Abstract
Reversible acetylation of nucleosomal histones H3 and H4 generally is believed to be correlated with potential transcriptional activity of eukaryotic chromatin domains. Here, we report that the extent of H4 acetylation within euchromatin and heterochromatic domains is linked with DNA replication rather than with transcriptional activity, whereas H3 acetylation remains fairly constant throughout the cell cycle. Compared with euchromatin, plant nucleolus organizers were more strongly acetylated at H4 during mitosis but less acetylated during S phase, when the nucleolus appeared to be (at least transiently) devoid of nucleosomes. Deposition-related acetylation of lysines 5 and 12 of H4 seems to be conserved in animals and plants and extended to K16 in plants. A possibly species-specific above-average acetylation at lysines 9/18 and 14 of H3 appeared in 4′,6-diamidino-2-phenylindole (DAPI)–stained heterochromatin fractions. These results were obtained by combining immunodetection of all acetylatable isoforms of H3 and H4 on mitotic chromosomes and nuclei in G1, early S, mid-S, late S, and G2 phases of the field bean with identification of specific chromatin domains by fluorescence in situ hybridization or DAPI staining. In addition, the histone acetylation patterns of distinct domains were compared with their replication and transcription patterns.
INTRODUCTION
The histone proteins H2A, H2B, H3, and H4 form octamers that constitute the nucleosome core particles in all eukaryotes. Their N-terminal tails are subject to post-translational modifications such as acetylation, phosphorylation, methylation, ubiquitination, glycosylation, and ADP ribosylation (reviewed in Smith et al., 1995; Spencer and Davie, 1999).
The reversible acetylation of N-terminal lysine residues at positions 5, 8, 12, and 16 of H4 and 9, 14, 18, and 23 of H3 mediates decondensation of the nucleosome structure (Loidl, 1988, 1994; Garcia-Ramirez et al., 1995), alters histone–DNA interactions (Hong et al., 1993), and facilitates access and binding of transcription factors to genes transcribed by RNA polymerases II or III (Lee et al., 1993; Vettese-Dadey et al., 1996).
A correlation between histone acetylation and potential transcriptional activity, initially proposed by Allfrey et al. (1964), has been proved in several cases (reviewed in Csordas, 1990; Turner, 1991, 1993; Loidl, 1994; Grunstein, 1997; Struhl, 1998). According to one attractive recent hypothesis, histone modifications may constitute a concerted code to “specify unique downstream functions” (Strahl and Allis, 2000; Turner, 2000).
After indirect immunolabeling with antibodies raised against acetylated isoforms of histone H4 (Turner and Fellows, 1989; Turner et al., 1989), mammalian metaphase chromosomes show intense acetylation of euchromatic R-bands and less intense acetylation of constitutive and facultative heterochromatin (Jeppesen and Turner, 1993). The patterns of histone H4 acetylation described for plant chromosomes (Houben et al., 1996, 1997; Belyaev et al., 1997; Vyskot et al., 1999) also reveal a below-average acetylation of late-replicating heterochromatin. However, whereas the most conserved histones H3 and H4 showed similar acetylation patterns along the mammalian chromosomes (Belyaev et al., 1996), the patterns for H3 and H4 differed conspicuously in field bean chromosomes (Belyaev et al., 1998).
Although H4 acetylation of mammalian nuclei appears to be confined to early replicating and actively transcribing euchromatin (Sadoni et al., 1999), and facultative heterochromatin is less acetylated than euchromatin in endosperm nuclei of Gagea lutea (Buzek et al., 1998), little is known about histone acetylation of specific chromosomal domains during defined interphase stages.
Treatment with trichostatin A, a specific inhibitor of histone deacetylase (Yoshida et al., 1990), several hours before mitosis mediated a switch to extensive acetylation of H4 (at lysines 5, 12, and 16) within the heterochromatin of field bean metaphase chromosomes, but H3 acetylation remained unchanged (Belyaev et al., 1997, 1998). This indicated that histone H4 acetylation of specific chromosomal domains may vary during interphase. Such alterations might be correlated with replication because newly replicated chromatin contains acetylated histones (Ruiz-Carrillo et al., 1975), which become deacetylated shortly after incorporation into chromatin (Jackson et al., 1976). Deposition-related acetylation of lysines 5 and 12 of H4, that is, incorporation of these acetylated isoforms into newly replicated chromatin, appears to be a highly conserved phenomenon (Sobel et al., 1995). Moreover, Idei et al. (1996) reported different histone H4 acetylation patterns of plant interphase nuclei; however, they were unable to relate the different patterns with either defined cell cycle stages or specific chromatin domains (except for the nucleolus).
