Abstract
Additional targets of CodY, a GTP-activated repressor of early stationary-phase genes in Bacillus subtilis, were identified by combining chromatin immunoprecipitation, DNA microarray hybridization, and gel mobility shift assays. The direct targets of CodY newly identified by this approach included regulatory genes for sporulation, genes that are likely to encode transporters for amino acids and sugars, and the genes for biosynthesis of branched-chain amino acids.
Bacteria have evolved a variety of mechanisms to accommodate gene expression to changes in nutritional availability. Some of these mechanisms are specific to a particular gene or operon. In other cases, regulatory proteins control large groups of genes of related function, such as the nitrogen metabolism genes regulated by the Ntr system in enteric bacteria (43) and by TnrA in Bacillus subtilis (17) and the carbon metabolism genes regulated by CcpA in gram-positive bacteria (13) and catabolic gene activator protein-cyclic AMP complex in gram-negative bacteria (59). Even broader forms of regulation are mediated by the leucine-responsive protein (Lrp) of gram-negative bacteria and the sigma-B protein of B. subtilis. Lrp and sigma-B control the transcription of operons that have diverse functions but have a common need to be expressed under a particular set of environmental conditions (50, 54). Lrp regulates the biosynthesis of leucine, isoleucine, valine, serine, glycine, and glutamate; the degradation of serine and threonine; transport of peptides, amino acids, and sugars; and production of fimbriae in response to the availability of leucine and serine (50). Sigma-B activates transcription of a host of genes when cells are exposed to excessive heat, ethanol, salt, or acid (54). Sigma-B responds through a complex, multibranched signal transduction pathway.
The B. subtilis CodY protein also has broad effects on gene expression. CodY is a GTP-binding repressor of several genes that are normally quiescent when cells are growing in a rich medium (57). A high concentration of GTP activates CodY as a repressor (57). When the growth rate of B. subtilis slows down because of limitation of the carbon or nitrogen or phosphorus source, the GTP level drops (39, 40), CodY loses repressing activity, and targets of CodY repression are transcribed. The known targets of CodY in B. subtilis include the genes that encode transport systems for dipeptides (dpp) (65) and γ-aminobutyrate (gabP) (16); catabolic pathways for acetate (acsA) (S. H. Fisher, personal communication), urea (ureABC) (71), histidine (hut) (18), arginine (rocABC and rocDEF) (B. Belitsky, personal communication), and branched-chain keto acids (the bkd operon) (12); an enzyme of surfactin synthesis (srfAA) (63); the transcription factor for DNA uptake genes (comK) (63); a ComA aspartyl phosphate phosphatase and its inhibitor (rapC-phrC) (37); motility and chemotaxis (hag, fla/che) (45; F. Bergara, C. Ibarra, J. Iwamasa, R. Aguilera, and L. M. Màrquez-Magaña, submitted for publication); and aconitase (citB) (33). CodY also regulates its own synthesis (56). Moreover, CodY is a highly conserved protein in the low-G+C group of gram-positive bacteria (57). In Lactococcus lactis, CodY represses expression of extracellular and intracellular peptidases and a peptide uptake system (23, 24). This range of targets suggests that CodY has a broad role in repressing, during rapid exponential growth phase, those genes whose products would allow the cell to adapt to poor nutritional conditions by swimming to a better environment, by taking up potential nutrients, and by metabolizing those nutrients to support continued growth. If so, it seems likely that many additional genes are under CodY control. Direct interaction of CodY with the regulatory regions of target genes has been demonstrated only for the dpp (62), srfAA (63), comK (63), cod (56), and citB (33) transcription units, however.
Some as yet unidentified CodY target genes in B. subtilis are likely to be involved in spore formation. When B. subtilis cells enter stationary phase, they have two choices. They can remain in a slow-growth or no-growth state or they can initiate sporulation (67). The onset of sporulation is dependent on nutrient limitation (60) and a consequent drop in the pool of GTP (40). Remarkably, CodY appears to be a major component of this regulation as well. Thus, sporulation of wild-type cells is inhibited in a medium that is highly enriched, but a codY null mutant grown in the same medium sporulates at high efficiency (57). The effect of a codY mutation can be mimicked by treating cells with a drug that causes a drop in the intracellular pool of GTP (20, 46), implying that in response to GTP excess, CodY represses at least one gene whose normal function is required for sporulation.
To assess the breadth of the CodY regulon, we used DNA microarray analysis to compare the pattern of transcripts found in a codY mutant to the pattern found in wild-type cells. Hundreds of genes organized in dozens of operons appeared to be directly or indirectly controlled by CodY. We then used antibody to CodY to detect segments of the B. subtilis chromosome that could be cross-linked to CodY in vivo. Combining the results of these two approaches, we identified many genes as candidates for direct targeting by CodY. For several of these candidates, we have confirmed the microarray results by assays of lacZ fusions to the promoter regions and have shown that CodY binds to the regulatory regions in vitro. The confirmed targets surprisingly include the operons for biosynthesis of branched-chain amino acids.
MATERIALS AND METHODS
Bacterial strains and their construction.
B. subtilis strains used in this study are listed in Table 1. Strains FU382 and FU383 were constructed by using plasmid pMUTIN2 (70) and primer pairs (GCCGAAGCTTGGATTCAGCATCTGCCGAAT/GCGCAGATCTCGGCAATGAATCAATCATGG and GCCGAAGCTTGATCACACAAGGAATCGATAG/GCGCAGATCTGATACAACCGTTTCCACAGA; coding sequences from the ilvB and ilvD genes, respectively, are underlined) as described previously (72). Isogenic codY+ and ΔcodY strains, each carrying pMUTIN-integrational disruptions, were constructed as follows. Strains PS29 (codY+) and PS37 (ΔcodY) were separately transformed with DNAs of the pMUTIN disruptants for ilvB, ilvD, ybgE, yufN, yufO, yurP, yurN, ykfA, and yhdG, selecting for erythromycin-resistant colonies (at 0.3 μg/ml) on tryptose-blood-agar base plates containing 10 mM glucose. The presence of gid::spc (a marker linked to codY), pMUTIN integration, and ΔcodY in the transformants was confirmed by resistance to spectinomycin (60 μg/ml) and erythromycin (0.15 μg/ml) and by the appearance of a PCR product in ΔcodY strains that is shorter by 250 bp than that of codY+ strains when amplified with the primer pair CCGGAATTCAATATGAGGAATGTTTAGGAGG/CGCGGATCCAACCCGAGAAATAAAGCTTATTG.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| PS29 | trpC2 gid::spc | 65 |
| PS37 | trpC2 gid::spc ΔcodY | 63 |
| PS56 | trpC2 abrB::(cat::tet) ΔamyE::(Φdpp′-lacZ neo) | P. Serror |
| PS83 | trpC2 abrB::(cat::tet) gid::spc ΔcodY ΔamyE::(Φdpp′-lacZ neo) | PS56 × DNA PS37 |
| FU382 | trpC2 ilvB::pMUTIN2 | This study |
| FU383 | trpC2 ilvD::pMUTIN2 | This study |
| YBGEd | trpC2 ybgE::pMUTIN2 | JAFANa |
| YUFNd | trpC2 yufN::pMUTIN2 | JAFAN |
| YUFOd | trpC2 yufO::pMUTIN2 | JAFAN |
| BFS1337 | trpC2 yurP::pMUTIN2 | Micadob |
| BFS1251 | trpC2 yurN::pMUTIN2 | Micado |
| BFS1807 | trpC2 ykfA::pMUTIN2 | Micado |
| YHDGd | trpC2 yhdG::pMUTIN2 | JAFAN |
| FU384 | trpC2 gid::spc ilvB::pMUTIN2 | This study |
| FU385 | trpC2 gid::spc ΔcodY ilvB::pMUTIN2 | This study |
| FU386 | trpC2 gid::spc ilvD::pMUTIN2 | This study |
| FU387 | trpC2 gid::spc ΔcodY ilvD::pMUTIN2 | This study |
| FU388 | trpC2 gid::spc ybgE::pMUTIN2 | This study |
| FU389 | trpC2 gid::spc ΔcodY ybgE::pMUTIN2 | This study |
| FU390 | trpC2 gid::spc yufN::pMUTIN2 | This study |
| FU391 | trpC2 gid::spc ΔcodY yufN::pMUTIN2 | This study |
| FU392 | trpC2 gid::spc yufO::pMUTIN2 | This study |
| FU393 | trpC2 gid::spc ΔcodY yufO::pMUTIN2 | This study |
| FU394 | trpC2 gid::spc yurP::pMUTIN2 | This study |
| FU395 | trpC2 gid::spc ΔcodY yurP::pMUTIN2 | This study |
| FU396 | trpC2 gid::spc yurN::pMUTIN2 | This study |
| FU397 | trpC2 gid::spc ΔcodY yurN::pMUTIN2 | This study |
| FU398 | trpC2 gid::spc ykfA::pMUTIN2 | This study |
| FU399 | trpC2 gid::spc ΔcodY ykfA::pMUTIN2 | This study |
| FU400 | trpC2 gid::spc yhdG::pMUTIN2 | This study |
| FU401 | trpC2 gid::spc ΔcodY yhdG::pMUTIN2 | This study |
| FU407 | trpC2 gid::spc amyE::(cat PyufN-lacZ) | This study |
| FU408 | trpC2 gid::spc ΔcodY amyE::(cat PyufN-lacZ) | This study |
JAFAN, Japan Functional Analysis Network for B. subtilis (http://bacillus genome.ad.jp/).
