktonic growth conditions. For each gene, the ratio was generated using the average of three independent experiments (independently grown and prepared samples). The variance associated with the average ratio for each gene was calculated by dividing the standard deviation associated with the ratio by the average of the three independent values. The data were then analyzed essentially as described previously (4), with the exception that the iterative outlier analysis was performed for three sets of data for each time point. The first set was genes whose variance was <0.7, the second set was those genes whose variance was <0.4, and the third set was those whose variance was <0.1. By separately analyzing those genes that had a low variance, we were able to identify through iterative outlier analysis the genes that had a small change that were otherwise masked by the more variable data sets. The genes identified in the outlier analysis of the three data sets were combined to construct a full list of genes that were outliers for each time point.
β-Galactosidase assays.
β-Galactosidase specific activity was measured in B. subtilis samples grown under both planktonic and BFC growth conditions. B. subtilis cells harboring a lacZ fusion were grown with shaking in biofilm medium until late exponential phase, as described above for the growth of cells for RNA isolation. The cells were then diluted to an OD600 of 0.1 in fresh biofilm medium, and an aliquot of the cells was retained, by centrifugation, as the planktonic sample. To generate the biofilm samples, 100 μl of the diluted culture was aliquoted in a PVC microtiter plate and placed at 37°C to allow biofilm formation. Twenty-four hours after the cells were transferred to the microtiter plate, the entire contents of four microtiter plate wells were collected and combined. At that time, the cells had formed maximal levels of biofilms (12). The samples were homogenized by vortexing them, and the number of cells was quantified by reading the OD600. The cells from the biofilm and planktonic samples were harvested by centrifugation and resuspended in Z buffer (23). The β-galactosidase activity of the cells was measured essentially as described previously (22, 23) and is presented as an n-fold change in β-galactosidase specific activity between the planktonic and biofilm samples.
Confocal microscopy.
Confocal microscopy of wild-type (BAL836) and ccpA (BAL837) strains was performed as described previously (12). The depth of the biofilm in the z plane was measured directly from the image captured from the computer, and the average depth of the biofilm was calculated by averaging the depths of at least two independent images of biofilms grown in three independent experiments (n = 6).
RESULTS
Identification of genes differentially expressed under BFC.
DNA microarrays, comprised of 4,074 of the 4,100 open reading frames of the B. subtilis genome (4), were used to monitor the differences in mRNA composition between cells grown under BFC and planktonic conditions. RNA from cells grown under BFC was harvested 8, 12, and 24 h after the transfer of planktonic B. subtilis cells to conditions optimal for biofilm formation. These cells transitioned from a planktonic state to a medium-air interface biofilm, with 7% of the cells in a biofilm at 8 h, 35% of the cells in a biofilm at 12 h, and 98% of the cells in a biofilm by 24 h. As the entire contents of the vessel used to grow the cells under BFC was harvested, the RNA from the BFC samples was a mixture of planktonic and biofilm cell RNAs. RNA was also isolated from the planktonic cells that were used to initiate biofilm formation. RNA samples from cells under BFC and planktonic growth conditions were fluorescently labeled with Cy5 and Cy3, respectively, through the generation of cDNA. The DNA microarrays were simultaneously hybridized with the planktonic growth condition and BFC cDNA samples to determine the ratio of gene expressions under these growth conditions. Those genes that had highly variable expression ratios were eliminated from further analysis (see Materials and Methods). Approximately 64% of the genes gave reproducible expression ratios, and as much as a 25-fold difference in the expression level of a gene was observed.
Iterative outlier analysis was applied to the expression ratios to determine which genes had significantly different expression under BFC versus planktonic conditions (4). A total of 519 genes were differentially expressed during the biofilm formation time course. More than 55% of these differentially expressed genes were expressed at only one time point, indicating a temporal control of gene expression. Sixty-one percent of the differentially expressed genes encode proteins with an assigned function, and 39% encode proteins of unknown function (http://www.mimg.ucla.edu/faculty/lazazzera/index.html?FacultyKey=788). Most of the functional gene categories for B. subtilis are represented among the differentially expressed genes, with a large number of genes involved in motility and chemotaxis, phage-related functions, membrane bioenergetics, glycolysis, and the tricarboxylic acid cycle being differentially expressed (http://www.mimg.ucla.edu/faculty/lazazzera/index.html?FacultyKey=788). Some of these differentially expressed genes have also been identified as differentially expressed during biofilm formation by other bacteria, suggesting conserved responses of bacterial cells to biofilm formation (see Discussion).