Transcriptionally active rDNA genes were shown to be devoid of nucleosomes (Sogo et al., 1984; Conconi et al., 1989, 1992; Dammann et al., 1993), but the presence of histones within the nucleolus and their degree of acetylation during the course of interphase is still an open question (Derenzini et al., 1985; Thiry and Muller, 1989; González-Melendi et al., 1998).
To learn whether the extent of acetylation at all acetylatable positions of the core histones H3 and H4 remains constant along the cell cycle for specific chromatin domains (nucleolus organizers, euchromatin, and two fractions of heterochromatin of the field bean), we developed a new approach. After immunodetection of histone isoforms on isolated meristematic nuclei sorted on the basis of their DNA content into G1, early S, mid-S, late S, and G2 fractions, defined chromatin domains of individual chromosomes are identified by fluorescence in situ hybridization (FISH) with specific probes. This approach has revealed distinct types of immunolabeling of specific chromatin domains depending on the isoform addressed and the cell cycle stage. We also compared the H4 acetylation patterns of nucleoli, euchromatin, and heterochromatin domains with the replication pattern and the potential transcriptional activity in these domains.
RESULTS
Acetylation Patterns of Histone H4 at Specific Chromatin Domains Are Modulated during the Cell Cycle
Mitosis
In accordance with our previous data (Houben et al., 1996; Belyaev et al., 1997), antisera recognizing histone H4 that was acetylated at lysines 5 (Figure 1D), 8, and 12 labeled the NOR of metaphase chromosomes of the field bean more intensely, and the interstitial heterochromatin less intensely, than they did the euchromatic regions.
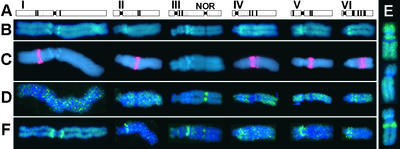
Figure 1.
The Six Chromosomes of the Field Bean Karyotype ACB.
(A) Scheme of Giemsa banding pattern, representing the heterochromatic regions.
(B) Fluorescence bands after staining with DAPI.
(C) FISH with tandemly repeated Fok elements (59 bp, red).
(D) Immunostaining of H4Ac5. Note that the acetylation is stronger at the NOR and weaker at the interstitial heterochromatin than at the euchromatin. The same pattern was obtained with antibodies against H4Ac8 and H4Ac12.
(E) Immunostaining of H4Ac16. Chromosome V is used to illustrate the three types of labeling during mitosis: 30% of the chromosomes showed an acetylation pattern identical to that obtained for H4Ac5 (top); 30% showed a uniform acetylation (middle), as described by Belyaev et al. (1997); and 40% revealed more strongly acetylated interstitial heterochromatin (bottom).
(F) Immunostaining of H3Ac14. Note the decreased acetylation of Fok element–containing (C) and the increased acetylation of Fok element–free, DAPI-positive (B) interstitial heterochromatic regions in comparison with euchromatin. The same pattern was obtained also for H3Ac9/18.
Antibodies against H4Ac16 were previously shown to label chromosomes uniformly, except for the NOR, which was more strongly labeled. Inspecting a higher number of chromosomes, we observed two additional patterns. Either the heterochromatin was more weakly labeled than euchromatin (30% of chromosomes), as seen for H4Ac5, 8, and 12, or it was more strongly labeled than euchromatin (40% of chromosomes; Figure 1E).
Incubation with the deacetylase inhibitor trichostatin A for 2 to 10 hr before mitosis resulted in highly intense acetylation of heterochromatin for the lysines K5, K12, and K16 (but not K8) of H4 (Belyaev et al., 1997). This agrees with the idea that deposition-related acetylation of K5 and K12 indeed might be highly conserved (Sobel et al., 1995) and that K16 in plants also might be acetylated in a deposition-related manner.
Interphase
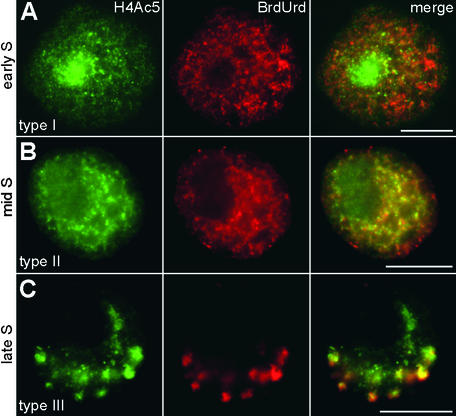
Immunodetection of H4Ac5 in interphase nuclei of the field bean revealed four distinct types and two subtypes of labeling patterns, which are shown in Figure 2 and described here:
Figure 2.
