Abstract
The synthesis of a functional nitrous oxide reductase requires an assembly apparatus for the insertion of the prosthetic copper. Part of the system is encoded by maturation genes located in Pseudomonas stutzeri immediately downstream of the structural gene for the enzyme. We have studied the transcriptional organization and regulation of this region and found a nosDFYL tatE operon structure. In addition to a putative ABC transporter, consisting of NosD, NosF, and NosY, the operon encodes a Cu chaperone, NosL, and a component of the Tat translocon, TatE. The nosD operon was activated in response to anaerobiosis and nitrate denitrification. The membrane-bound regulator NosR was required for operon expression; in addition, DnrD, a regulator of the Crp-Fnr family, enhanced expression under anaerobic conditions. This establishes a likely signal transduction sequence of NO → DnrD → nosR/NosR → nosD operon. DnrD-dependent expression was also observed for the nnrS operon (located immediately downstream of the nosD operon), which encodes a putative heme-Cu protein (NnrS) and a member of the short-chain dehydrogenase family (ORF247). The NosF protein, encoded within the nosD operon, exhibits sequence similarity to ABC-type ATPases. It was fused to the Escherichia coli maltose-binding protein and overexpressed in soluble form. The fusion protein was purified and shown to have ATPase activity. NosF is the first maturation factor for which a catalytic function has been demonstrated in vitro.
The multicopper enzyme nitrous oxide reductase (N2OR) undergoes a maturation process for the insertion of its prosthetic metal (32, 39, 44, 48, 49). Several accessory proteins are required for the biosynthesis of the catalytically active enzyme, some of which are encoded by genes located downstream of the N2OR structural gene, nosZ. Expression of nosZ is dependent on the multitopic, membrane-bound regulator NosR (10). At least three proteins, encoded by the maturation genes nosD, -F, and -Y, form a putative assembly complex which extends to both sides of the cytoplasmic membrane. From sequence similarity, it was inferred that nosF may encode an ATP/GTPase and that a step in the biosynthesis of N2OR is energy dependent (49). A lacZ reporter gene fusion had been used to deduce a location in the cytoplasm for NosF (9). NosD belongs to a family of proteins with a carbohydrate binding and sugar hydrolase domain (8) whose significance is still unclear. NosY is a six-helix transmembrane protein; it is presumed that, together with NosF and periplasmic NosD, it forms an ABC-type transporter. The biosynthesis of N2OR also involves a number of additional factors which are nonessential or can be functionally replaced by other cellular processes (44).
Catalyzed assembly of the prosthetic Cu in N2OR seems to be obligatory for the CuZ center, representing the catalytic site in the form of a tetranuclear CuS cluster (6, 35), rather than for the binuclear CuA center, which represents the electron entry site (27). Mutational inactivation of any of the nosDYF maturation genes results in an enzyme without the CuZ center (35, 45, 48). The components of the core maturation complex have not been isolated, due to the unsolved functional role, impeding the development of proper assays. Also, information about the transcriptional organization and regulation of the maturation genes is lacking. We found that the maturation genes are organized in a polycistronic nosDFYL tatE operon, which is followed by the nnrS operon. The principal factors which regulate these operons were identified. Further, we have expressed the nosF gene and show with the purified protein that it is an ATPase. NosF exhibits structural similarity to the ATPase of maltose or histidine ABC transporters.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The Escherichia coli strains DH10B (Gibco BRL) and XL-1Blue (Stratagene) were used as hosts for cloning and expression, respectively. MK21, a spontaneous Smr mutant (47) of Pseudomonas stutzeri ATCC14405, was the parent strain for the following mutants: MK418 (nosR::Tn5); MK402 and MK404 (both nosD::Tn5) (49); MKR2 (fnrA::kan) (11); MRD235 (dnrD::kan) (42); and MK118 (ΔnarL::kan) (20). The plasmids used in this study were BRH97 for nnrS promoter analysis (18); pBTE1, carrying a 1.017-kb fragment with tatE cloned into pBluescript II SK(+) (22); cDEN1, a pJA1 cosmid clone of a Sau3A genomic library with the nos genes (5); pMal-c2, an expression vector based on malE of E. coli under the control of ptac (New England Biolabs) (29); and pNS200, a pBR325 derivative carrying the maturation genes of the nos gene region (49).
Strains of P. stutzeri were grown in a synthetic asparagine- and citrate-containing medium at 30°C. Aerobic and denitrifying cultures were established as previously described (11) unless otherwise stated. The following antibiotics were used at the indicated concentrations (in micrograms per milliliter of medium): ampicillin, 100; gentamicin, 30; kanamycin, 50; and streptomycin, 200.
Recombinant DNA techniques.
Plasmid DNA was prepared by a modified alkaline cell lysis method (16). Spin columns with a silica membrane (Roche Diagnostics) were used for plasmid DNA purification and preparative isolation from agarose slabs. Recombinant plasmids were transformed into E. coli by electroporation (15). For DNA manipulation, we followed standard protocols (38) or the instructions for commercial products provided by the manufacturers. Restriction endonucleases and other enzymes were purchased from MBI Fermentas.
RNA analysis.