Identification of transcription factors that affect gene expression under BFC.
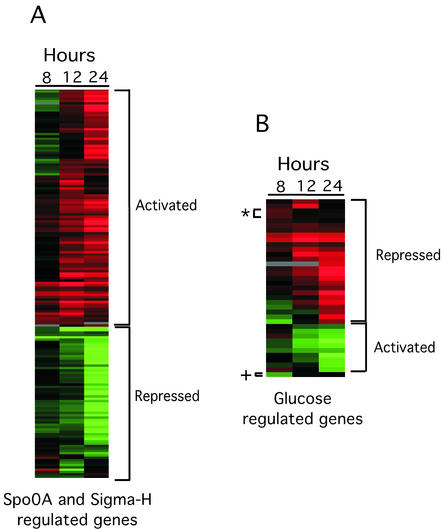
To identify environmental signals or transcription factors that could be regulating the genes differentially expressed under BFC and therefore biofilm formation, we compared the genes differentially expressed under BFC with genes identified as differentially expressed through other genomewide expression-profiling studies (2, 4, 5, 10, 17, 24, 25, 26, 34, 35). We hypothesized that, if a transcription factor regulates biofilm formation, a significant percentage of the genes controlled by that transcription factor would be differentially expressed under BFC. As a test of this hypothesis, we compared the genes differentially expressed under BFC with those identified through genomewide transcriptional profiling as regulated by Spo0A (10), a transcription factor that is known to be required for biofilm formation in both laboratory and wild isolates of B. subtilis (3, 12). Twenty-two percent of Spo0A-regulated genes were differentially expressed under BFC (Table 1). As Spo0A is required for biofilm formation, we predicted that Spo0A-activated genes would be expressed at higher levels and that Spo0A-repressed genes would be expressed at lower levels in the biofilm samples. Sixty-eight percent of the differentially expressed biofilm genes regulated by Spo0A followed this pattern (Table 1), with maximal expression or repression 12 and 24 h after incubation under BFC (Fig. 1). Indeed, expression of spo0A itself was maximal at 24 h.
TABLE 1.
Transcription factors and environmental signals that regulate gene expression under BFC in B. subtilis
| Regulator | No. (%) of genes differentially expresseda | Time of maximal activity (h)b | No. (%) of genes with same patternc | Reference(s) |
|---|---|---|---|---|
| Glucose | 68/205 (33) | 8 | 40/68 (59) | 24, 35 |
| Oxygen | 37/121 (31) | 8 | 22/37 (59) | 34 |
| LytS | 2/6 (33) | 8 | 2/2 (100) | 17 |
| ResE | 27/57 (47) | 8 | 24/27 (89) | 34 |
| Sigma-H | 153/433 (35) | 12, 24 | 114/153 (74.5) | 4 |
| Sigma-W | 15/49 (31) | 8 | 15/15 (100) | 5 |
| Spo0A | 162/732 (22) | 12, 24 | 110/162 (68) | 10 |
| YbdK | 2/9 (22) | 8 | 2/2 (100) | 17 |
| YcbA | 4/19 (21) | 8 | 4/4 (100) | 17 |
| YfiJ | 12/31 (39) | 8 | 12/12 (100) | 17 |
Number (percent) of genes known to be controlled by the indicated regulator that were differentially expressed under BFC.
Time under BFC at which the genes activated by the regulator are maximally expressed and the genes repressed by the regulator are least expressed.
Number (percent) of genes differentially expressed under BFC and known to be controlled by the regulator that are maximally expressed or repressed at the time indicated in the previous column. For example, we found 40 of the 68 genes which are regulated by glucose and differentially expressed under BFC to have maximal expression at 8 h under BFC.
FIG. 1.