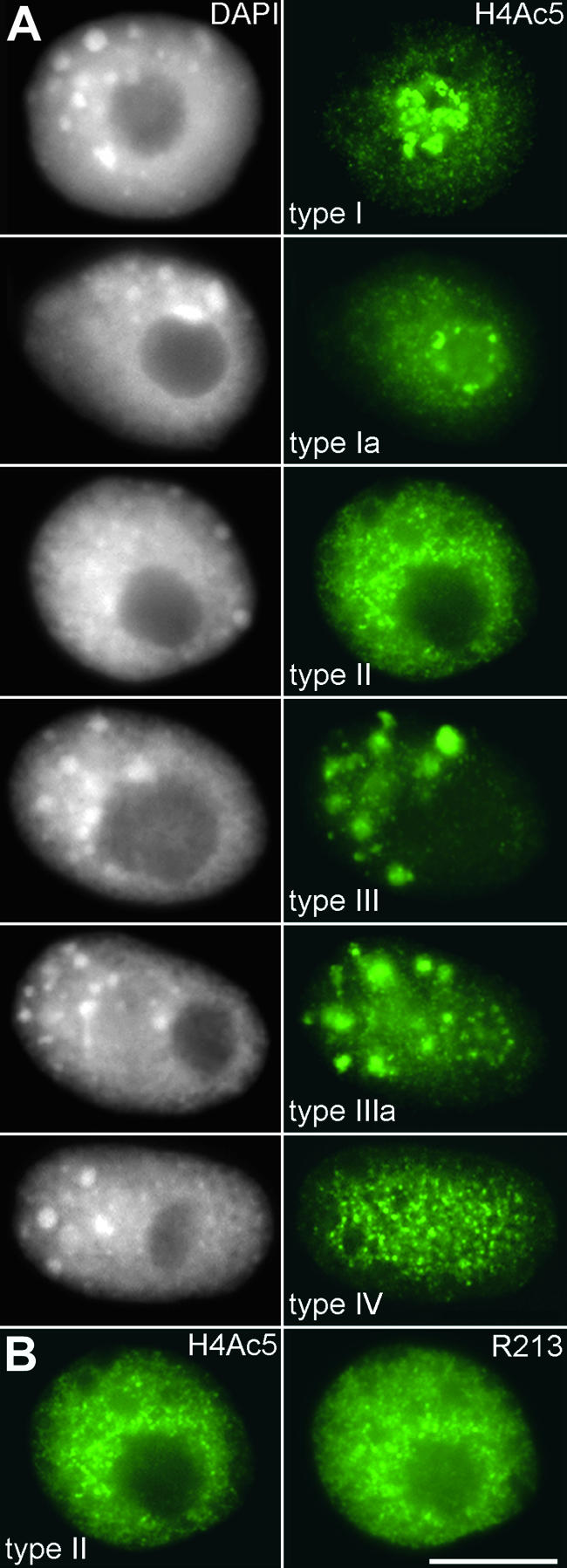
Types of Immunolabeling Pattern of Field Bean Interphase Nuclei Obtained with Antiserum against Histone H4Ac5.
(A) Nuclei counterstained with DAPI (left) and after immunolabeling of H4Ac5 (right). Note the different labeling intensities of nucleoli (strong in types I, Ia, and IIIa but absent in types II and III) and the additional “empty” spots (types I, Ia, II, and IV) or “bright” signal spots (types III and IIIa) in chromatin.
(B) Type II nucleus with nucleolus free of H4Ac5 (left) and also nearly free of immunosignals after subsequent labeling with antiserum R213 (right), which recognizes H4 regardless of acetylation. The absence of this label indicates depletion of H4 and therefore the absence of complete nucleosomes within nucleoli of these types of nuclei; the same was true for nucleoli of type III nuclei. Bar = 10 μm.
Type I shows the most intense signals within the nucleolus. The remaining chromatin is more weakly labeled by dispersed signals, and several unlabeled regions (“empty spots”) are visible. This type corresponds to the metaphase labeling pattern.
Subtype Ia differs from type I by weaker labeling of the nucleolus, the signals locating preferentially at the nucleolar periphery.
Type II shows nearly no signals within the nucleolus, stronger labeling of the average chromatin than in type I, and again, clear empty spots.