Micado, Microbial Advanced Database Organization (http://locus.jouy.inra.fr/micado).
B. subtilis strains FU407 and FU408 were constructed as follows. The yufN promoter region was amplified by PCR by using chromosomal DNA of strain 168 as a template and the primer pair 5′-AGTCGCTAGTCTAGAGAAAACGCACTGCTTGC-3′ and 5′-GATCGCGGATCCTTAGATCACAAGTGACATCG-3′ (the underlined sequences are upstream and downstream from the promoter region, respectively). The PCR product was doubly digested with XbaI and BamHI and then ligated to the large XbaI-BamHI fragment of plasmid pCRE-test2 (47) from which Pspac had been removed. The ligated DNA was used for transformation of E. coli strain DH5α to ampicillin resistance (50 μg/ml). After the sequence of the cloned DNA was confirmed, the plasmid was linearized with PstI and used to transform strains PS29 (codY+) and PS37 (ΔcodY) to chloramphenicol resistance. In the resulting transformants, the yufN-lacZ fusion had integrated at the amyE locus by double-crossover recombination.
Growth of cells and extraction of RNA for microarray analysis.
Strains PS29 (codY+) and PS37 (ΔcodY) were grown in minimal medium (72) supplemented with glucose (0.5%), glutamine (0.2%), and a mixture of 16 other amino acids (only histidine, tyrosine, and asparagine were omitted) (1) until the optical density at 600 nm (OD600) reached 1.0. To examine the effects of the 16-amino-acid mixture on gene expression in wild-type cells, strain 168 (trpC2) was grown in the glucose- and glutamine-supplemented minimal medium (minimal glucose-glutamine medium), as described above, with and without the 16 amino acids, until the OD600 reached 0.5.
Extraction of RNA from 100-ml portions of the cultures (73), preparation of fluorescently labeled cDNA (52), and hybridization to microarrays (35, 73) were as described previously. The arrays were spotted with PCR products corresponding to 4,005 B. subtilis open reading frames as well as the human β-actin gene and calf thymus DNA as negative controls (73). Fluorescence intensity was determined by using the GMS 418 Array Scanner (Affymetrix) and ImaGene software (version 3) (BioDiscovery, Inc.). Each spot was tested in duplicate, and the hybridization results were averaged for the two samples. Background was defined as the average intensities of eight spots of calf thymus DNA and four spots of the β-actin gene.
Cultures of strains PS56 (abrB) and PS83 (abrB ΔcodY) were grown at 37°C in DS medium (19), a nutrient broth-based medium in which cells grow rapidly and then sporulate after entering stationary phase. Samples were removed when the absorbance at 600 nm reached 0.5 (mid-exponential phase). (AbrB is a repressor of early stationary-phase gene expression [68] whose targets overlap with those of CodY [65].) Additional samples were harvested during early stationary phase. RNA was harvested from each culture and prepared for hybridization as described by Britton et al. (5). The arrays were spotted with 4,074 PCR products corresponding to B. subtilis open reading frames as well as with four Escherichia coli genes as negative controls (5). The hybridization results were scanned by using a GenePix 4000B scanner (Axon Instruments, Inc.) and analyzed with GenePix 3.0 software (Axon Instruments, Inc.) (5). The entire procedure was carried out four times, and the results were averaged.
Chromatin immunoprecipitation-microarray (ChIP-to-chip) experiments.
An overnight culture of B. subtilis strain PS56 (abrB) on Luria-Bertani agar was used to inoculate a 50-ml culture in DS medium to give an initial OD600 of 0.05. Cross-linking with formaldehyde, extraction and shearing of DNA, and immunoprecipitation generally followed the protocol of Quisel et al. (55) with differences in details noted. When the cells growing at 37°C had reached an OD600 of 0.4 to 0.6, the cultures were treated for 30 min with formaldehyde (1% final concentration) in 10 mM sodium phosphate buffer (pH 7.0). Glycine was added to 125 mM final concentration, and the cultures were incubated for an additional 5 min. Cells were washed twice with 40 ml of phosphate-buffered saline (pH 7.3) (2), resuspended in 1 ml of IP buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 0.5% Triton X-100) supplemented with 50 μl of 1× protease inhibitor cocktail (Roche) and 10 mg of lysozyme, and incubated at 37°C for 20 min. DNA in the lysate was sheared by sonication (Branson 250 microtip sonicator) to give an average fragment size of 300 to 1,000 bp. Ten microliters of the supernatant fluid of subsequent centrifugation was removed and saved for later analysis (total DNA). The remainder of the supernatant fluid was precleared by incubation with one-tenth volume of 50% protein A-Sepharose slurry (Sigma) for 1 h at 4°C. After centrifugation, CodY and CodY-DNA complexes in the supernatant fluid were immunoprecipitated overnight at 4°C by using a rabbit antibody that is highly specific to CodY (57), followed by incubation with 50 μl of a 50% protein A-Sepharose slurry (1 h at 4°C). Complexes were washed four times (15 min each) with 1 ml of IP buffer. The slurry was resuspended in 150 μl of elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, and 1% sodium dodecyl sulfate). The 10-μl total-DNA sample was mixed with 150 μl of elution buffer. To reverse formaldehyde-induced cross-links, the immunoprecipitated and total-DNA samples were incubated at 65°C overnight. Supernatants were collected and treated at 37°C for 2 h with 150 μl of Tris-EDTA buffer (TE) containing glycogen (0.27 mg/ml) and proteinase K (100 μg/ml). The DNA was purified by phenol-chloroform extraction, precipitated with isopropanol, and washed with 70% ethanol. Immunoprecipitated DNA was resuspended in 25 μl of TE, and the total DNA sample was resuspended in 100 μl of TE.
PCR amplification of DNA, differential fluorescence labeling, hybridization to microarrays, and array scanning were done according to the protocols at http://microarrays.org/protocols.html. The entire procedure was carried out five times, and the results were averaged. The enrichment factor for a given gene was calculated as the ratio of hybridized immunoprecipitated DNA to hybridized total DNA, normalized by using Resolver software (Rosetta).
Cell growth and β-galactosidase assays.
B. subtilis cells (codY+ and ΔcodY) with lacZ fusions integrated in the target genes or integrated at the amyE locus were grown overnight at 30°C on tryptose-blood-agar base plates containing 10 mM glucose and erythromycin (0.3 μg/ml) and spectinomycin (60 μg/ml). Strains FU407 and 408 were grown in the same medium with chloramphenicol (5 μg/ml) and spectinomycin. The overnight cultures were used to inoculate 50 ml of minimal glucose-glutamine medium with 16 amino acids, as described above, and were incubated with shaking at 37°C. At various times, 1-ml samples were withdrawn and β-galactosidase activity was determined as previously described (72).
Purification of CodY and gel mobility shift assays.
E. coli strain KS272 carrying pKT1, a plasmid in which a C-terminal, six-histidine-tagged version of the codY gene is under the control of the araBAD promoter (33), was grown in Luria broth containing ampicillin (50 μg/ml) until the absorbance at 600 nm reached 0.7. l-Arabinose was added to give a final concentration of 0.2%, and incubation was continued for an additional 4 to 5 h. After sonication, the soluble extract was treated with streptomycin sulfate (62) to remove ribosomes and nucleic acids, and the soluble fraction was mixed with Talon Co+ beads (Clontech). After several washes, CodY-His6 protein was eluted with increasing concentrations of imidazole. The preparation was free of contaminating proteins, as determined by Coomassie blue staining of a sodium dodecylsulfate-polyacrylamide gel.