Expression profiles of genes regulated by Spo0A, sigma-H, and glucose under BFC. (A) Heat map showing the expression profiles of 165 Spo0A- and sigma-H-regulated genes differentially expressed under BFC. (B) Heat map showing the expression profiles of 40 glucose-regulated genes differentially expressed under BFC. These genes were ordered using a hierarchical clustering algorithm (Gene Cluster; Eisen Lab) so that those with similar expression patterns were grouped together. Hybridization ratios are displayed calorimetrically: shades of green indicate that a gene had a higher RNA level under planktonic growth conditions (ratio, <1.0), and shades of red indicate that a gene had a higher RNA level under BFC (ratio, >1.0). Activated and repressed refer to regulation by Spo0A, sigma-H, or glucose. Exceptions are indicated as follows: *, two glucose-activated genes; +, one glucose-repressed gene.
Sigma-H is known to be required to form the complex fruiting-body structures seen in biofilms formed by wild isolates of B. subtilis (3). We examined whether sigma-H affected gene expression in a laboratory isolate of B. subtilis under BFC. Although expression of sigH itself did not appear to change, 153 of the 433 sigma-H-regulated genes (4) were identified as differentially expressed under BFC (Table 1). To address the question of whether sigma-H was activated under BFC, we examined the expression profiles of the sigma-H genes throughout the biofilm formation time course. Seventy-nine percent of the 153 genes indicated that sigma-H-activated and -repressed genes were expressed at higher or lower levels, respectively, in the biofilm sample 12 and 24 h after incubation under BFC (Fig. 1). Thus, it appeared that sigma-H had a major effect on gene expression by the laboratory strain of B. subtilis during biofilm formation.
To find additional transcription factors and environmental signals that might affect biofilm formation, we compared the genes identified as differentially expressed under BFC to genes identified through other genomewide transcriptional-profiling experiments as regulated by ComK (2, 25), ComA (26; N. Comella and A. D. Grossmann, unpublished data), DegU (26), PhoP (26), sigma-W (5), ResE (34), 24 two-component regulatory systems (17), glucose (24, 35), oxygen (34), and cysteine (1). Since 22% of the genes identified as regulated by Spo0A (10) were differentially expressed under BFC and Spo0A is involved in biofilm formation by B. subtilis, we considered further those transcription factors or environmental signals for which >20% of the genes regulated by that specific factor were differentially expressed under BFC. Furthermore, we considered a transcription factor or environmental signal to be controlling the expression of genes under BFC only if the majority of the genes regulated by the specific factor and differentially expressed under BFC had the same pattern of expression over the time course. For example, 100% of sigma-W-controlled genes that were identified as differentially expressed under BFC were maximally expressed in the biofilm sample at 8 h (Table 1). This suggested that the sigma-W-induced genes that were expressed during biofilm formation were probably expressed due to increased sigma-W activity rather than some other factor. Based on these criteria, LytS, ResE, YbdK, YcbA, and YfiJ were also identified as having a significant effect on gene expression during biofilm formation, with maximal activity 8 h after incubation under BFC (Table 1). Of these transcription factor genes, both sigW and ycbA were differentially expressed under BFC. In addition, the genes that are inhibited or activated by the presence of glucose or the depletion of oxygen were maximally down- or up-regulated, accordingly, at 8 h in the biofilm formation time course (Table 1).
Determination of the role of glucose in regulating biofilm formation.
To determine whether the comparative DNA microarray analysis had identified any transcription factors or environmental signals that regulated biofilm formation, we examined the effect that glucose had on biofilm formation. Glucose was identified as regulating the largest number of genes under BFC after sigma-H and Spo0A (Table 1). Forty genes whose expression is regulated by glucose were differentially expressed under BFC. The pattern of expression of glucose-regulated genes suggests that glucose was being depleted from the medium. Genes activated by the presence of glucose were maximally up-regulated at 8 h in the biofilm formation time course, and glucose-repressed genes were maximally up-regulated at 24 h in the time course (Fig. 1). This is consistent with glucose being present 8 h and absent 24 h after transfer to BFC. The medium used to grow the cells under BFC contained 0.1% glucose.
To confirm via a second method that glucose-repressed genes were induced under BFC, we monitored the expression of two glucose-repressed genes, qcrA and hutP, by measuring the levels of β-galactosidase from strains containing either a qcrA- or hutP-lacZ transcriptional fusion. The levels of β-galactosidase activity were measured in cells grown under planktonic conditions and in cells incubated for 24 h under BFC. We observed 9.6- and 8.4-fold inductions in the expression of qcrA and hutP, respectively, under biofilm versus planktonic conditions. These data support the conclusion that glucose-regulated genes are differentially expressed under biofilm and planktonic conditions.