Type III shows unlabeled nucleoli; the chromatin on average is weakly labeled but contains several bright signal spots instead of empty spots.
Subtype IIIa differs from III by additional signals within the nucleolus, often forming a ring at the nucleolar periphery.
Type IV represents an intermediate between types I and II. It has empty spots but shows no difference in labeling intensity of nucleoli and the remaining chromatin.
Comparable labeling patterns were observed after immunodetection of H4Ac12, H4Ac16, and tetraacetylated H4 (not shown), though we noted that H4Ac16-labeled nuclei with bright spots always contained labeled nucleoli (subtype IIIa). Nuclei of types III and IIIa (that is, those with bright signal spots outside the nucleoli) were not seen when labeled with antibodies to H4Ac8.
Type II and III nuclei (those with unlabeled nucleoli) appeared to have less acetylated histone H4 inside the nucleolus than in the extranucleolar chromatin. To determine whether this reflects a lower overall amount of H4 in such nucleoli, we sequentially labeled the nuclei with antibodies recognizing acetylated H4 and with antibodies recognizing histone H4 regardless of its acetylation status (R213). The nucleoli of types II and III remained less intensely labeled than the surrounding chromatin, even after labeling with R213 (Figure 2B). This result was independent of the order in which the antisera were added. If the H4 tail in the nucleoli is not inaccessible for R213, then this observation suggests that nucleoli in type II and III nuclei have less H4 than in other types of nuclei.
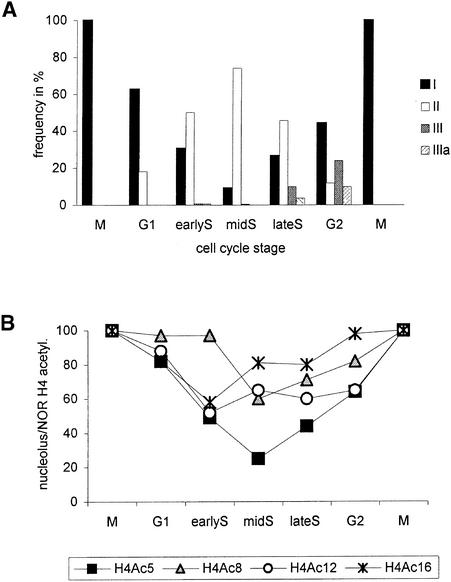
Variable Frequency of Histone H4 Labeling Patterns during the Cell Cycle
To ascertain whether the different histone H4 labeling patterns appeared with a constant frequency throughout the cell cycle, we flow-sorted formaldehyde-fixed field bean nuclei from unsynchronized root tip meristems according to their DNA content into fractions covering G1, early S, mid-S, late S, and G2 cell cycle phases (Figure 3). Several hundred nuclei from each fraction were immunolabeled with the specific antibodies. After immunodetection of H4Ac5, only nuclei of the intermediate types Ia and IV showed a similar, low frequency in all fractions (Table 1). Many of the nuclei in G1 (63%) and G2 (44%) contained both strongly labeled nucleoli and unlabeled regions (empty spots) within weakly labeled chromatin (type I), whereas in mid-S phase, the majority (74%) of the nuclei showed unlabeled nucleoli and strongly labeled chromatin (type II). Type III and IIIa nuclei with bright instead of empty spots in weakly labeled chromatin were observed only in late S and (early) G2 (Table 1 and Figure 4A). That not all nuclei of a fraction show the same pattern typical for the corresponding cell cycle stage might result primarily from nuclei showing an intermediate type such as Ia, IIIa, or IV. Missorting or “contamination” of G1 (or G2) fractions by nuclei from differentiated cells surrounding meristems cannot be totally excluded. However, the shape of the histogram (high and narrow G1 peak, separated clearly by S phase from a somewhat lower and broader G2 peak, and an absence of peaks for higher ploidy levels) (Figure 3) and the distribution of labeling types indicate that missorted and nonmeristematic nuclei should account for only a minor proportion within the sorted fractions.
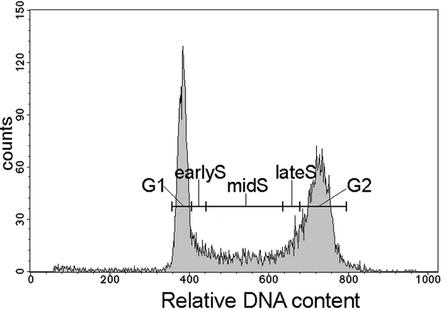
Figure 3.
Histogram of Relative DNA Content of Unsynchronized Field Bean Root Tip Nuclei after DAPI Staining and Flow-Cytometric Analysis.