The regulatory regions of genes to be tested were amplified by PCR by using B. subtilis chromosomal DNA as a template and two primers, one of which was radioactively labeled. The PCR products ranged from 199 to 525 bp in length. Primer labeling with T4 polynucleotide kinase and [γ-32P]ATP has been described previously (34). In some cases, enough was known about the transcription unit to design a probe that would be sure to include any likely regulatory sites. When there was insufficient knowledge, we prepared probes that extended from the beginning of the coding sequence to a position several hundred base pairs upstream that overlapped with the end of the neighboring coding sequence.
Labeled DNA was mixed with increasing amounts of CodY protein in a 10-μl reaction mixture that contained 20 mM Tris-Cl [pH 8.0], 50 mM sodium glutamate, 10 mM MgCl2, 5 mM EDTA, 0.05% (vol/vol) Nonidet P-40 (Igepal; Sigma Chemical Co.), 5% (vol/vol) glycerol, and 250 ng of calf thymus DNA. Where indicated, GTP was also present at 2 mM. After 20 min of incubation at room temperature, the samples were loaded on a 12% polyacrylamide gel that was running at 110 V. Subsequent electrophoresis in 35 mM HEPES and 43 mM imidazole (pH 7.4) was at 150 V for 90 to 150 min. The gels were dried under vacuum and exposed to a phosphorimager screen before analysis with a Molecular Dynamics Storm 860 Imager and ImageQuant version 1.2 Macintosh software.
RESULTS
Transcript analysis.
CodY was originally defined as a factor that exerts repression of the dpp and hut operons in a minimal medium containing an excess of glutamine and 16 other amino acids (1, 18, 65). For cells grown in such a medium, we identified, by whole genome microarray analysis, 124 genes located in about 70 apparent transcription units that were overexpressed by a factor of 3 or more in a codY null mutant compared to a wild-type strain. The 54 genes whose transcript level was most highly derepressed by the codY mutation (as well as other genes previously identified as being under CodY control) are listed in Table 2. (Genes whose highest level of hybridization was less than twofold greater than the background were excluded from this analysis.) Most of these genes were repressed in wild-type cells by addition of the amino acid mixture (Table 2), but not all genes repressed by the amino acids were derepressed by a codY mutation (data not shown). Notably, the biosynthetic pathways for arginine, cysteine, methionine, and the branched-chain amino acids were strongly repressed by the amino acid mixture (reference 42 and data not shown), but of these, only the pathway for branched-chain amino acid biosynthesis (and one of two genes for asparagine synthetase [asnH]) were derepressed in a codY mutant (Table 2). Another 27 genes located in 20 transcription units appeared to be dependent on CodY for their expression. A few of those genes are included in Table 2. There is no prior evidence for positive regulation by CodY. The results of these microarray experiments are available at the KEGG Expression Database website (http://www.genome.ad.jp/kegg/expression/).
TABLE 2.
Selected results of transcript analysisa
| Gene designation | Transcript ratio
|
ChIP enrichment factorb | Functional assignmentc | ||
|---|---|---|---|---|---|
| ΔcodY/codY+, MM + AAs | Wild type in MM/wild type in MM + AAs | ΔcodY/codY+, DS medium | |||
| Previously known targets | |||||
| acsA | 4.4 | 2.3 | 2.4 | 2.10 | Acetyl CoA synthetase |
| dppA | 55 | 7.1 | 7.1 | 2.78 | d-Ala-aminopeptidase |
| dppB | 65 | 7.1 | 4.8 | ∼1.0e | Dipeptide permease |
| dppC | 20 | 5.4 | 6.4 | ∼1.0 | Dipeptide permease |
| dppD | 26 | 7.5 | 5.7 | ∼1.0 | Transport ATPase |
| dppE | 28 | 7.0 | 9.2 | ∼1.0 | Dipeptide binding protein |
| srfAA | 1.0 | 3.6 | 0.55 | ∼1.0 | Surfactin synthetase, competence factor |
| comK | 2.5 | 1.5 | 1.3 | 1.63 | Competence transcription factor |
| ureA | 3.7 | 5.4 | 1.4 | 1.40 (ywmG) | Urease subunit |
| ureB | 2.6 | 2.4 | 2.8 | 1.23 | Urease subunit |
| ureC | 0.9 | 0.7 | 2.5 | 1.25 | Urease subunit |
| hag | 1.0 | 1.5 | 0.38 | ∼1.0 | Flagellin |
| bkdR | 0.4 | 0.9 | NDd | ∼1.0 | Regulatory protein for branched- chain, keto acid dehydrogenase operon |
| ptb | 1.3 | 0.4 | ND | ∼1.0 | Phosphate butyryltransferase |
| bcd | 1.1 | 0.9 | ND | ∼1.0 | Leucine dehydrogenase |
| buk | 0.4 | 0.7 | ND | ∼1.0 | Butyrate kinase |
| lpdV | 1.3 | 0.8 | ND | ∼1.0 | BCKA dehydrogenase subunit |
| bkdAA | 1.1 | 0.9 | ND | ∼1.0 | BCKA dehydrogenase subunit |
| bkdAB | 0.9 | 0.6 | ND | ∼1.0 | BCKA dehydrogenase subunit |
| bkdB | 0.6 | 0.9 | ND | ∼1.0 | BCKA dehydrogenase subunit |
| hutP | 11.1 | 1.3 | 1.3 | ∼1.0 | Histidine utilization regulator |
| rapA | 2.8 | 2.5 | 0.9 | 2.24 | Spo0F∼P phosphatase |
| rapC | 0.7 | 0.8 | 0.9 | ∼1.0 | ComA∼P phosphatase |
| rocA | 12 | 0.4 | 0.6 | ∼1.0 | Pyrolline-5-carboxylate dehydrogenase |
| rocB | 37 | 2.2 | 0.9 | 1.65 | Probable citrullinasef |
| rocC | 24 | 1.4 | 1.0 | ∼1.0 | Arginine permease |
| rocD | 11 | 1.1 | 0.53 | ∼1.0 | Ornithine transaminase |
| rocE | 4.0 | 1.0 | 0.43 | ∼1.0 | Arginine permease |
| rocF | 2.0 | 0.8 | 0.47 | ∼1.0 | Arginase |
| gabP | 1.0 | 1.2 | 1.5 | 1.31 | γ-Aminobutyrate permease |
| codV | 1.0 | 0.8 | 2.5 | 2.56 | Recombinase |
| clpQ | 1.0 | 0.7 | 2.0 | ∼1.0 | Chaperone-type ATPase |
| clpY | 1.3 | 1.5 | 1.0 | ∼1.0 | Protease |
| codY | 1.3 | 1.2 | 2.9 | ∼1.0 | GTP-dependent regulatory protein |
| citB | 1.4 | 11.3 | 1.2 | ∼1.0 | Aconitase |
| Newly identified targets | |||||
| appD | 17 | 22 | 1.2 | 1.54 | Transport ATPase |
| appF | 28 | 27 | 1.2 | ∼1.0 | Transport ATPase |
| appA | 18 | 54 | 2.1 | ∼1.0 | Oligopeptide binding protein |
| appB | 30 | 22 | 1.5 | ∼1.0 | Oligopeptide permease |
| appC | 3.7 | 5.9 | 1.2 | ∼1.0 | Oligopeptide permease |
| glpF | 0.19 | 0.4 | 0.9 | ∼1.0 | Glycerol uptake facilitator |
| glpT | 0.20 | 0.6 | 0.40 | ∼1.0 | Glycerol-3-phosphate permease |
| guaB | 0.28 | 1.0 | 0.24 | 1.86 | IMP dehydrogenase |
| guaC | 0.23 | 0.32 | ND | ∼1.0 | GMP reductase |
| ilvB | 38 | 21 | 21 | 2.1 (ysnD) | Acetolactate synthase subunit |
| ilvH | 44 | 11 | ND | ∼1.0 | Acetolactate synthase subunit |
| ilvC | 49 | 25 | 11 | ∼1.0 | Ketol-acid reductoisomerase |
| leuA | 47 | 26 | 12 | 1.16 | 2-Isopropylmalate synthase |
| leuB | 46 | 38 | 51 | ∼1.0 | 3-Isopropylmalate dehydrogenase |
| leuC | 46 | 16 | 51 | ∼1.0 | 3-Isopropylmalate dehydratase subunit |
| leuD | 2.4 | 1.9 | 6.6 | 1.17 | 3-Isopropylmalate dehydratase subunit |
| ilvD | 20 | 11 | 3.6 | 3.2 (ypgR) | Dihydroxy-acid dehydratase |
| ilvA | 5.1 | 4.2 | 1.8 | ∼1.0 | Threonine dehydratase |
| kinB | 7 | 2.8 | 3.3 | 1.76 | Spo0F kinase/PICK> |
| ybgE | 17 | 1.3 | 42 | 2.63 | Branched-chain amino acid Aminotransferase |