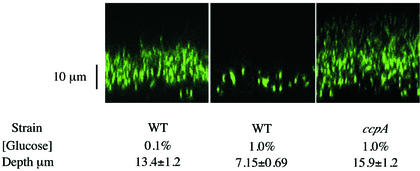
The correlation between the induction of glucose-repressed genes and biofilm formation led us to hypothesize that biofilm formation is inhibited by the presence of glucose. To test this hypothesis, we quantified the level of biofilm formation by cells grown in biofilm growth medium containing 1 or 0.1% glucose. Using a microtiter plate assay to quantify biofilm formation, we observed twofold-lower levels (P < 0.01) of biofilm formation in the presence of 1% glucose than in the presence of 0.1% glucose (Table 2). The effect of glucose on biofilm formation was also assayed by CSLM of B. subtilis cells expressing GFP. In the presence of 1% glucose, the cells formed a biofilm with an average depth of 7.15 ± 0.69 μm, while in the presence of 0.1% glucose, the cells formed a biofilm with an average depth of 13.4 ± 1.2 μm (Fig. 2). The difference in the levels of biofilm formation did not appear to be due to a negative effect of 1% glucose on growth, as comparable numbers of cells were reached under both conditions (data not shown). These data together indicate that glucose is a negative regulator of biofilm formation.
TABLE 2.
Catabolite repression of B. subtilis biofilm formation
| Strain | Biofilm formation with addition ofa:
|
|||||
|---|---|---|---|---|---|---|
| Glucose
|
Fructose
|
Acetoin
|
||||
| 0% | 0.1% | 1.0% | 0.1% | 1.0% | 20 mM | |
| WT | 1.73 (±0.19) | 2.5 (±0.18) | 1.32 (±0.13) | 2.25 (±0.16) | 0.59 (±0.09) | 5.03 (±0.44) |
| ccpA | 1.34 (±0.058) | 2.3 (±0.31) | 3.4 (±0.34) | ND | ND | ND |
Biofilm formation by B. subtilis in medium containing the indicated supplement was quantified using the microtiter plate assay after growth of the biofilms for 48 h. The data shown are the averages of between 3 and 43 independent experiments, and the numbers in parentheses represent the standard errors of the mean. ND, not determined. The percentages are the levels (wt/vol) of supplement added to the biofilm growth medium. WT, strain BAL218; ccpA, strain BAL795.
FIG. 2.
Effect of catabolite repression on the structure of B. subtilis biofilms. Shown are representative CSLM images of biofilms of B. subtilis expressing GFP in the xz plane. The depth of biofilm (± standard error of the mean), calculated from a minimum of three independent experiments (n = 6 to 10), is indicated. WT, strain BAL218; ccpA, strain BAL795.
The inhibitory effect of glucose on biofilm formation would predict that high levels of biofilm formation would be observed in the absence of glucose. However, we consistently observed lower levels of biofilm formation (30% lower; P < 0.05) in biofilm growth medium lacking glucose than in medium containing 0.1% glucose, as quantified by the microtiter plate assay (Table 2), indicating that glucose stimulates biofilm formation. It was previously reported that glucose, through metabolism to acetoin, could stimulate Spo0A activity under some sporulation development conditions (30). To determine whether the stimulation of biofilm formation by glucose could be due to its metabolism to acetoin, we quantitated the levels of biofilm formation by cells grown in the presence and absence of 20 mM acetoin. Biofilm formation was stimulated 2.9-fold (P < 0.05) in the presence of acetoin, consistent with the metabolism of glucose to acetoin stimulating biofilm formation (Table 2).
Role of the catabolite control protein CcpA in regulating biofilm formation.
As high levels of glucose inhibited biofilm formation, this suggested that biofilm formation might be subject to catabolite repression. To test this model, we determined whether biofilm formation by B. subtilis was inhibited by high levels of another catabolite sugar, fructose (9). As quantified by the microtiter plate assay, cells grown in 1% fructose exhibited a fourfold-lower (P < 0.01) level of biofilm formation than cells grown in the presence of 0.1% fructose (Table 2). The difference in the levels of biofilm formation was not due to differences in the growth of the cells, as similar numbers of cells were reached under both conditions (data not shown). These data are consistent with biofilm formation being subject to catabolite repression.