The gates (representing G1, early S, mid-S, late S, and G2 phases) used for sorting are as indicated.
Table 1.
Histone H4Ac5 Labeling Patterns of Field Bean Nuclei during Interphase
| Cell Cycle Stage
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1
|
Early S
|
Mid-S
|
Late S
|
G2
|
||||||
| Labeling Typea | % | n | % | n | % | n | % | n | % | n |
| I | 63 | 258 | 30 | 112 | 9 | 35 | 27 | 120 | 44 | 217 |
| Ia | 14 | 56 | 12 | 43 | 11 | 43 | 8 | 38 | 5 | 24 |
| II | 18 | 75 | 50 | 184 | 74 | 284 | 46 | 207 | 12 | 58 |
| III | 0 | 0 | 1 | 3 | 1 | 2 | 10 | 45 | 24 | 118 |
| IIIa | 0 | 0 | 1 | 2 | 0 | 0 | 4 | 17 | 10 | 48 |
| IV | 5 | 23 | 6 | 23 | 5 | 19 | 5 | 25 | 5 | 25 |
| Σ | 100 | 412 | 100 | 367 | 100 | 383 | 100 | 452 | 100 | 490 |
For description of labeling types see Figure 2A and text.
Figure 4.
Variation of Histone H4 Acetylation Patterns during the Cell Cycle.
(A) Proportion of nuclei of labeling types I, II, III, and IIIa after immunodetection of H4Ac5 in different cell cycle stages. Because types Ia and IV revealed a nearly constant frequency, ranging from 5 to 14% and 5 to 6%, respectively, they therefore were omitted (cf. with Table 1).
(B) Relative frequency of nuclei showing acetylation of lysines 5, 8, 12, and 16 of H4 inside nucleoli during the cell cycle.
A similar but less pronounced decrease of nucleolus labeling during S phase was observed after use of antibodies against H4Ac8 (minimum in mid-S), H4Ac12, and H4Ac16 (both with a minimum in early S; see Figure 4B). However, after immunostaining of H4Ac16, the proportion of type IIIa nuclei was greater in G2 (almost 50%), and some (15%) were found even in G1.
The temporal acetylation pattern of histone H4 of euchromatin was opposite that of the nucleolus organizers or nucleoli. Euchromatin was most intensely labeled (type II) for all acetylatable lysines, particularly during early and mid-S phase. To compare directly histone acetylation and DNA replication patterns, the cells were pulse-treated for 30 min with 5-bromo-2′-deoxyuridine (BrdUrd) before fixation and isolation of the nuclei. Immunodetection of BrdUrd and subsequently of H4Ac5 yielded similar labeling patterns. The high degree of colocalized signals (except for the nucleolus) shown in Figure 5 indicates that the most intense H4 acetylation occurs during or shortly after replication.
Figure 5.
Correlation of Histone Acetylation (H4Ac5) and DNA Replication during S Phase.
After 30 min of BrdUrd pulse, the nuclei were isolated, flow-sorted, and double-immunolabeled for H4Ac5 (green, left) and BrdUrd (red, middle).
(A) Early S phase.
(B) Mid-S phase.
(C) Late S phase.
Note the large degree of colocalization of both immunosignals in early S (except for the NOR), mid-S, and late S nuclei. The bright spots in (C) represent late-replicating heterochromatin.  .
.
Acetylation of Histone H4 of Heterochromatic Regions: Strongest during Replication
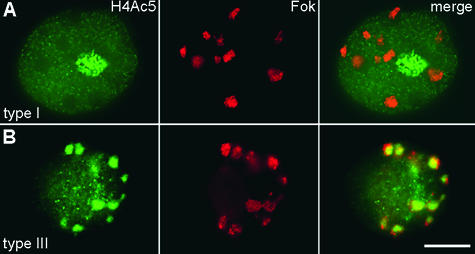
FISH with the tandem repetitive Fok element (contained within ∼75% of the Giemsa-banded interstitial heterochromatic regions of the field bean; see Figures 1A and 1C, and Fuchs et al., 1998) after immunodetection of H4Ac5 revealed that the 10 large Fok element sites exclusively colocalize with less intensely acetylated chromatin regions (empty spots) of labeling types I, Ia, II, and IV (see Figure 6A). The empty spots without Fok signals probably correspond to the interstitial heterochromatic regions, which contain repeats other than Fok elements. In type III and IIIa nuclei, bright spots (indicating strongly acetylated chromatin) were observed instead of empty spots. As Figure 6B shows, all major Fok element positions colocalize with the most strongly acetylated regions of type III and IIIa nuclei.