| yccC | 15 | ND | 1.5 | ∼1.0 | Similar to asparaginase |
| ycgA | 13 | 1.9 | 2.7 | 1.6 | Unknown |
| ycgM | 6.3 | 3.4 | 0.63 | 2.9 | Proline oxidase |
| yhdG | 116 | 26 | 11 | 1.86 | Amino acid transporter |
| yhjC | 92 | 4.3 | 6.3 | 1.57 | Unknown |
| ykfA | 13 | 2.8 | 5.6 | ∼1.0 | Similar to microcin |
| ykfB | 16 | 4.5 | 8.5 | ∼1.0 | l-Ala-d/l-Glt epimerase |
| ykfC | 14 | 4.4 | 8.4 | ∼1.0 | γ-d-glutamyl-l-amino acid peptidase |
| ykfD | 14 | 4.9 | 5.6 | ∼1.0 | Transporter |
| yufN | 21 | 8.2 | 13 | 1.65 | Substrate binding protein |
| yufO | 55 | 15 | 14 | ∼1.0 | Probable transport ATPase |
| yufP | 14 | 4.7 | 8.7 | ∼1.0 | Permease |
| yufQ | 14 | 9.4 | 13 | ∼1.0 | Permease |
| yuiC | 23 | 2.0 | 4.7 | 1.72 | Unknown |
| yuiB | 18 | 3.3 | 6.9 | 2.32 | Unknown |
| yurJ | 120 | 1.1 | 8.5 | ∼1.0 | Probable transport ATPase |
| yurP | 312 | 7.2 | 20 | 1.52 | Similar to glutamine-fructose transaminase |
| yurO | 346 | 5.4 | 41 | ∼1.0 | Similar to sugar binding protein |
| yurN | 723 | 3.7 | 33 | ∼1.0 | Similar to sugar permease |
| yurM | 58 | 1.7 | 18 | 1.94 | Similar to sugar permease |
| yurL | 55 | 1.9 | 13 | ∼1.0 | Similar to ribokinase |
| yusC | 0.30 | 4.8 | 0.61 | 1.85 | Similar to transport ATPase |
| yxbC | 45 | 5.6 | 4.0 | 1.96 | Unknown |
| yxbB | 65 | 11 | 10 | 1.74 | Unknown |
| yxbA | 99 | 5.7 | 7.3 | ∼1.0 | Unknown |
| yxnB | 78 | 10 | 10 | ∼1.0 | Unknown |
| asnH | 5.6 | 2.5 | 7.6 | ∼1.0 | Asparagine synthetase |
| yxaM | 51 | 4.2 | 5.9 | ∼1.0 | Similar to antibiotic resistance protein |
Previously known and newly identified targets of CodY and genes whose transcript level was highly affected by a codY mutation during growth in minimal medium (MM) with or without added amino acids (AAs) or in DS medium.
ChIP enrichment data were taken from the data of Table 3 and from additional data not shown. They are presented here for comparative purposes.
Functional assignments are from SubtiList (http://genolist.pasteur.fr/SubtiList/) or from literature cited in the text. CoA, coenzyme A.
ND, transcript level was not significantly above background in either the mutant or wild-type sample.
∼1.0, enrichment factor for precipitation of specific genes by antibody to CodY (see text and Table 3) was not statistically different from 1.0.
B. Belitsky (personal communication).
For cells grown in the nutrient broth-based DS medium, 187 genes in 84 apparent transcription units were overexpressed in a codY mutant during exponential phase and an additional 79 genes in 43 apparent transcription units were overexpressed in a codY mutant only during stationary phase (when CodY is less active). One hundred thirty-two genes in 62 transcription units were underexpressed in a codY mutant during exponential growth phase. The full data set for these experiments can be viewed at the website of the Losick laboratory (http://mcb.harvard.edu/losick/). For the experiments on cells grown in DS medium, both the codY+ and ΔcodY strains carried a deletion in the abrB gene to avoid missing genes whose transcription is repressed by AbrB as well as by CodY. In fact, the vast majority of the genes that were overexpressed in a codY mutant in medium containing amino acids were also overexpressed in a codY abrB double mutant in DS medium. An apparent discrepancy in the behavior of the rocABC and rocDEF operons in the two different growth conditions can be rationalized. Both of these operons are dependent for their expression on RocR, a positive regulator that is activated by arginine or ornithine (21). The defined medium contains a high concentration of arginine, but DS medium does not. For genes that were underexpressed in a codY mutant, we saw very little correlation between the results obtained in minimal glucose-glutamine medium with 16 amino acids and DS medium.
The microarray analysis did not identify all targets of CodY. Of the previously known CodY targets, only the acsA gene and the dpp and ure operons were overexpressed in codY mutant cells grown both in minimal medium containing amino acids and broth medium. Other known targets either were not expressed above background levels in cells grown in one of the media tested or were not overexpressed in a codY mutant in one or both media. These genes included srfAA, comK, hag, the bkd cluster, hutP, rapA, rocABC, rocDEF, gabP, rapC, the cod operon, and citB. Several of these transcription units require positive regulators that might not be active under the conditions tested. For instance, bkd expression requires BkdR (12), gabP requires TnrA (16), srfAA, comK, and rapA require ComA∼P (25, 48, 49), and hag requires sigma-D (45). The citB gene, on the other hand, is strongly repressed by CcpC in cells in glucose-glutamine-containing medium and during rapid exponential growth phase in DS medium (31, 33, 34); the effect of a codY mutation on citB expression can be detected only in a ccpC mutant strain (33).
The hutP gene was overexpressed in a codY mutant in minimal medium containing amino acids (Table 2), but the other genes of the hut operon were not detectably transcribed (data not shown). Transcription of genes downstream of hutP depends on an antitermination event that requires histidine (51), one of three amino acids not present in the mixture used.
Among the previously unsuspected targets of CodY, the most highly affected by a codY mutation included the appDFABC, ykfABCD, and ilvBHC-leuABCD operons, the ilvD, ilvA, ybgE, and yhdG genes, the yufNOPQ cluster, and the yurPONML cluster (Table 2). Experiments described below indicate that some but not all of these genes and operons are direct targets of CodY binding.
Expression of lacZ fusions.
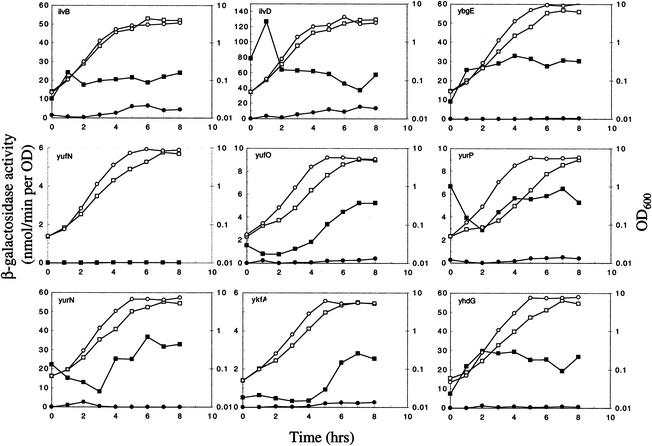
To confirm the transcript analysis for a subset of the newly identified potential target genes (ilvB, ilvD, ybgE, yhdG, ykfA, yufN, yufO, yurP, and yurN), we constructed isogenic codY+ and ΔcodY strains carrying integrational disruptions created through single-crossover recombination with plasmid pMUTIN2 derivatives possessing short coding regions from the 5′ ends of the genes. Such integration resulted in the transcriptional fusion of the upstream region of each disrupted gene to the E. coli lacZ gene. To monitor gene expression, samples taken at various times during growth in minimal glucose-glutamine medium with 16 amino acids were assayed for β-galactosidase activity. As shown in Fig. 1, lacZ fusions to the ilvB, ilvD, ybgE, yhdG, ykfA, yufO, yurP, and yurN promoter regions were all at least partially derepressed in ΔcodY strains during exponential growth and stationary phase. (RNA for the DNA microarray analysis of cells in defined medium containing a mixture of amino acids was prepared from cells harvested at an OD600 of 1.0, i.e., near the end of the rapid exponential growth phase.)