The catabolite control protein CcpA of B. subtilis is a transcriptional regulator that mediates catabolite repression of many genes in B. subtilis in response to glucose and fructose (9, 15). To determine whether the inhibition of biofilm formation in the presence of high levels of glucose was due to CcpA, we quantified the level of biofilm formation by a ccpA mutant strain grown in the presence of different concentrations of glucose by the microtiter plate assay. In the presence of 1% glucose, the ccpA mutant strain exhibited a 2.5-fold-higher (P < 0.01) level of biofilm formation than the wild-type strain (Table 2). Biofilm formation by a ccpA mutant strain in the presence of 1% glucose was also assayed by CSLM of strains expressing GFP. Under these conditions, the ccpA mutant strain formed biofilms with an average depth of 15.9 ± 1.2 μm (Fig. 2), which is twofold greater (P < 0.01) than the biofilms formed by the wild-type strain. These data support the model that glucose inhibits biofilm formation by stimulating CcpA to inhibit a gene(s) that is involved in biofilm formation.
DISCUSSION
DNA microarrays of the B. subtilis genome were used to follow the changes in gene expression that occurred as B. subtilis cells transitioned from a planktonic state to a biofilm state. We used these data to identify several transcription factors and environmental signals that appeared to affect gene expression during this transition. Many of these transcription factors are regulated by an environmental stress, indicating that biofilm formation by B. subtilis is stimulated by nonoptimal growth conditions. Through quantitative biofilm assays and microscopic analysis, we were able to show that one of the environmental signals, catabolite repression, inhibited biofilm formation. This inhibition was mediated by the transcription factor CcpA, which appears to repress a gene that limits the depth of the mature B. subtilis biofilm. This indicates that biofilm formation is inhibited by the presence of a rapidly metabolized carbon source. The similarity of the genes that were expressed and the signals that regulate these genes in B. subtilis under BFC to those in the divergent Pseudomonas aeruginosa and Escherichia coli biofilms suggest that there may be conserved responses of bacterial cells to biofilm formation.
DNA microarrays identified transcription factors that affect biofilm formation.
By comparing the results of the genomewide transcriptional-profiling study presented here with those of other B. subtilis genomewide transcriptional-profiling studies, we were able to identify several transcription factors and environmental signals that appeared to regulate genes during the transition from a planktonic cell to a biofilm. This is not a complete list of transcription factors that regulate gene expression under BFC, as even those transcription factors analyzed may not have appeared to regulate gene expression under BFC due to differences in the growth media or the strains used in the different genomewide transcriptional-profiling studies. However, two of the transcription factors, Spo0A and sigma-H, and one of the environmental signals, catabolite repression, identified through this approach affect biofilm formation. It is not yet known whether the other transcription factors identified also have regulatory roles in biofilm formation. Preliminary data indicate that sigma-W is not essential for biofilm formation (M. Hamon, N. Stanley, and B. Lazazzera, unpublished data). Although many of the transcription factors identified are maximally active after 8 h under BFC, when only 7% of the cells in the culture are in the biofilm, these transcription factors could be activating genes in the planktonic cells that direct these cells to form a biofilm. Indeed, it was previously postulated that oxygen depletion, which appears to occur at 8 h under BFC, may be an important signal for biofilm formation, as biofilm formation was observed only in standing cultures, which would be more oxygen limiting than slowly shaking cultures (12). ResE, which appears to regulate gene expression under BFC, is part of a two-component regulatory system that is activated by anaerobiosis to regulate the expression of proteins involved in bioenergetics (34). It will be interesting to determine whether ResE is active in planktonic cells and whether it could be regulating the decision to transition from a planktonic cell to a biofilm.
Biofilm formation by B. subtilis is induced by nonoptimal growth conditions.
From the transcription factors identified in this study as affecting the expression of genes under BFC, it would appear that biofilm formation by B. subtilis is stimulated by nonoptimal growth conditions. A large proportion of the genes identified as differentially expressed during biofilm formation are regulated by Spo0A and/or sigma-H (4, 10). Spo0A is required for cells to transition from an attached monolayer to a mature biofilm (3, 12) and is maximally active under conditions of starvation and high cell density (11). In addition, sigma-H is required for the formation of the complex fruiting-body structures seen on biofilms formed by wild isolates of B. subtilis (3) and also is activated by starvation (14). This suggests that starvation, and possibly high cell density, stimulates biofilm formation by B. subtilis.