Figure 6.
Histone H4 Acetylation of Interstitial Heterochromatin Changes during the Cell Cycle.
(A) FISH with Fok elements (red, middle) performed after H4Ac5 immunolabeling (green, left) shows that heterochromatin domains coincide with empty spots representing underacetylation in type I nuclei, that is, during G1 and G2 (cf. with Table 1); the same is true for nuclei of types Ia, II, and IV.
(B) In nuclei of type III (and IIIa), which appear during late S and early G2 (cf. with Table 1 and Figure 4A), late-replicating heterochromatin domains colocalize with bright spots of H4Ac5 labeling. 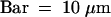 .
.
To be sure that neither bright FISH signals nor acetylation signals escaped detection by epifluorescence microscopy, we also performed optical sectioning of type III nuclei by using a confocal laser microscope after FISH with Fok elements and immunostaining for H4Ac5. As seen from Figure 7, all bright FISH and acetylation signals proved to be detectable by both techniques in individual field bean nuclei.
Figure 7.
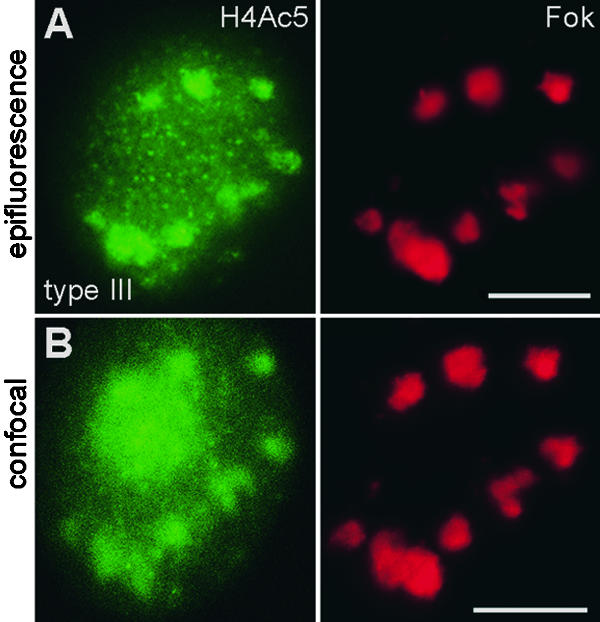
Images of a Type III Nucleus of Late S to Early G2 Phase after Immunodetection and FISH.
(A) and (B) Immunodetection of H4Ac5 (green, left) followed by FISH with Fok elements (red, right) as observed under (A) epifluorescence and (B) confocal microscopy overlaying 13 optical sections through the nucleus. H4Ac5 immunosignals were captured.
(A) Before FISH.
(B) After FISH.
The major immuno- and FISH signals are identical within both images. The green signal covering the nucleolus in (B) is autofluorescence, which frequently appeared when images were taken after FISH, although it was absent from the same nuclei when checked before FISH, as in (A). 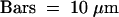 .
.
Because bright spots for H4 acetylation appeared only in late S and G2 nuclei and because the interstitial heterochromatin of the field bean was found to replicate latest in S phase (Döbel et al., 1978; Fuchs et al., 1998), presumably the H4 of the prominent interstitial heterochromatic domains becomes strongly acetylated at K5 during or shortly after replication. This agrees with our data from experiments combining H4Ac5 immunodetection and BrdUrd pulse labeling of early, mid, and late S-phase nuclei (Figure 5) and with the current view of deposition-related acetylation (Sobel et al., 1995).
During G2, H4Ac5 in heterochromatic domains is deacety-lated to an extent clearly less than that in euchromatin. This process is finished at least 2 hr before mitosis (Belyaev et al., 1997); the deacetylated state then lasts until the next replication.
A similar temporal pattern of acetylation was observed for K12 of H4, but K8 in heterochromatin was never acetylated as strongly as or more strongly than euchromatin. Because strongly acetylated heterochromatin at K16 was found in 50% of G2 nuclei, in 40% of mitotic chromosomes (Figure 1E), and in 15% of G1 nuclei but in only 2% of early S and mid-S nuclei, deacetylation of K16 presumably is delayed in comparison with K5 and K12, both of which were highly acetylated within the heterochromatin only in late S and part of G2 but not during mitosis and G1.