FIG. 1.
Expression of lacZ fusions to promoters of putative CodY target genes in codY+ and ΔcodY strains. Strains were grown in minimal glucose-glutamine medium containing a mixture of 16 additional amino acids (see Materials and Methods), and samples were removed at indicated times after inoculation for assays of β-galactosidase activity. Isogenic codY+ and ΔcodY strains carrying each gene disruption were FU384 and FU385 for ilvB, FU386 and FU387 for ilvD, FU388 and FU389 for ybgE, FU390 and FU391 for yufN, FU392 and FU393 for yufO, FU394 and FU395 for yurP, FU396 and FU397 for yurN, FU398 and FU399 for ykfA, and FU400 and FU401 for yhdG. Circles and squares denote codY+ and ΔcodY strains, respectively, whereas open and closed symbols represent the OD600 and β-galactosidase activity, respectively.
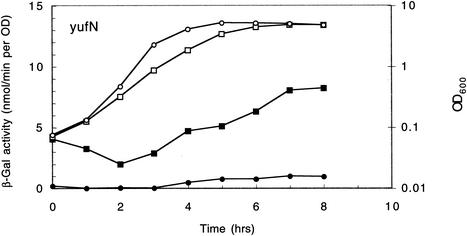
By contrast, the strain carrying a pMUTIN2-derived lacZ fusion to yufN (and a yufN disruption) seemed to behave anomalously. No β-galactosidase activity was detected in either the codY+ or ΔcodY strain at any stage of growth (Fig. 1). To test the possibility that expression of the yufN gene depends on its own product, we constructed codY+ and ΔcodY strains carrying at the amyE locus a lacZ transcriptional fusion to the intergenic region upstream of yufN. In this case, the yufN-lacZ fusion was clearly expressed and derepressed in the ΔcodY background (Fig. 2). Thus, the results of the lacZ fusion experiments with respect to ilvB, ilvD, ybgE, yhdG, ykfA, yufN, yufO, yurP, and yurN coincided well with those of the DNA microarray analysis.
FIG. 2.
Expression of a yufN-lacZ fusion integrated at the amyE locus. Strains FU407 (codY+) and FU408 (ΔcodY) were grown in minimal glucose-glutamine medium containing 16 additional amino acids (see Materials and Methods). Samples removed at the indicated times after inoculation of the culture were assayed for β-galactosidase activity. Circles and squares denote codY+ and ΔcodY strains, respectively, whereas open and closed symbols represent the OD600 and β-galactosidase activity, respectively.
Immunoprecipitation of CodY-DNA complexes.
Formaldehyde-induced in vivo cross-linking of protein to DNA has been used with immunoprecipitation by specific antibodies to demonstrate binding of proteins to specific DNA regions in both eukaryotes and prokaryotes (38, 66). The power of this method can be extended by combining the selectivity of chromatin immunoprecipitation (ChIP) with the global analysis provided by a DNA microarray (chip) (29, 36, 58). To identify segments of B. subtilis DNA that interact with CodY in vivo, we treated cells with formaldehyde to create protein-DNA cross-links and then used antibody to CodY to select those DNA regions that were specifically cross-linked to CodY (see Materials and Methods). The putative binding sites were then revealed by hybridization to a DNA microarray of the B. subtilis genome (ChIP-to-chip). A calculated enrichment factor denotes the extent to which the relative abundance of a given gene was augmented by immunoprecipitation.
As summarized in Table 3, 68 regions of the chromosome were preferentially selected by immunoprecipitation with antibody to CodY. Forty-two of these regions contained at least one gene whose transcript level was significantly affected by a codY mutation when cells were grown in minimal medium or DS medium or both. The nature of the method does not permit unambiguous assignment of the codY binding site to a specific gene, however. That is, fragmentation of the DNA may separate the CodY binding site from the coding sequence that it regulates. Since the microarrays were spotted with PCR products corresponding to coding sequences but not intergenic regions, the analysis of the immunoprecipitated DNA might fail to identify some target sites and might identify a neighboring gene in addition to or even instead of the actual target. For instance, in the ureA-ywmG-ywmF region, we detected the ywmG gene but not ureA by immunoprecipitation, even though ureA is a known target of CodY (71) (Tables 2 and 3). As a result, we made educated guesses about the likely target gene in some cases (e.g., ilvB). On the basis of this analysis, we determined that the ilvB, ilvD, ilvA, ybgE, yhdG, yurM, and yufN genes are likely to be direct targets of CodY, whereas the ykfABCD and yufOPQ operons may be indirectly regulated by CodY. The case of yurP was uncertain; the yurPON cluster was highly derepressed in a codY mutant but the ChIP-to-chip enrichment factor was only 1.52.
TABLE 3.
Targets of CodY revealed by ChIP-to-chip analysis
| Region detecteda | Enrichment factorsb | Likely target genec | Transcript ratio of likely target gened
|
Function of likely target gene/operon | |
|---|---|---|---|---|---|
| ΔcodY/codY+, DS medium | ΔcodY/codY+, MM + AAs | ||||
| Biosynthesis | |||||
| ilvB-ysnD-ysnE | 2.1 | ilvB | 21.3 | 38.1 | BCAA biosynthesis |
| ilvD-ypgR-ypgQ | 3.2 | ilvD | 3.6 | 19.5 | BCAA biosynthesis |
| ybgA-ybgB-ybgE-ybgF | 3.54 (ybgB), 2.63 (ybgE) | ybgE | 42.2 | 17 | BCAA transaminase |
| ggaB-ggaA-tagH | 3.1 | ggaA | 1.1 | 0.6 | Teichoic acid synthesis |
| rrnO-yaaC-guaB | 2.4 (yaaC), 1.86 (guaB) | guaB | 0.24 | 0.27 | IMP dehydrogenase |
| Catabolism | |||||
| tyrS-acsA-acuA | 2.1 | acsA | 2.4 | 4.4 | Acetyl CoA synthetase |
| opuAC-amhX-ycgA | 2.27 (amhX), 1.6 (ycgA) | amhX | 34.3 | 2.5 | Aminohydrolase |
| yhjN-aprE-yhfO | 1.94 (aprE), 1.74 (yhfO) | aprE | 1.5 | 2.3 | Subtilisin E |
| cgeE-cgeD-cgeC | 2.05 | cgeD | 0.9 | 1.4 | Spore coat maturation |
| gid-codV-clpQ | 1.93 (gid), 2.56 (codV) | codV | 2.5 | 1.0 | Site-specific recombinase (cod operon) |
| proG-dppA-dppB | 3.06 (proG), 2.78 (dppA) | dppA | 7.1 | 55 | Dipeptide permease |
| metE-ispA-ykoB | 3.21 | ispA | 1.8 | NDe | Intracellular protease |
| yktD-nprE-ylaA | 1.73 | nprE | 0.29 | 0.7 | Neutral protease |
| trpS-oppA-oppB | 1.51 | oppA | 0.55 | 0.8 | Oligopeptide permease |
| rocG-rocA-rocB-rocC | 1.65 | rocA | 0.60 | 11.8 | Arginine catabolism |
| rocR-rocD-rocE | 1.52 | rocD | 0.53 | 4.1 | Arginine catabolism |
| ycgL-ycgM-ycgN | 4.18 (ycgL), 2.9 (ycgM) | ycgM | 0.63 | 6.3 | Proline catabolism |
| ureA-ywmG-ywmF | 1.4 | ureA | 1.4 | 3.7 | Urea catabolism |