Cell sample preparation, RNA isolation, electrophoresis, nucleic acid transfer, and hybridization were done as described previously (43), but the time for precipitation of mRNA was reduced from overnight to 3 h. The following primers were used for the amplification of digoxigenin-labeled probes for Northern blot analysis: 5′-GTATCAAGGCCAGTTCACCA-3′ and 5′-TCATCAGGATGCCGTAGTTC-3′ (nosD) and 5′-TATGCGC TGCTCGCCATTCC-3′ and 5′-CGCCGATCAACGCCATCAAC-3′ (nnrS). The gene probe for nosD was amplified from the plasmid pNS200, and the probe for nnrS was amplified from the cosmid cDEN1. To obtain a signal from the tatE transcript, it was necessary to use an RNA probe. This probe was generated from plasmid pBTE1 (22), using an RNA-labeling kit (Roche Diagnostics) and the primers 5′-GTATCAGCGTCTGGCAACTCC-3′ and 5′-ctaatacgacgactcactatagggagaTCAGCTCCTGCTTGACGC-3′ (the lowercase letters indicate 5′ extension for the promoter sequence of T7 RNA polymerase). PCR-derived probes for the genes nirS and narG were prepared by the incorporation of digoxigenin-dUTP as described elsewhere (20). For membrane detection, the dioxetane derivative CDP-Star (Roche Diagnostics) was used as the chemiluminescent substrate.
The 5′ end of the nnrS transcript was mapped by primer extension (2). Total RNA was isolated from MK21 and MK402 which had been cultivated for 1 h under nitrate-denitrifying conditions. Reverse transcription was initiated from the 5′ γ-32P-labeled primer 5′-TCGCCGATCGATAACTTGCACGGAAGG-3′, which is complementary to the coding strand at positions 9119 to 9093 (18). The nucleotide sequence was obtained by the dideoxy chain termination method using the same primer with plasmid pBRH97 as the template.
Construction of plasmid pMal4F for nosF expression.
nosF was amplified by PCR from plasmid pNS200 with the primers 5′-tagacaggatccATGAACGCCGTCGAGATCCA-3′ and 5′-ttcactgtcgacTCATAGACGGCCCTCCTGAGC-3′ (the lowercase letters indicate 5′ added extensions to generate restriction sites for BamHI and SalI). PCR amplification was done with Fast Start Taq DNA polymerase (Roche Diagnostics). After restriction, the fragment was ligated in plasmid pMal-c2 in frame to the 3′ end of malE to give pMal4F. malE encodes the maltose-binding protein (MBP). The amino acids ISEFGS were added at the N terminus of NosF as a consequence of the cloning procedure. The integrity of the expression vector was verified by automatic fluorescence-based cycle sequencing using the Thermo-Sequenase cycle-sequencing kit (Amersham Biosciences) according to the instructions of the manufacturer.
Conditions for expression and purification of the NosF hybrid protein.
E. coli strain XL-1Blue was transformed with pMal4F, selecting for blue-white screening and ampicillin resistance. The strain was cultivated on a gyratory shaker (240 rpm) at 37°C in Luria-Bertani medium containing 0.2% glucose and 100 μg of ampicillin per ml of medium. After the culture had reached an optical density at 600 nm of 0.5, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.1 mM. Growth continued for 4 h at 22°C. The cells were harvested by centrifugation, and the pellet was stored at −22°C. The frozen cells were thawed, suspended in 30 ml of chromatography buffer (50 mM Tris-HCl, pH 7.4, 200 mM NaCl), and disrupted twice in a French press at 110 MPa. After centrifugation for 20 min at 15,000 × g, the supernatant was applied to a 5-ml column of amylose resin (New England Biolabs) equilibrated at 4°C with the buffer described above. The column was washed with 12 volumes of buffer and eluted with 10 mM maltose in chromatography buffer. Ten 2-ml fractions were collected and analyzed for protein content. If necessary, the fractions were concentrated using a Vivaspin 50 concentrator (Sartorius). MBP-NosF could be stored on ice for up to 4 days without loss of ATPase activity.
ATPase assay.
ATPase activity was determined colorimetrically using Na2HPO4 as the standard for following inorganic phosphate (Pi) release (7). The reaction mixture contained 37.5 to 150 μg of MBP-NosF in 30 μl of ATPase buffer (100 mM Tris-HCl, pH 8.0, 8 mM ATP). It was equilibrated for 3 min at 37°C before the reaction was started by the addition of MgCl2 (final concentration, 5 mM). The reaction was stopped after 2 min by transferring the mixture to reaction tubes containing 40 μl of 7.5% sodium dodecyl sulfate (SDS) (Sigma-Aldrich).
Analytical and immunochemical techniques.
SDS-polyacrylamide gel electrophoresis (PAGE) with a 10% acrylamide gel was used for protein separation. Immunochemical detection of MBP-NosF was done with anti-MBP antiserum (New England Biolabs) and protein A-horseradish peroxidase conjugate (Bio-Rad) according to the instructions of the manufacturers. Protein mass spectrometry was done with a Bruker Biflex IV instrument following in-gel trypsin digestion. Protein determination was done by the Bradford dye-binding assay. Inhibitors and nucleotides were purchased from Sigma-Aldrich; ATP and ADP were obtained from Boehringer Mannheim.
Nucleotide sequence accession numbers.
EMBL databank accession numbers for the nos gene region are as follows: nosR, Z13988; nosZ, M22628; nosDFY, X53676; nosL, Z69589; and tatE, nnrS, and ORF274, Z73914.
RESULTS
Kinetics of nosD transcription on downshift to nitrate denitrification.