Histone H4 Acetylation: Nearly Absent from rDNA during S Phase and Not Directly Correlated with Transcriptional Activity
Only chromosome pair III of the field bean karyotype ACB harbors the genes for the 5.8, 18, and 25S rRNAs. Interphase nuclei therefore contain one or two nucleoli. In G1, 60 to 70% of the nuclei have only one nucleolus, compared with 85 to 90% in G2. These results suggest the nucleoli have a tendency to fuse as the cell cycle progresses.
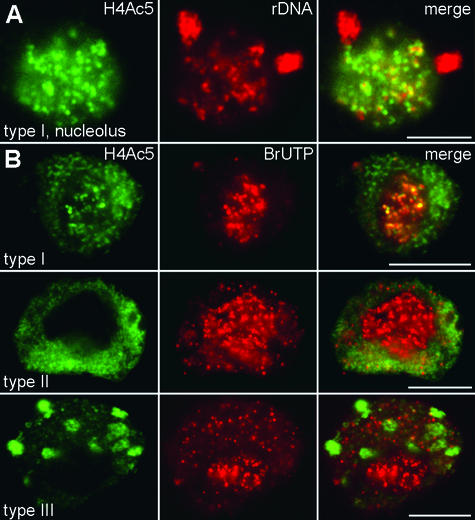
After FISH with labeled rDNA, what we observed most frequently in isolated nuclei were two perinucleolar signal clusters. Signals inside the nucleoli appeared as intensely fluorescing small dots or faint threadlike or diffuse signals (Figure 8A).
Figure 8.
Histone Acetylation and Transcriptional Activity of rDNA during Interphase.
(A) FISH with rDNA (red, middle) after immunolabeling of H4Ac5 (green, left) from a type I nucleus (only the nucleolus is shown) characteristic for G1 and G2 stages. Perinucleolar knobs containing inactive rDNA and some foci of condensed rDNA inside nucleoli are free of H4Ac5, as shown after merging of both signals. Most of H4Ac5 immunosignals are confined to faint, diffuse rDNA signals.
(B) Immunostaining of H4Ac5 (left), BrUTP incorporation (middle), and merging of both signals (right) for type I (G1), II (mid-S), and III (late S) nuclei. The transcriptional activity of rDNA is not correlated with H4 acetylation. Nucleoli are heavily labeled already after 4 min of BrUTP incorporation (red), irrespective of their acetylation status (green). Although BrUTP signals outside nucleoli are much weaker, no transcription signals were detected within heterochromatin domains (neither within empty spots in type II nuclei nor within bright spots in type III nuclei).
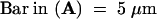 ;
; 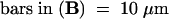 .
.
During mitosis, the NOR is more strongly acetylated than the euchromatin (Figure 1D; Belyaev et al., 1997). After H4Ac5 immunolabeling of type I nuclei (with strongly immunolabeled nucleoli), FISH with rDNA revealed that at least part of the rDNA inside the nucleoli, but not the perinucleolar rDNA, was colocalized with H4Ac5 immunosignals, as seen in Figure 8A. However, intense signals for rDNA as well as for H4Ac5 often were found in separate positions inside the nucleoli of type I nuclei.
To compare histone H4 acetylation with transcriptional activity, we labeled nascent RNA transcripts with 5-bromouridine-5′-triphosphate (BrUTP). As demonstrated in Figure 8B, after 4 min of BrUTP incorporation into isolated nuclei, all nuclei revealed intensely labeled nucleoli, regardless of the degree of H4 acetylation within the nucleoli. Type I nuclei showed a partial colocalization of BrUTP and acetylation signals inside nucleoli. The remaining chromatin in the field bean cells—unlike that in the observations made with mammalian cells (Jackson et al., 1993; Wansink et al., 1993; Sadoni et al., 1999)—was less densely labeled in all types of nuclei. Types II and III nuclei (representative of most of the S-phase cells) showed no H4 acetylation signals within nucleoli; that is, there was no association of acetylated H4 and intranucleolar rDNA (Figures 2A, 4A, 4B, and 8B). Heterochromatin domains were free of BrUTP signals. This became clear from overlaying BrUTP and H4Ac5 signals in type III nuclei (see bright spots in Figure 8B) but also was true for the empty spots of types I and II nuclei. This confirmed the transcriptional inactivity of the interstitial field bean heterochromatin (Houben et al., 1994) .