| uxaC-yjmB-yjmC-yjmD | 2.39 (yjmB), 2.18 (yjmC) | yjmB | 1.1 | ND | Hexuronate catabolism? |
| yoaB-yoaC-yoaD | 1.79 (yoaC), (2.81 yoaD) | yoaC | 1.2 | ND | Similar to xylulokinase |
| ycbE-ycbF-ycbG | 1.88 | ycbF | 2.4 | ND | Glucarate dehydratase? |
| Regulation | |||||
| spoIVB-spo0A-recN | 1.92 | spo0A | 1.3 | ND | Transcription factor |
| patB-kinB-kapB | 1.76 | kinB | 3.3 | 7.0 | Histidine kinase |
| yjoB-rapA-phrA | 2.24 | rapA | 0.9 | 2.8 | Spo0F∼P phosphatase |
| yqcI-yqcJ-rapE-phrE | 3.16 | rapE | 5.4 | 5.8 | Spo0F∼P phosphatase |
| ywhK-rapF-phrF | 2.32 (rapF), 3.17 (phrF) | rapF | 2.2 | 2.0 | Aspartyl∼P phosphatase |
| yddK-rapI-phrI-yddM | 1.8 | rapI | 0.02 | ND | Aspartyl∼P phosphatase |
| yozJ-rapK-phrK | 1.84 | rapK | 1.2 | 3.7 | Aspartyl∼P phosphatase |
| ykuV-rok-yknT | 1.83 | rok | 1.0 | 1.1 | Competence regulator |
| yveK-slr-pnbA | 2.65 | slr | 1.5 | 1.3 | Regulatory protein |
| yrhI-yrhH-yrzI | 1.79 (yrhI), 2.86 (yrhH) | yrhI | 1.4 | ND | Transcription regulator? |
| xkdA-xre-xkdB-xkdC | 1.42 (xre), 1.97 (xkdB) | xkdB | 1.2 | 1.7 | PBSX phage regulator? |
| Transport | |||||
| sunA-sunT-yolF | 4.18 | sunT | 1.5 | ND | Lantibiotic transporter |
| yrrI-glnQ-glnH | 1.77 | glnQ | 1.3 | 2.8 | Glutamine transport |
| nifZ-braB-ezrA | 1.94 | braB | 0.36 | 0.5 | BCAA transport |
| yjaZ-appD-appF | 2.39 (yjaZ), 1.54 (appD) | appD | 1.2 | 17 | Oligopeptide transport |
| gltT-yhfh-yhfI | 2.17 | gltT | 0.42 | 0.4 | H+-Na+/glutamate symporter |
| ydiD-gcp-ydiF-ydiG | 1.46 (gcp), 1.53 (ydiF) | ydiF | 1.5 | 0.7 | Sugar transporter? |
| ytmM-ytmL-ytmK | 3.9 (ytmM), 1.95 (ytmL) | ytmL | 0.9 | ND | Amino acid transporter? |
| citA-yhdF-yhdG-yhdH | 2.71 (yhdF), 1.86 (yhdG) | yhdG | 11.5 | 116 | Amino acid transporter? |
| yurL-yurM yurN | 1.94 | yurM | 18.3 | 58.7 | Sugar permease? |
| yufM-yufN-yufO | 1.65 | yufN | 13.2 | 21.4 | ABC transporter? |
| Other | |||||
| ybbI-ybbJ-ybbK | 1.91 | ybbJ | 1.2 | ND | Unknown |
| ydcK-ydcL-ydcM-ydcN | 2.24 (ydcL), 2.55 (ydcM), 2.55 (ydcN) | ydcL | ND | 1.8 | Prophage integrase? |
| yddS-yddT-ydeA | 1.79 | yddT | 1.1 | ND | Unknown |
| ydhQ-ydhR-ydhS | 1.78 | ydhR | 1.0 | 1.0 | Similar to fructokinase |
| yfmC-yfmB-yfmA | 1.67 | yfmC | 1.8 | 1.0 | Unknown |
| yfmI-yfmH-yfmG | ND | yfmG | 5.5 | 6.5 | Unknown |
| yhjC-yhjD-yhjE | 1.57 | yhjC | 6.3 | 93 | Unknown |
| pit-ykaA-ykbA | 3.24 | ykaA | 1.0 | ND | Unknown |
| splB-ykwB-mcpC | 2.42 | ykwB | 4.4 | 25.7 | Unknown |
| ylbO-ylbP-ylbQ | 1.85 | ylbP | 1.5 | 3.0 | Unknown |
| yndJ-yndK-yndL-yndM | 1.69 (yndK), 1.79 (yndL) | yndK | 1.3 | 0.8 | Unknown/PICK> |
| yoaC-yoaD-yoaE | 1.78 (yoaC), 2.81 (yoaD) | yoaD | 2.4 | ND | Similar to phosphoglycerate dehydrogenase |
| yobQ-yobR-yobS | 1.91 | yobR | 2.3 | 3.4 | Unknown |
| odhA-yojO-yojN | 1.83 | yojO | 1.5 | 0.8 | Unknown |
| yqgB-yqgA-yqfZ | 1.86 | yqgA | 1.1 | 1.1 | Unknown |
| yqjZ-yqjY-yqjX | 1.99 | yqjZ | 1.6 | 2.5 | Unknown |
| yuaG-yuaF-yuaE | 1.78 | yuaF | 1.5 | 2.1 | Unknown |
| yuiC-yuiB-yuiA-yumB | 2.32 (yuiB), 3.39 (yuiA) | yuiB | 6.9 | 17.8 | Unknown |
| yurP-yurQ-yurR | 1.52 | yurP | 19.5 | 312 | Unknown |
| yusC-yusD-yusE | 1.85 | yusC | 0.61 | 0.3 | Unknown |
| yvaV-yvaW-yvaX-yvaY | 1.7 (yvaW), 3.29 (yvaX), 1.67 (yvaY) | yvaX | 0.92 | 4.2 | Unknown |
| yvdB-yvdA-yvcT | 1.76 (yvdB), 3.16 (yvdA) | yvdA | 1.3 | 2.3 | Carbonic anhydrase? |
| yxbC-yxbB-yxbA | 1.96 (yxbC), 1.74 (yxbB) | yxbC, yxbB | 3.9 (yxbC), 10.1 (yxbB) | 35 (yxbC), 65 (yxbB) | Unknown |
| Unknown | |||||
| pepT-yxjJ-yxjI | 2.24 | yxjJ | 1.1 | ND | Unknown |
| yycO-yycN-rapG | 1.83 | yycN | 1.2 | 0.7 | Unknown |
Gene clusters are listed within which the underlined genes were preferentially precipitated by antibody to CodY, after in vivo cross-linking, as detected by hybridization to a genomic array.
The enrichment factor indicates the extent to which a gene was preferentially precipitated compared to its abundance in total DNA.
The likely target gene was either the enriched gene or a neighboring, promoter-proximal gene whose transcription is strongly affected by a codY mutation.
Transcript ratios are from the data of Table 2 and additional data not shown; they are included here for comparative purposes. MM + AAs, minimal medium with added amino acids.
ND, transcript not detectable in either mutant or wild-type sample.
The ChIP-to-chip analysis revealed 26 apparent CodY target genes whose transcript level was below detection or did not appear to be influenced dramatically by a codY mutation. These genes may depend on a positive regulator that is inactive under the conditions tested or may be expressed in a CodY-dependent manner only under growth conditions other than those used here.
Only a few CodY targets identified by ChIP-to-chip analysis (gltT, guaB, and braB) were consistently and significantly underexpressed in a codY mutant. Their regulatory regions may be sites of direct, positive regulation by CodY.
Gel mobility shift assays of CodY binding.
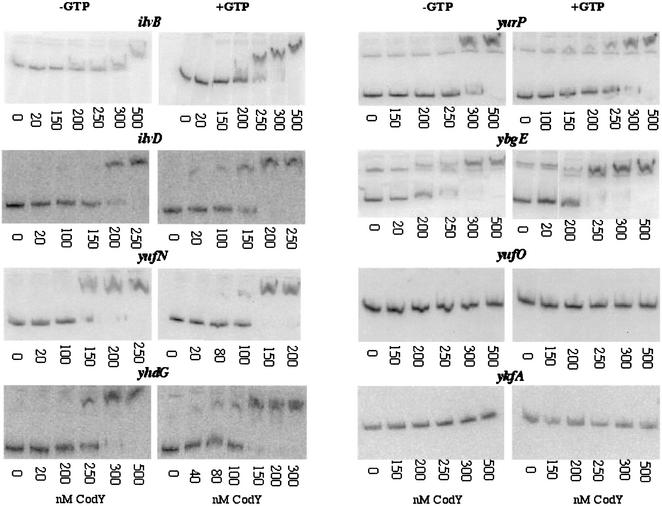
To test whether CodY binds directly to the regulatory regions of putative target genes, we prepared radioactive double-stranded DNA probes corresponding to the upstream regions of several of the genes. Gel mobility shift analysis showed that CodY can interact in vitro with probes for the ilvB, ilvD, ybgE, yufN, yhdG, and yurP genes (Fig. 3). By contrast, probes for the ykfA and yufO genes did not interact with CodY even at very high protein concentrations (Fig. 3). Thus, these two transcription units, which are strongly responsive to a codY mutation (Table 1 and Fig. 1), are probably indirect targets of CodY. The in vitro binding results are consistent with the absence of ykfA and yufO among the genes that were cross-linked to CodY in vivo. A likely explanation is that ykfA and yufO lie immediately downstream of transcription units that are highly regulated by CodY (dpp and yufN, respectively). Although the neighboring operons are in both cases separated by an apparent transcription terminator, this terminator must be leaky, allowing read-through into the ykfA operon by dpp transcription and into the yufO operon by yufN transcription. The fact that yufO and the ykfA operon are only indirect targets of CodY does not diminish the physiological significance of their response to CodY (see Discussion).