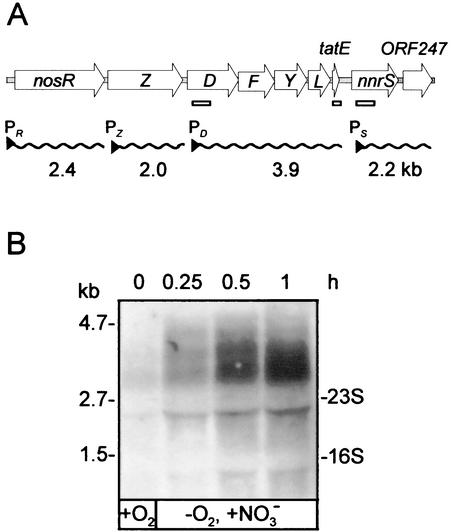
The regulation of the maturation genes and their transcriptional organization have not yet been investigated, although the promoters of nosR, nosZ, and nosD were characterized previously (9). Since nosD is the initial gene of the maturation gene cluster (Fig. 1A), we have studied the time course of its expression on shifting cells to nitrate denitrification (Fig. 1B). In a well-aerated culture (shaking frequency, 240 rpm) of strain MK21 (representing the wild-type background), only traces of nosD mRNA were detected. Upon the onset of nitrate denitrification by lowering the oxygen transfer rate (the shaking frequency was reduced to 120 rpm) and adding nitrate at a final concentration of 0.1%, the amount of nosD transcript increased steadily. At 30 to 60 min after induction, saturation was reached. The data established the time window for RNA sampling in subsequent induction experiments. We noticed mRNA instability on Northern blots. The size of the largest signal, ∼3.9 kb, exceeded by far the size of a monocistronic nosD transcript and led us to investigate the formation of a polycistronic message, making use of nosD mutants.
FIG. 1.
Physical map of the nos gene region and kinetics of the nosD operon transcription. (A) Physical map and transcriptional organization. The wavy lines indicate transcripts from the respective promoters identified in this work or previously for nosR (43) and nosZ (10). Promoters are represented by triangles and labeled P with a qualifying subscript. The open bars show the locations and sizes of probes used in Northern hybridization. (B) Time-resolved appearance of the nosD transcript on shift of an aerobic culture to nitrate denitrification. Strain MK21 (representing wild-type traits) was grown aerobically (shaking frequency, 240 rpm) and probed for a nosD transcript by Northern blot analysis (+O2). The culture was induced for denitrification by lowering the shaking frequency to 120 rpm (shift to low pO2) and adding 1 g of sodium nitrate per liter (−O2, +NO3−). The transcript was followed for 1 h after the downshift. The 0.6-kb nosD probe, isolation of mRNA, and conditions for hybridization were as specified in Material and Methods.
Evidence for a nosDFYL tatE operon.
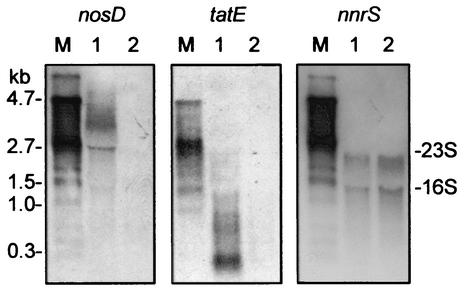
To probe for the cotranscription of nosD with other genes, mRNA was isolated from strain MK21 and two Tn5 insertion mutants, MK402 and MK404, 60 min after the shift from aerobic respiration to nitrate denitrification. MK402 is a nosD promoter mutant, whereas MK404 carries the insertion within the nosD coding region. The phenotype of both mutants is the synthesis of an N2OR with only the CuA center metallated. Northern blot analysis was done with the probes for nosD, tatE, and nnrS as described in Materials and Methods. Both mutants gave the same response. Figure 2 illustrates representative results obtained with the mutant MK402. With the probes for nosD and tatE, we found transcripts in MK21 but not in MK402, whereas probing for nnrS resulted in a transcript of ∼2.2 kb in both the wild-type background and the nosD mutant. The mutation in nosD was therefore polar on tatE but not on nnrS. This indicated a nosDFYL tatE operon and an independent promoter for nnrS. We exclude the presence of further transcriptional start sites within this operon for nosF, nosY, or nosL, because probing of total RNAs from MK402 and MK404 with these genes did not reveal any transcripts (data not shown).
FIG. 2.
Evidence for a nosDFYL tatE operon by mutational analysis. Total RNAs were prepared from MK21 (lanes 1) and the nosD::Tn5 mutant MK402 (lanes 2). Cells were shifted to nitrate-denitrifying conditions for 60 min (shaking frequency, 120 rpm; 0.1% NaNO3) prior to RNA isolation. Transcripts were detected by Northern hybridization with the DNA or RNA probes indicated at the top. The RNA molecular size marker I (M) was from Roche Diagnostics.
tatE mRNA could be detected only by the use of a riboprobe (see Materials and Methods) (Fig. 2). We presume that formation of a low-mass species is related to mRNA processing or is a result of modulating the translational efficiency of the polycistronic multifunctional nosD operon. Processing of tatE is unlikely to be detectable in the 3.9-kb operon transcript because of the small size of the tatE gene, 171 bp.
The independent promoter for nnrS was confirmed by primer extension analysis (Fig. 3). We located the transcript start site at position 9194 of the published sequence (18). The promoter carries a sequence motif, CTGAT-N4-ATCAA (positions 9230 to 9243), centered at −42.5 bp upstream of the transcript start site and with strong similarity to the canonical Fnr box, TTGAT-N4-ATCAA. We assume that this regulatory motif is involved in DnrD-dependent transcription of nnrS (see below). DnrD and its homologues, Nnr and Dnr, are thought to bind to an Fnr-type recognition motif (for a review, see reference 46). The primer extension with RNA isolated from MK402 also corroborated the findings from Northern blot analysis with this mutant. The size determined for the nnrS mRNA indicated that nnrS was transcribed together with the gene located downstream, orf247.
FIG. 3.