Further support for biofilm formation by B. subtilis being stimulated by nonoptimal growth conditions is the finding that biofilm formation is inhibited in the presence of a rapidly metabolized carbon source. Similar observations have also been made concerning E.coli biofilm formation, which is also subject to catabolite repression (16). The catabolite control protein CcpA mediated this inhibition in B. subtilis, and ccpA mutants formed thicker biofilms than wild-type strains in the presence of glucose. This suggests that in the presence of a preferred carbon source, B. subtilis cells prefer to be in a planktonic state. One possible model to explain the regulation of the depth of the biofilm is that, under conditions of catabolite repression, CcpA represses a gene that either decreases the rate of attachment of cells to a biofilm or increases the rate of detachment of cells from the biofilm.
The hypothesis that biofilm formation occurs under suboptimal growth conditions is supported by stimulation of biofilm formation in the presence of acetoin. Acetoin is a metabolic by-product of glucose metabolism and as such an indicator of the depletion of a rapidly metabolized carbon source. Although the mechanism by which acetoin stimulates biofilm formation by B. subtilis is unknown, it is possible that it is through the activation of Spo0A in a manner similar to acetoin activation of sporulation (30).
Although it is not yet known whether the other transcription factors identified as regulating genes expressed under BFC affect biofilm formation, the signals that regulate some of these transcription factors suggest that the cells may be perceiving stressful or nonoptimal growth conditions under BFC. Many genes stimulated by oxygen depletion, or the anaerobic gene regulator ResE, were up-regulated 8 h after the transfer of cells to BFC. At the same time, genes controlled by sigma-W are induced; sigma-W is activated by alkaline conditions or other extracytoplasmic stress (5). The signal that regulates YfiJK, a two-component regulatory system, is not known, but YfiJK inhibits transcription of several genes involved in amino acid biosynthesis and uptake (17). These genes are maximally expressed 24 h after the transfer of cells to BFC, suggesting that at that time the cells may be starved for one or more amino acids.
Genes differentially expressed in biofilm cells of B. subtilis and other bacteria.
In addition to generating information about the transcription factors that regulate the transition from a planktonic state to a biofilm, this study also provided information about the nature of the genes that are differentially expressed in the biofilm cells. Six percent of the B. subtilis genome was identified as differentially expressed 24 h after transition to BFC, the time point when 98% of the cells are within a biofilm. It was proposed that there would be global changes in the gene expression profiles of cells growing within a biofilm versus cells growing in a planktonic state (8, 29). However, microarray analysis of P. aeruginosa biofilms indicates that a relatively small percentage of genes (1%) have altered expression between biofilm and planktonic cells (31). It is likely that the 6% of B. subtilis genes that are differentially expressed is an overrepresentation of the number of genes differentially expressed due to biofilm formation, as some of these genes have probably changed due to changes in the media. It appears that, similar to P. aeruginosa, a small percentage of B. subtilis genes change in response to biofilm formation.
One of the major classes of genes differentially expressed under BFC were those involved in motility. Thirty-five flagellar and chemotaxis genes were expressed at lower levels 24 h after transition to BFC; the majority of these genes are repressed through sigma-H activation, and their expression profiles are shown in Fig. 1 (http://www.mimg.ucla.edu/faculty/lazazzera/index.html?FacultyKey=788). The repression of genes involved in the synthesis of the flagella has also been seen in biofilms of P. aeruginosa and E. coli (29, 31). This suggests that inhibition of flagellar-gene transcription may be a general phenomenon for bacteria in biofilms.
PBSX is a defective B. subtilis prophage (32); 17 genes in the B. subtilis genome involved in prophage production were more highly expressed in the biofilm, with maximal expression after 24 h of incubation under BFC. It has been proposed that prophage production may have a role in generating genetic diversity in the biofilm (31). The higher expression levels of prophage genes in biofilm cells were also apparent from the P. aeruginosa study (31). The induction of prophage genes may represent another general phenotype of bacteria in biofilms.