FIG. 3.
Gel mobility shift analysis of CodY binding to putative target regulatory regions. The regulatory regions of potential target genes were amplified by PCR by using radioactive primers, incubated with purified CodY-His6 protein at various concentrations, and analyzed by nondenaturing polyacrylamide gel electrophoresis (see Materials and Methods for details). The lengths of the PCR products in base pairs were as follows: ilvB, 453; ilvD, 525; ybgE, 446; yhdG, 345; yufN, 492; yufO, 199; ykfA, 340; and yurP, 321. In each panel, the position of unshifted DNA is seen in the leftmost lane, which contained no CodY. Where indicated, GTP was included in the reaction mixture at a concentration of 2 mM.
Binding of CodY to the dpp promoter region is slightly stimulated by GTP (57). (Efficient repression of dpp transcription by CodY has a more stringent requirement for GTP [57].) GTP also stimulated binding of CodY to the ilvB, ilvD, yufN, yhdG, yurP, and ybgE regulatory regions (Fig. 2).
DISCUSSION
Our results show that ChIP-to-chip, when combined with global transcriptional analysis, is a powerful method for locating previously unknown, direct targets of a bacterial regulatory protein. An alternative strategy would be to combine ChIP-to-chip results with a sequence scan that searches for a conserved binding site for the regulatory protein (15). In the case of CodY, such a search was not possible, because no conserved binding site is yet known.
Even the combination of ChIP-to-chip analysis and transcript profiling did not identify all CodY targets, however. Transcriptional profiling was expected to miss some targets because many stationary-phase genes regulated by CodY are also subject to control by other regulatory proteins. As a result, multiple mutations are needed in some cases to reveal fully the role of CodY (33, 65). More surprisingly, some genes whose regulatory regions bind CodY in vitro and whose expression is subject to CodY-mediated repression in vivo failed to be enriched for by immunoprecipitation. Examples are srfAA, hutP, and citB. Perhaps these targets have affinities for CodY that are relatively low, or perhaps binding of CodY under the growth conditions tested was prevented by interference by other regulators that bind to overlapping sites.
The newly identified targets of CodY unexpectedly include genes for amino acid biosynthesis (the ilvB operon and the ilvA, ilvD, and ybgE genes). All previously recognized and many newly identified CodY targets are genes whose products permit the cell to search for, take up, and metabolize secondary nutritional sources or to sporulate. The cell's rationale in coregulating biosynthesis of amino acids and catabolism of secondary nutrients (including some amino acids) may be the following. During rapid growth in rich medium, cells utilize preformed amino acids and repress the relevant biosynthetic pathways. When the external amino acid supply becomes limited, cells turn on de novo biosynthesis at the same time that they hunt for other carbon and nitrogen sources. This principle should hold for all amino acids and has been at least partially verified experimentally (reference 42, Table 2, and data not shown). However, only the isoleucine-leucine-valine biosynthetic pathway proved to be under CodY control. Either the branched-chain amino acids are preferentially consumed or the cell has evolved to tie its control of stationary-phase gene expression specifically to the availability of branched-chain amino acids. In fact, preliminary experiments establish that the ability of CodY to bind to many of its targets in vitro is stimulated by branched-chain amino acids (R. Shivers and A. L. Sonenshein, unpublished results). This finding fits well with the observation of Guédon et al. (23) that dipeptides containing branched-chain amino acids are particularly effective in activating CodY in vivo in L. lactis. These authors, in fact, suggested that the amino acids might be direct effectors of CodY (23). Given the central position of the branched-chain amino acids in cellular metabolism, such regulation would not be surprising and is, in fact, reminiscent of the role of Lrp in E. coli (50). The B. subtilis genome encodes seven homologs of Lrp, none of which is yet known to be a global regulator (3, 4, 11). It is conceivable that CodY in gram-positive bacteria is the functional equivalent of Lrp in enteric bacteria. A central role for the branched-chain amino acid biosynthetic pathway in B. subtilis metabolism is also suggested by its susceptibility to direct or indirect repression by CcpA, a global regulator that responds to glucose availability (41).
Other newly identified targets of CodY also have interesting features. The ykfABCD operon, which seems to be controlled via the dpp promoter, encodes an l-alanine-d/l-glutamate epimerase (YkfB), a γ-d-glutamyl-l-diamino acid peptidase (YkfC), and a transport protein (YkfD) (61). YkfA is related to a microcin-resistance protein in E. coli (22, 61). These proteins may be involved in recycling of peptidoglycan degradation products. If so, the coregulation of the ykfABCD and dpp operons would be reasonable, since the dpp operon encodes a d-aminopeptidase and a dipeptide uptake system (9, 44).
On the basis of homology searches, the yufN, yufO, yufP, and yufQ genes appear to encode, respectively, the lipid-linked substrate binding protein, the ATP-binding component, and the permease proteins of an ABC transporter; the yurPONML cluster is likely to be involved in sugar transport and metabolism, and the YhdG protein is similar to amino acid transporters.
Since CodY appears to be responsible for the inhibition of sporulation that occurs when nutrients are in excess (57), we anticipated that one or more key sporulation genes would be revealed as CodY targets. In fact, at least three participants in regulation of early sporulation gene expression through the Spo0A phosphorelay (6, 26) can be found among the CodY targets. The kinB gene encodes a membrane-associated histidine kinase that can serve as the first enzyme of the Spo0A phosphorelay (69). This gene was enriched in the ChIP-to-chip analysis and was overexpressed in a codY mutant in both minimal medium containing 16 amino acids and DS medium. The kinB gene is unlikely to be the only sporulation-related CodY target, however, because kinB is not by itself essential for sporulation (69).
The rapA-phrA and rapE-phrE operons also proved to be likely CodY targets. The genes of these operons encode Rap phosphatases for Spo0F∼P, an intermediate component of the phosphorelay, and inhibitors of the phosphatases (28, 30, 53). These proteins appear to determine the time at which enough Spo0A∼P accumulates to cause cells to choose the sporulation pathway (48, 53). PhrA is essential for sporulation (53), indicating that its derepresed expression in a codY mutant might be sufficient to unleash sporulation under conditions of nutrient excess.
The spo0A region was enriched in the ChIP-to-chip analysis, but its transcription was barely detectable and was not derepressed in a codY mutant under the conditions tested. The spo0A gene has two promoters, however. A low-level vegetative promoter provides a basal level of spo0A mRNA during growth (10). A second, more active promoter is induced when cells enter stationary phase, and transcription from this promoter is essential for sporulation (10, 64). Under the growth conditions we have used, transcription from the vegetative promoter would have predominated. The sporulation promoter of spo0A not only requires sigma-H for its activity but is also repressed by Soj, SinR, and ScoC (7). Thus, the lack of any detectable change in spo0A expression in a codY mutant is not surprising. Preliminary in vitro experiments indicate that the sporulation promoter region of spo0A does indeed include a binding site for CodY (K. Tachikawa, M. Ratnayake-Lecamwasam, and A. L. Sonenshein, unpublished). Therefore, spo0A may be the critical gene whose repression by CodY ties initiation of sporulation to nutrient depletion.
The CodY regulon partially overlaps with the RelA and ScoC regulons. The dpp operon is induced by activation of the stringent response (57), presumably because the activity of RelA (stringency factor) leads to a drop in the GTP pool (27). A survey of global transcription after exposure to norvaline, an inhibitor of isoleucyl- and leucyl-tRNA synthetases, showed RelA-dependent induction of the ilvB operon, appD, ureA, gabP, rapA, spo0A, yurP, and yxbC (14), all of which are CodY targets (Tables 1 and 2). Other stringency-induced genes may not be targets of CodY.
ScoC (also known as Hpr) negatively regulates extracellular enzyme production and sporulation and positively regulates other genes (32). Caldwell et al. (8) noted some overlap among genes that are regulated by ScoC and CodY. The hutP, comK, rapA, and hag genes, the bkd cluster, and the ureABC operon are all underexpressed in a scoC mutant but overexpressed in a codY mutant (reference 8 and Tables 1 and 2). On the other hand, the glnQ gene is overexpressed in both scoC and codY mutants. While not all of these genes may be direct targets of either regulatory protein, there are probably cases where the two proteins bind simultaneously to the same regulatory region, either in cooperation or in competition.