Organization of the nnrS promoter. (A) Mapping of the 5′ terminus of the nnrS transcript. Primer extension reactions were done with total RNAs extracted from MK21 (lane 1) and MK402 (lane 2), which had been cultured under nitrate-denitrifying conditions. The reaction products were separated on a sequencing gel together with a sequence ladder generated with the primer indicated in panel B. The complementary sequence of the transcription initiation site is shown. (B) Features of the nnrS promoter. The 5′ end of the transcript is labeled +1. Putative sites for binding polymerase (−10) and DnrD (−42.5) are boxed. The oligonucleotide that was used for primer extension is underlined. The first few N-terminal amino acids of the NnrS protein are shown in one-letter code. RBS, ribosome-binding site.
The regulators NosR and DnrD are required for activating the nosD operon.
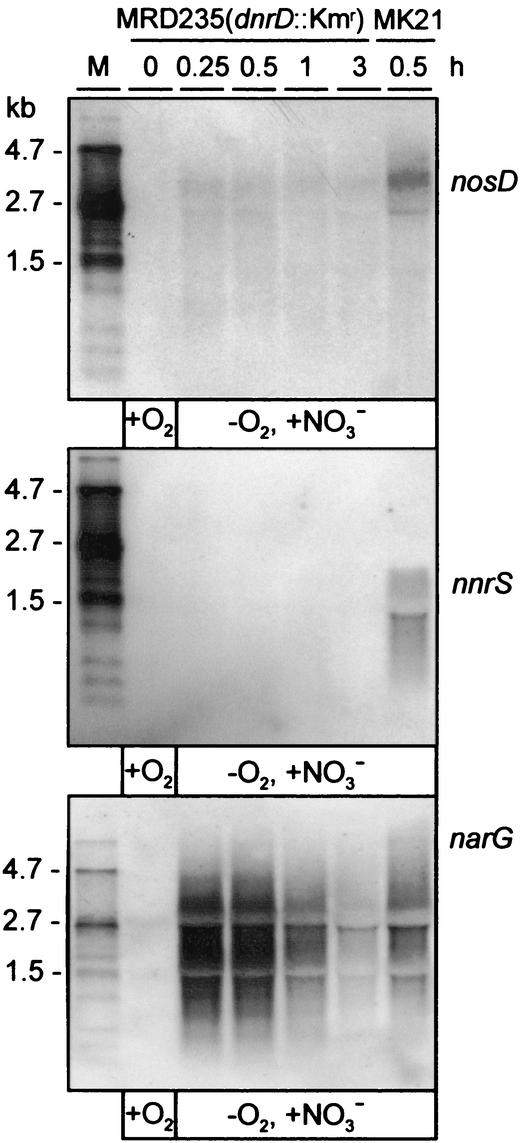
The appearance of the nosDFYL tatE transcript in response to denitrifying conditions led to the question of which regulatory proteins are involved in the transcriptional control of this operon. NosR had been shown to be required for nosZ transcription (10), but it remained open whether other nos genes are targets of NosR. It was also shown previously that NO is a key signal for the expression of denitrification genes in P. stutzeri and that processing of this signal involves the DnrD regulator, a member of the Crp-Fnr family (43). Therefore, we analyzed the nosR mutant MK418 and the dnrD mutant MRD235 for the presence of the nosDFYL tatE transcript. The absence of NosR abolished transcription of the nosD operon under both aerobic (i.e., the low-level transcription seen in Fig. 1B) and nitrate-denitrifying conditions (Fig. 4). To probe cells for the successful shift to the denitrifying state, the same mRNA was analyzed for transcripts from the nirS operon, which revealed the polycistronic and monocistronic species nirSTB and nirS, respectively (21). Activation of the nirS promoter is independent of NosR, and detection of the nirS mRNA thus proved the metabolic switch to denitrification.
FIG. 4.
Transcription of the nosD operon is dependent on NosR. Cells were conditioned as for Fig. 1. Total RNAs were prepared from the nosR::Tn5 mutant MK418 and from MK21 and analyzed at the indicated times by Northern blotting with gene probes for nosD (top) and nirS (bottom). M, RNA marker.
The absence of DnrD also affected nosDFYL tatE transcription. Under denitrifying conditions, the amount of mRNA from the nosD operon was substantially lowered in MRD235 compared to the wild type (Fig. 5). DnrD thus acted as an enhancer of transcription under denitrifying conditions. Since the nirS operon is under the control of DnrD, the successful shift of MRD235 to nitrate denitrification was proven in this case by probing for the expression of the nitrate reductase operon by hybridization with narG.
FIG. 5.
DnrD enhances the expression of the nosD operon and is essential for nnrS expression. Total RNAs from MRD235 and MK21 were subjected to Northern hybridization with the gene probes for nosD, nnrS, and narG. The cells were conditioned as for Fig. 1. M, RNA marker.
Transcription of the nnrS operon was DnrD dependent and induced by nitrate-denitrifying conditions. It reached a maximum after 15 min of induction (data not shown). We observed that transcription of the nnrS operon under nitrate-denitrifying conditions was completely arrested in the dnrD mutant (Fig. 5). Other than with the nosD operon, no effect of NosR on transcription of the nnrS operon was observed; MK418 exhibited the same levels of nnrS mRNA as MK21.
We also probed total RNA obtained from nitrate-induced cells for nosD operon transcription by the fnrA mutant MKR2 and the narL mutant MRL118. These mutants are defective in the anaerobic regulator FnrA (11) and the nitrate-dependent regulator (20) of the two-component system, NarXL, respectively. Since nosD transcripts were found in both mutants (data not shown), it excludes these regulators from the signal transduction pathway.
Construction of the nosF expression vector.