One set of genes that we would expect to be differentially expressed under BFC is genes involved in exopolysaccharide production, as such genes are required for biofilm formation in several bacteria (27). One cluster of genes, yveK-yvfE, predicted to be involved in exopolysaccharide production and required for the complex biofilm architecture formed by wild isolates of B. subtilis (3), was not differentially expressed in the gene expression profiling study presented here. However, the laboratory strain of B. subtilis used in this study does not form the complex architecture formed by the wild isolates. The genes involved in formation of the extrapolymeric matrix of the laboratory isolates of B. subtilis are unknown and may be included among the genes of unknown function differentially expressed under BFC.
The work described here demonstrates the use of DNA microarrays to identify environmental and transcription factors involved in regulating biofilm formation by B. subtilis. Similar approaches could be used to analyze other physiological processes in organisms for which detailed knowledge of the regulation of gene transcription is available. The analysis of the genes differentially regulated after 24 h of incubation under BFC has highlighted the similarity between biofilm-specific gene expression in B. subtilis and that in other bacteria. Likewise, the identification of catabolite repression as an inhibitor of biofilm formation in B. subtilis and E. coli (16) suggests that common regulatory mechanisms may control the transition of planktonic cells into a biofilm.
Acknowledgments
We thank Melanie Hamon and Mathew Schibner for advice and help with the CSLM, performed at the UCLA Brain Research Institute confocal microscope facility. We also thank Robert Gunsalus and members of the Lazazzera laboratory for critical reading of the manuscript.
This work was supported in part by the University of California Academic Senate Council on Research of the Los Angeles Division, the UCLA foundation, and a Frontiers of Science seed grant from the Howard Hughes Medical Institute (B.A.L.) and by Public Health Services Grant GM50895 (A.D.G.). N.R.S. was supported by a long-term postdoctoral fellowship awarded by the EMBO.
REFERENCES
- 1.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 3.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Winston Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 6.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP) gene. 173:33-38. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R., Korber, and H. M. Lappin-Scott. 1995. Microbial biofilm. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 8.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deutscher, J., E. Kuster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr-CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol. Microbiol. 15:1049-1053. [DOI] [PubMed] [Google Scholar]
- 10.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 12.Hamon, M. A., and B. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199-1209. [DOI] [PubMed] [Google Scholar]
- 13.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., Chichester, England.
- 14.Healy, J., J. Weir, I. Smith, and R. Losick. 1991. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor Sigma-H in Bacillus subtilis. Mol. Microbiol. 5:477-487. [DOI] [PubMed] [Google Scholar]
- 15.Henkin, T. M. 1996. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, D. W., J. W. Simecka, and T. Romeo. 2002. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuchma, S. L., and G. A. O'Toole. 2000. Surface-induced and biofilm-induced changes in gene expression. Curr. Opin. Biotechnol. 11:429-433. [DOI] [PubMed] [Google Scholar]
- 19.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterisation of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemon, K. P., and A. D. Grossman. 1998. Localisation of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 21.Levin, P. A., R. L. Schwartz, and A. D. Grossman. 2001. Polymer stability plays an important role in positional regulation of FtsZ. J. Bacteriol. 183:5449-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnuson, R., J. M. Soloman, and A. D. Grossman. 1994. Biochemical and genetic characterisation of a competence pheromone from B. subtilis. Cell 77:207-216. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 25.Ogura, M., H. Yamaguchi, K. Kobayashi, N. Ogasawara, Y. Fujita, and T. Tanaka. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogura, M., H. Yamaguchi, K.-I. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. arsid15824633 [DOI] [PMC free article] [PubMed]
- 27.O'Toole, G. A., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 28.Perego, M., G. B. Speigelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator. abrB:regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 29.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 32.Wood, H. E., M. T. Dawson, K. M. Devine, and D. J. McConnell. 1990. Characterisation of PBSX, a defective prophage of Bacillus subtilis. J. Bacteriol. 172:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wray, L. V., Jr., and S. H. Fisher. 1994. Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: regulation of transcription initiation and inhibition of histidine transport. J. Bacteriol. 176:5466-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida, K., K. Kobayashi, Y. Miwa, C.-M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youngman, P., H. Poth, B. Green, K. York, G. Olmedo, and K. Smith. 1989. Methods for genetic manipulation, cloning, and functional analysis of sporulation genes in Bacillus subtilis. American Society for Microbiology, Washington, D.C.
- 37.Yu, J., L. Hederstedt, and P. J. Piggot. 1995. The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization. J. Bacteriol. 177:6751-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]