Acknowledgments
We thank B. Belitsky for helpful discussions and criticism of the manuscript.
V.M. was a fellow of the European Molecular Biology Organization, and R.P.S. was a predoctoral trainee of the U.S. Public Health Service (T32 GM07310). The research described was supported by a Grant-in-Aid for Scientific Research on Priority Area from the Ministry of Education, Science, Sports, and Culture of Japan to Y. Fujita and by research grants from the U.S. Public Health Service to R. Losick (GM18568) and A. L. Sonenshein (GM42219).
REFERENCES
- 1.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Belitsky, B. R., M. C. Gustafsson, A. L. Sonenshein, and C. von Wachenfeldt. 1997. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol. 179:5448-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beloin, C., R. Exley, A. L. Mahe, M. Zouine, S. Cubasch, and F. Le Hegarat. 2000. Characterization of LrpC DNA-binding properties and regulation of Bacillus subtilis lrpC gene expression. J. Bacteriol. 182:4414-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 7.Burkholder, W. F., and A. D. Grossman. 2000. Regulation of the initiation of endospore formation in Bacillus subtilis, p. 151-166. In Y. Brun and L. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 8.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheggour, A., L. Fanuel, C. Duez, B. Joris, F. Bouillenne, B. Devreese, G. Van Driessche, J. Van Beeumen, J. M. Frere, and C. Goffin. 2000. The dppA gene of Bacillus subtilis encodes a new D-aminopeptidase. Mol. Microbiol. 38:504-513. [DOI] [PubMed] [Google Scholar]
- 10.Chibazakura, T., F. Kawamura, and H. Takahashi. 1991. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J. Bacteriol. 173:2625-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dartois, V., J. Liu, and J. A. Hoch. 1997. Alterations in the flow of one-carbon units affect KinB-dependent sporulation in Bacillus subtilis. Mol. Microbiol. 25:39-51. [DOI] [PubMed] [Google Scholar]
- 12.Debarbouille, M., R. Gardan, M. Arnaud, and G. Rapoport. 1999. Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2002. Carbohydrate uptake and metabolism, p. 129-150. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.) Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 14.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferson, A. E., L. V. Wray, Jr., and S. H. Fisher. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol. Microbiol. 22:693-701. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 18.Fisher, S. H., K. Rohrer, and A. E. Ferson. 1996. Role of CodY in regulation of the Bacillus subtilis hut operon. J. Bacteriol. 178:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouet, A., and A. L. Sonenshein. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freese, E., J. E. Heinze, and E. M. Galliers. 1979. Partial purine deprivation causes sporulation of Bacillus subtilis in the presence of excess ammonia, glucose and phosphate. J. Gen. Microbiol. 115:193-205. [DOI] [PubMed] [Google Scholar]
- 21.Gardan, R., G. Rapoport, and M. Debarbouille. 1995. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J. Mol. Biol. 249:843-856. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Pastor, J. E., J. L. San Millan, M. A. Castilla, and F. Moreno. 1995. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 177:7131-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 24.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn, J., L. Kong, and D. Dubnau. 1994. The regulation of competence transcription factor synthesis constitutes a critical control point in the regulation of competence in Bacillus subtilis. J. Bacteriol. 176:5753-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoch, J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441-465. [DOI] [PubMed] [Google Scholar]
- 27.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 184:3923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa, S., L. Core, and M. Perego. 2002. Biochemical characterization of aspartyl phosphate phosphatase interaction with a phosphorylated response regulator and its inhibition by a pentapeptide. J. Biol. Chem. 277:20483-20489. [DOI] [PubMed] [Google Scholar]
- 29.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, M., R. Grau, and M. Perego. 2000. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J. Bacteriol. 182:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jourlin-Castelli, C., N. Mani, M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865-878. [DOI] [PubMed] [Google Scholar]
- 32.Kallio, P. T., J. E. Fagelson, J. A. Hoch, and M. A. Strauch. 1991. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266:13411-13417. [PubMed] [Google Scholar]
- 33.Kim, H.-J., S.-I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. 2003. Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, H.-J., A. Roux, and A. L. Sonenshein. 2002. Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes. Mol. Microbiol. 45:179-190. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 99:4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazazzera, B. A., I. G. Kurtser, R. S. McQuade, and A. D. Grossman. 1999. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J. Bacteriol. 181:5193-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, D. C., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 39.Lopez, J. M., A. Dromerick, and E. Freese. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146:605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez, J. M., C. L. Marks, and E. Freese. 1979. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta 587:238-252. [DOI] [PubMed] [Google Scholar]
- 41.Ludwig, H., C. Meinken, A. Matin, and J. Stulke. 2002. Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J. Bacteriol. 184:5174-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mader, U., G. Homuth, C. Scharf, K. Buttner, R. Bode, and M. Hecker. 2002. Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability. J. Bacteriol. 184:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magasanik, B. 1996. Regulation of nitrogen utilization, p. 1344-1356. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 44.Mathiopoulos, C., J. P. Mueller, F. J. Slack, C. G. Murphy, S. Patankar, G. Bukusoglu, and A. L. Sonenshein. 1991. A Bacillus subtilis dipeptide transport system expressed early during sporulation. Mol. Microbiol. 5:1903-1913. [DOI] [PubMed] [Google Scholar]
- 45.Mirel, D. B., W. F. Estacio, M. Mathieu, E. Olmsted, J. Ramirez, and L. M. Marquez-Magana. 2000. Environmental regulation of Bacillus subtilis σD-dependent gene expression. J. Bacteriol. 182:3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitani, T., J. Heinze, and E. Freese. 1977. Induction of sporulation in Bacillus subtilis by decoyinine or hadacidin. Biochim. Biophys. Acta 77:1118-1125. [DOI] [PubMed] [Google Scholar]
- 47.Miwa, Y., and Y. Fujita. 2001. Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 183:5877-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mueller, J. P., and A. L. Sonenshein. 1992. Role of the Bacillus subtilis gsiA gene in regulation of early sporulation gene expression. J. Bacteriol. 174:4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakano, M. M., L. A. Xia, and P. Zuber. 1991. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J. Bacteriol. 173:5487-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman, E. B., R. T. Lin, and R. D'Ari. 1996. The leucine/Lrp regulon, p. 1513-1525. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 51.Oda, M., N. Kobayashi, A. Ito, Y. Kurusu, and K. Taira. 2000. cis-Acting regulatory sequences for antitermination in the transcript of the Bacillus subtilis hut operon and histidine-dependent binding of HutP to the transcript containing the regulatory sequences. Mol. Microbiol. 35:1244-1254. [DOI] [PubMed] [Google Scholar]
- 52.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perego, M., and J. A. Hoch. 1996. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:1549-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price, C. W. 2002. The general stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.) Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 55.Quisel, J., D. Lin, and A. D. Grossman. 1999. Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4:665-672. [DOI] [PubMed] [Google Scholar]
- 56.Ratnayake-Lecamwasam, M. 2001. Ph.D. thesis. Tufts University, Boston, Mass.
- 57.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 59.Saier, M. H., Jr., T. M. Ramseier, and J. Reizer. 1996. Regulation of carbon utilization, p. 1325-1343. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 60.Schaeffer, P., J. Millet, and J.-P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt, D. M., B. K. Hubbard, and J. A. Gerlt. 2001. Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as L-Ala-D/L-Glu epimerases. Biochemistry 40:15707-15715. [DOI] [PubMed] [Google Scholar]
- 62.Serror, P., and A. L. Sonenshein. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843-852. [DOI] [PubMed] [Google Scholar]
- 63.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siranosian, K. J., and A. D. Grossman. 1994. Activation of spo0A transcription by sigma H is necessary for sporulation but not for competence in Bacillus subtilis. J. Bacteriol. 176:3812-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 66.Solomon, M. J., P. L. Larsen, and A. Varshavsky. 1988. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53:937-947. [DOI] [PubMed] [Google Scholar]
- 67.Sonenshein, A. L. 1989. Metabolic regulation of sporulation and other stationary phase phenomena, p. 109-130. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of procaryotic development. American Society for Microbiology, Washington, D.C.
- 68.Strauch, M. A. 1993. AbrB, a transition state regulator, p. 757-764. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 69.Trach, K. A., and J. A. Hoch. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol. Microbiol. 8:69-79. [DOI] [PubMed] [Google Scholar]
- 70.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 71.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]
- 73.Yoshida, K., K. Kobayashi, Y. Miwa, C.-M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]