In an attempt to characterize biochemically the components encoded by the maturation gene cluster, we addressed the question of whether the nosF product indeed constitutes an ATPase as predicted from sequence comparison. For this purpose, nosF was overexpressed in E. coli. To facilitate purification, we employed a fusion strategy with the MBP of E. coli. Insertion of nosF into the plasmid pMal-c2 immediately downstream of the malE gene generated the expression vector pMal4F (see Materials and Methods). The fusion of nosF to malE in frame and the absence of point mutations were both verified by sequencing. pMal4F resulted in the overexpression of the MBP-NosF fusion protein in the host, E. coli XL-1Blue.
Purification and properties of NosF.
For the purification of the NosF hybrid protein, E. coli XL-1Blue(pMal4F) was grown at 37°C under aerobic conditions. The most favorable conditions to maximize the amount of soluble protein were induction of the cells with 0.1 mM IPTG and incubation for 4 h at 22°C. The soluble cell fraction of the induced cells exhibited a prominent band in SDS-PAGE at ∼77 kDa, the mass expected for MBP-NosF. This band was absent from uninduced cells but represented by far the most abundant protein in the induced cells. The identity of this band as MBP-NosF was proven with anti-MBP antiserum by Western blot analysis and by matrix-assisted laser desorption ionization-time of flight mass spectrometry. MBP-NosF was purified from cell extract by single-step affinity chromatography on an amylose matrix. A typical preparation yielded 15 mg of MBP-NosF from a 2-liter culture. The preparation was judged to be 95% homogenous based on SDS-PAGE.
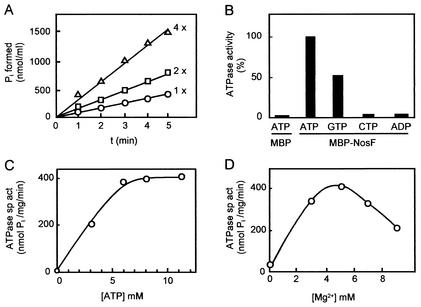
To determine whether MBP-NosF catalyzed ATP hydrolysis, we followed the release of Pi by a colorimetric assay (7). Under standard conditions, a specific activity of ∼400 nmol of Pi mg of protein−1 min−1 was determined. The reaction proceeded linearly with time and was proportional to the amount of MBP-NosF (Fig. 6A). MBP has no nucleotide-binding domain and does not bind or hydrolyze ATP. Nonetheless, the possibility was considered that the ATPase activity was due to the MBP moiety, for instance, by binding traces of the MalK ATPase of the host strain. The MBP protein was expressed in strain XL-1Blue from plasmid pMal-c2 and purified identically to the procedure for MBP-NosF. No notable ATP hydrolysis was found with MBP alone (Fig. 6B). Contamination with a nonspecific phosphatase as the source of ATP hydrolysis was also excluded, since ADP as a substrate did not result in Pi release. The reaction was largely specific for ATP. GTP was hydrolyzed at a rate of ∼50%, and CTP was hydrolyzed at <5% of the activity with ATP. A similar selectivity in the utilization of these nucleotides has been found for other ABC-type ATPases (1, 26, 34).
FIG. 6.
Enzymatic properties of NosF. (A) NosF-catalyzed ATPase activity proceeds linearly in time and is proportional to the protein concentration. ATPase activity was assayed in 90 μl of reaction buffer; the reaction was started by the addition of MgCl2; 37.5 (○), 75 (□), and 150 (▵) μg of MBP-NosF were used in the assays. (B) ATP hydrolysis is specific for ATP and is due to the NosF moiety of the MBP-NosF fusion protein; 150 μg of MBP or 50 μg of MBP-NosF in 30 μl of reaction buffer and 8 mM ATP, ADP, GTP, or CTP were used. (C) Dependence of enzyme activity on ATP concentration. (D) Dependence of ATPase activity on Mg2+ concentration. The assays were performed as described in Materials and Methods.
The ATPase activity of NosF was dependent on Mg2+ in the assay buffer, with the highest activity found in the presence of 5 mM Mg2+ at 8 mM ATP (Fig. 6C and D). Cu and Ca could not replace Mg. A Km value for ATP of 3 mM and a value of 10 mM for GTP were calculated under standard conditions. The ATPase activity of NosF was moderately pH dependent in the alkaline range. The highest activity resulted between pHs 8.0 and 8.5, while at pHs 7.5 and 9.3, the activity was lowered to 80%.
The sensitivity of NosF to inhibitors was investigated. ATPase activity was measured in standard assay buffer after a 30-min incubation period on ice with the inhibitor. Both typical and atypical behaviors of NosF with respect to ABC-type ATPases were found. The sulfhydryl reagent N-ethylmaleimide (NEM) at 0.1 mM lowered the activity by about half, whereas the substrate analog AMP-PNP inhibited NosF completely. Ouabain, a specific inhibitor for Na+ and K+ ATPases, affected ATP hydrolysis only slightly at 3 mM. Vanadate as an analog of Pi is thought to trap ADP in the transition state of ATP hydrolysis (12). Incubation with 10 mM orthovanadate reduced the ATPase activity of NosF by about half. Vanadate also impairs the ATPase activity of the purified ABC domain of the galactose importer MglA of Salmonella enterica serovar Typhimurium (36) and the MalFGK2 (13) and HisQMP2 complexes (28) but shows no effect on purified HisP (34) and MalK (19, 33). A 90% decrease in NosF-catalyzed ATPase activity was caused by 10 mM NaN3. Azide is an inhibitor of F0F1 ATPases and also acts on the oleandomycin exporter OleB of Streptomyces antibioticus (1) and weakly on MalK (33), but not on HisQMP2 (28), HisP (34), or the hemolysin exporter HlyB of E. coli (26).
DISCUSSION
A nosF knockout mutant results in an N2OR without the CuZ center (49). Here, we have shown that NosF has the activity of an ATPase, and we deduce from this that CuZ assembly involves an energy-requiring step driven by ATP hydrolysis. The 5′ in-frame fusion of nosF to malE resulted in a soluble, overexpressed MBP-NosF hybrid. MBP has been found to be superior to other fusions in keeping passenger proteins in a soluble state (25). Further, the affected C-terminal fusion has an additional beneficial effect on protein solubility (37).
NosF shows similarity to the ATPases of ABC transporters (49). To corroborate NosF as a member of this protein family, the crystal structure of MalK (14) was used to predict the secondary structure of NosF (Fig. 7). NosF shows a high degree of correspondence with MalK in the numbers, positions, and extents of β-sheets and α-helices, in spite of a low overall sequence identity. Nearly the same correspondence was observed when the crystal structure of HisP (23) was used as the template for secondary-structure prediction. Characteristic features of ABC-type ATPases, such as the Walker motifs A and B, the ABC signature, and the switch region, are located in NosF in the same positions as in the template. MalK belongs to a special group among the ABC-type ATPases because of its C-terminal domain of 149 amino acids, which has a regulatory function and acts on MalT, the activator of mal gene expression (for a review, see reference 4). NosF also has a C-terminal extension of 81 amino acids, but this region has no structural correspondence to MalK (Fig. 7). No clue to the function of this NosF domain exists. A further distinguishing feature of NosF is that its ABC signature, YSKGM, does not fully correspond to the conserved region of ABC-type ATPases, which have the consensus sequence LSGGQ. Recent findings based on vanadate-catalyzed photocleavage support the notion that this motif contacts the γ-phosphate of ATP (17).
FIG. 7.
Prediction of secondary structure of NosF. The protein was modeled on the crystal structure of MalK (Protein Data Bank file 1G29) using the ExPASy server of the Swiss Institute of Bioinformatics (http://www.expasy.ch). Extensions of predicted α-helices are shown by boxes; those of β-sheets are shown by arrows. The secondary structural elements of NosF are numbered according to those of MalK from Thermococcus litoralis (14). Distinct motifs of ABC-type ATPases are shaded. The C-terminal amino acids of NosF lack the structural elements of the C-terminal regulatory domain of MalK and could not be modeled from the template. Positionally identical residues of the two sequences are shown in boldface.
NEM is thought to inhibit the ATPase activity of ABC transporters by the covalent modification of the cysteine residue located in the P-loop (the Walker A motif). NosF lacks this residue; nevertheless, its ATPase activity was affected by NEM. The only two cysteine residues of NosF are located in and immediately behind the switch region. Thus, by modifying these residues, NEM might affect protein integrity. The switch region is thought to be involved in the signal transduction process between the transmembrane component and the ATPase of ABC transporters (40).
The nosF gene is expressed as part of the nosDFYL tatE operon. Although the nosD transcription start point is defined (9), no specific regulatory motifs are evident in the promoter region. Putative NarL binding sites for the nitrate response regulator, noted previously, appear fortuitous in light of the present results of NosR- and DnrD-dependent regulation. In aerobic cells, a small amount of N2OR is found, i.e., nos genes are transcribed constitutively at a low level. Under anaerobic N2O-respiring or nitrate-denitrifying conditions, nos gene expression is activated and leads to a manifold increase in cellular N2OR content. Transcription of nosZ requires NosR, a multidomain transmembrane protein and putative metalloprotein (45). We have shown here that NosR also activates the transcription of the nosD operon. Transcription of nosR, in turn, is activated in response to NO and depends on the DnrD regulator, a member of the Crp-Fnr family (43). We suggest a signal transduction pathway where NO derived from nitrate denitrification leads to activation of DnrD, which results in nosR expression and subsequent activation of the nosD operon: NO → DnrD → nosR/NosR → nosD.
It remains to be established to what extent this positive control circuit involves a sequence of direct interactions. The basal nos transcription allows the cell to also acquire N2OR in the absence of DnrD on long-term exposure to anaerobic conditions. The question of an additional signal derived from the redox status or the oxygen tension remains open. No Fnr-binding sites exist in the nosD promoter, and a mutation in fnrA did not affect nosD transcription.
Two other functions, encoded by nosL and tatE, are part of the nosD operon. NosL is a Cu protein; it was tentatively assigned a metallochaperone function directed at N2OR (32). nosL is consistently found as part of nosDFY gene clusters, emphasizing a role in the maturation process (44). TatE is a component of the Tat translocation machinery and assures complete processing and export of N2OR (22, 44). It was unexpected to find tatE cotranscribed with genes for N2OR maturation. A different stoichiometry required for the different functions encoded in polycistronic messages can be achieved by posttranscriptional control mechanisms, as exemplified for the ATPase operon of E. coli (30, 31) or the histidine transport operon of S. enterica serovar Typhimurium (41). The nosD operon thus may be subjected to regulation of translational efficiency or the stability of distinct mRNA segments. Probing for tatE has revealed a low-mass mRNA species, which is seen within this context.
The nnrS gene in P. stutzeri is adjacent to the maturation gene cluster but not part of the nosD operon. Nevertheless, it is expressed in response to denitrifying conditions and its transcription depends on DnrD. The nnrS gene product (previously called ORF396) had been predicted to be a 12-helix membrane protein (18). Recently, the homologous protein was purified from the purple bacterium Rhodobacter sphaeroides 2.3.4 and was shown to be a heme-Cu protein by optical and electron paramagnetic resonance spectroscopy (3). Mutational inactivation of nnrS resulted in an altered chemotactic behavior toward nitrate. In R. sphaeroides, nnrS is under the control of NnrR, another member of the Crp-Fnr regulator family. A putative NnrR recognition motif, CTGAT-N4-ATCAA, is located 122 bp upstream of the translation start site of nnrS (3). We found in silico a striking association of nnrS orthologues with nos genes, but orthologues also exist in the nondenitrifying Vibrio cholerae (accession no. Q9KPN6) and Pseudomonas putida (http://www.tigr.org), which suggests a function for NnrS that is not limited to denitrification. Northern blot analysis of P. stutzeri showed that nnrS is transcribed with orf247, located downstream, resulting in a 2.2-kb transcript. The orf247 product, composed of 247 amino acids, belongs to the family of short-chain dehydrogenases (18). Members of this family have an average of 250 residues per subunit, an N-terminal GXXXGXG coenzyme-binding pattern, and an active-site pattern of YXXXK and catalyze NAD(P)(H)-dependent oxidation-reduction reactions (24).
Acknowledgments
We thank E. Härtig for providing the RNA probe, U. Schiek and S. Berker for the primer extension experiment, H. Körner for mass spectrometric analysis, and E. Schneider for a critical reading of the manuscript.
The work was supported by the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie.
REFERENCES
- 1.Aparicio, G., A. Buche, C. Méndez, and J. A. Salas. 1996. Characterization of the ATPase activity of the N-terminal nucleotide binding domain of an ABC transporter involved in oleandomycin secretion by Streptomyces antibioticus. FEMS Microbiol. Lett. 141:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bartnikas, T. B., Y. S. Wang, T. Bobo, A. Veselov, P. Scholes, and J. P. Shapleigh. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem-copper protein. Microbiology 148:825-833. [DOI] [PubMed] [Google Scholar]
- 4.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, C., and W. G. Zumft. 1992. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for denitrification. J. Bacteriol. 174:2394-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, K., M. Tegoni, M. Prudêncio, A. S. Pereira, S. Besson, J. J. Moura, I. Moura, and C. Cambillau. 2000. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat. Struct. Biol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 7.Chifflet, S., A. Torriglia, R. Chiesa, and S. Tolosa. 1988. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal. Biochem. 168:1-4. [DOI] [PubMed] [Google Scholar]
- 8.Ciccarelli, F. D., R. R. Copley, T. Doerks, R. B. Russell, and P. Bork. 2002. CASH-a β-helix domain widespread among carbohydrate-binding proteins. Trends Biochem. Sci. 27:59-62. [DOI] [PubMed] [Google Scholar]
- 9.Cuypers, H., J. Berghöfer, and W. G. Zumft. 1995. Multiple nosZ promoters and anaerobic expression of nos genes necessary for Pseudomonas stutzeri nitrous oxide reductase and assembly of its copper centers. Biochim. Biophys. Acta 1264:183-190. [DOI] [PubMed] [Google Scholar]
- 10.Cuypers, H., A. Viebrock-Sambale, and W. G. Zumft. 1992. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J. Bacteriol. 174:5332-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuypers, H., and W. G. Zumft. 1993. Anaerobic control of denitrification in Pseudomonas stutzeri escapes mutagenesis of an fnr-like gene. J. Bacteriol. 175:7236-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson, A. L. 2002. Mechanism of coupling of transport to hydrolysis in bacterial ATP-binding cassette transporters. J. Bacteriol. 184:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, A. L., S. S. Laghaeian, and D. E. Mannering. 1996. The maltose transport system of Escherichiacoli displays positive cooperativity in ATP hydrolysis. J. Biol. Chem. 271:4858-4863. [PubMed] [Google Scholar]
- 14.Diederichs, K., J. Diez, G. Greller, C. Müller, J. Breed, C. Schnell, C. Vonrhein, W. Boos, and W. Welte. 2000. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC-transporter of the archaeon Thermococcus litoralis. EMBO J. 19:5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feliciello, I., and G. Chinali. 1993. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia coli. Anal. Biochem. 212:394-401. [DOI] [PubMed] [Google Scholar]
- 17.Fetsch, E. E., and A. L. Davidson. 2002. Vanadate-catalyzed photocleavage of the signature motif of an ATP-binding cassette (ABC) transporter. Proc. Natl. Acad. Sci. USA 99:9685-9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glockner, A. B., and W. G. Zumft. 1996. Sequence analysis of an internal 9.72-kb segment from the 30-kb denitrification gene cluster of Pseudomonas stutzeri. Biochim. Biophys. Acta 1277:6-12. [DOI] [PubMed] [Google Scholar]
- 19.Greller, G., R. Horlacher, J. DiRuggiero, and W. Boos. 1999. Molecular and biochemical analysis of MalK, the ATP-hydrolyzing subunit of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 274:20259-20264. [DOI] [PubMed] [Google Scholar]
- 20.Härtig, E., U. Schiek, K.-U. Vollack, and W. G. Zumft. 1999. Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J. Bacteriol. 181:3658-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Härtig, E., and W. G. Zumft. 1999. Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeri during the transition from aerobic respiration to denitrification: evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system. J. Bacteriol. 181:161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heikkilä, M. P., U. Honisch, P. Wunsch, and W. G. Zumft. 2001. Role of the Tat transport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 183:1663-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, L.-W., I. X. Wang, K. Nikaido, P.-Q. Liu, G. F.-L. Ames, and S.-H. Kim. 1998. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature 396:703-707. [DOI] [PubMed] [Google Scholar]
- 24.Jörnvall, H., J.-O. Höög, and B. Persson. 1999. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 445:261-264. [DOI] [PubMed] [Google Scholar]
- 25.Kapust, R. B., and D. S. Waugh. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8:1668-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koronakis, V., C. Hughes, and E. Koronakis. 1993. ATPase activity and ATP/ADP-induced conformational change in the soluble domain of the bacterial protein translocator HlyB. Mol. Microbiol. 8:1163-1175. [DOI] [PubMed] [Google Scholar]
- 27.Kroneck, P. M. H., W. A. Antholine, J. Riester, and W. G. Zumft. 1988. The cupric site in nitrous oxide reductase contains a mixed-valence [Cu(II), Cu(I)] binuclear center: a multifrequency electron paramagnetic resonance investigation. FEBS Lett. 242:70-74. [DOI] [PubMed] [Google Scholar]
- 28.Liu, C. E., and G. F.-L. Ames. 1997. Characterization of transport through the periplasmic histidine permease using proteoliposomes reconstituted by dialysis. J. Biol. Chem. 272:859-866. [DOI] [PubMed] [Google Scholar]
- 29.Maina, C. V., P. D. Riggs, A. G. Grandea, B. E. Slatko, L. S. Moran, J. A. Tagliamonte, L. A. McReynolds, and C. di Guan. 1988. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene 74:365-373. [DOI] [PubMed] [Google Scholar]
- 30.McCarthy, J. E. 1990. Post-transcriptional control in the polycistronic operon environment: studies of the atp operon of Escherichia coli. Mol. Microbiol. 4:1233-1240. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy, J. E., B. Gerstel, B. Surin, U. Wiedemann, and P. Ziemke. 1991. Differential gene expression from the Escherichia coli atp operon mediated by segmental differences in mRNA stability. Mol. Microbiol. 5:2447-2458. [DOI] [PubMed] [Google Scholar]
- 32.McGuirl, M. A., J. A. Bollinger, N. Cosper, R. A. Scott, and D. M. Dooley. 2001. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J. Biol. Inorg. Chem. 6:189-195. [DOI] [PubMed] [Google Scholar]
- 33.Morbach, S., S. Tebbe, and E. Schneider. 1993. The ATP-binding cassette (ABC) transporter for maltose/maltodextrins of Salmonella typhimurium. Characterization of the ATPase activity associated with the purified MalK subunit. J. Biol. Chem. 268:18617-18621. [PubMed] [Google Scholar]
- 34.Nikaido, K., P.-Q. Liu, and G. F.-L. Ames. 1997. Purification and characterization of HisP, the ATP-binding subunit of a traffic ATPase (ABC transporter), the histidine permease of Salmonella typhimurium. Solubility, dimerization, and ATPase activity. J. Biol. Chem. 272:27745-27752. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen, T., B. C. Berks, J. Sanders-Loehr, D. M. Dooley, W. G. Zumft, and A. J. Thomson. 2000. The catalytic center in nitrous oxide reductase, CuZ, is a copper sulfide cluster. Biochemistry 39:12753-12756. [DOI] [PubMed] [Google Scholar]
- 36.Richarme, G., A. El Yaagoubi, and M. Kohiyama. 1993. The MglA component of the binding protein-dependent galactose transport system of Salmonella typhimurium is a galactose-stimulated ATPase. J. Biol. Chem. 268:9473-9477. [PubMed] [Google Scholar]
- 37.Sachdev, D., and J. M. Chirgwin. 1998. Order of fusions between bacterial and mammalian proteins can determine solubility in Escherichia coli. Biochem. Biophys. Res. Commun. 244:933-937. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Saunders, N. F. W., J. J. Hornberg, W. N. M. Reijnders, H. V. Westerhoff, S. de Vries, and R. J. M. van Spanning. 2000. The NosX and NirX proteins of Paracoccus denitrificans are functional homologues: their role in maturation of nitrous oxide reductase. J. Bacteriol. 182:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speiser, D. M., and G. F.-L. Ames. 1991. Salmonella typhimurium histidine periplasmic permease mutations that allow transport in the absence of histidine-binding proteins. J. Bacteriol. 173:1444-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stern, M. J., E. Prossnitz, and G. F.-L. Ames. 1988. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol. Microbiol. 2:141-152. [DOI] [PubMed] [Google Scholar]
- 42.Vollack, K.-U., E. Härtig, H. Körner, and W. G. Zumft. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 31:1681-1694. [DOI] [PubMed] [Google Scholar]
- 43.Vollack, K.-U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183:2516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wunsch, P., M. Herb, H. Wieland, U. M. Schiek, and W. G. Zumft. 2003. Requirements for CuA and Cu-S center assembly of nitrous oxide reductase deduced from complete periplasmic enzyme maturation in the nondenitrifier Pseudomonas putida. J. Bacteriol. 185:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zumft, W. G. 2002. Nitric oxide signaling and NO dependent transcriptional control in bacterial denitrification by members of the FNR-CRP regulator family. J. Mol. Microbiol. Biotechnol. 4:277-286. [PubMed] [Google Scholar]
- 47.Zumft, W. G., K. Döhler, and H. Körner. 1985. Isolation and characterization of transposon Tn5-induced mutants of Pseudomonas perfectomarina defective in nitrous oxide respiration. J. Bacteriol. 163:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zumft, W. G., and P. M. H. Kroneck. 1996. Metal-center assembly of the bacterial multicopper enzyme nitrous oxide reductase. Adv. Inorg. Biochem. 11:193-221. [Google Scholar]
- 49.Zumft, W. G., A. Viebrock-Sambale, and C. Braun. 1990. Nitrous oxide reductase from denitrifying Pseudomonas stutzeri: genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. Eur. J. Biochem. 192:591-599. [DOI] [PubMed] [Google Scholar]