Histone H3 Acetylation Patterns in Interphase Nuclei Differ from Those of H4 and Are Nearly Invariant during Cell Cycle Progression
Labeling field bean chromosomes with antisera recognizing histone H3 acetylated at lysine positions 14 (H3Ac14; Figure 1F) and 9/18 (H3Ac9/18) looked different from the pattern obtained after labeling of histone H4Ac5 (Figure 1D). Besides the NOR, the Fok element–free interstitial heterochromatin also was more strongly acetylated, whereas Fok element–containing heterochromatin (Figure 1C) was again less acetylated than euchromatin. H3Ac23 immunolabeling was uniform along the chromosomes, except for the NOR, which was less strongly labeled (Belyaev et al., 1998).
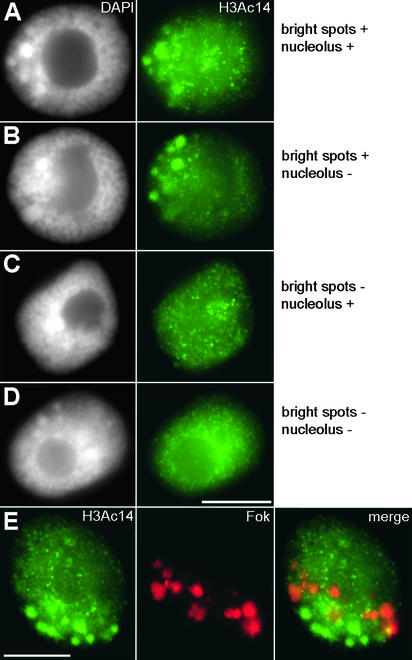
Immunodetection of H3Ac14 in interphase nuclei revealed two main patterns of labeling that differed by the presence or absence of intensely labeled spots in chromatin (Figure 9). These spots, mostly colocalizing with regions stained brightly by 4′,6-diamidino-2-phenylindole (DAPI), were not uniformly distributed throughout the nucleus but instead were clustered at one nuclear pole. Because most of the Fok element–free but DAPI-positive heterochromatin is located close to the centromeres (Figure 1B; Fuchs et al., 1998), this corresponds to the so-called Rabl orientation (Rabl, 1885). The number of these spots varied from five to 18, with a median number of eight. Empty spots were present in (almost) all nuclei. In many nuclei, however, they were not easily recognizable because of weak average chromatin labeling. In nuclei without bright spots, the average chromatin labeling was stronger, and empty spots were more easily detectable (Figures 9C and 9D). Nucleoli were either labeled or unlabeled, whether bright spots were present or not. The labeling of the nucleolus, if present, was generally less intense than in type I nuclei after H4Ac5 labeling. The signals of the nucleoli usually appeared as small dots in the center or as a ring at the periphery of the nucleolus.
Figure 9.
H3Ac14 Labeling Patterns of Field Bean Interphase Nuclei and Their Correlation with Heterochromatic Domains.
(A) Nucleus with bright signal spots for H3Ac14 and labeled nucleolus. This type represents the majority (63 to 74%) of nuclei in all interphase stages.
(B) Nucleus with bright signal spots but without intense labeling of the nucleolus.
(C) Nucleus without bright signal spots; the nucleolus is somewhat more strongly labeled than the remaining chromatin.
(D) Nucleus with neither bright signal spots nor intensely labeled nucleolus.
DAPI staining (left) and immunodetection (right). Note the presence of more weakly labeled areas in all nuclei and the correlation of bright signal spots with areas of positive DAPI fluorescence. The frequencies of these types in the course of interphase are given in Table 2.
(E) Same type of nucleus as in (B) after immunodetection of H3Ac14 (left), FISH with Fok elements (middle), and merging of both (right). Fok element sites (red) occupy the less acetylated areas and do not colocalize with the bright signal spots for H3Ac14, which represent Fok element–free heterochromatin.
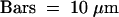 .
.
Comparable labeling patterns of interphase nuclei were obtained with the antiserum that recognized H3Ac9/18. After immunodetection of H3Ac23, interphase chromatin was more or less homogeneously labeled and the nucleoli were slightly less labeled. Neither bright spots nor empty spots were found.
H3Ac14 labeling on sorted nuclei revealed that the pattern with bright spots in chromatin and labeled nucleoli was the one seen most frequently (63 to 74%) in all cell cycle stages. Between 72 and 85% of nuclei (with either labeled or unlabeled nucleoli) showed bright spots; only between 10 and 18% of the nuclei revealed unlabeled nucleoli during all cell cycle stages (Table 2). Together these data show that, unlike histone H4 acetylation, the variability of H3 acetylation patterns was much less pronounced and not clearly dependent on the cell cycle stage.
Table 2.
Histone H3Ac14 Labeling Patterns of Field Bean Nuclei during Interphase
| Cell Cycle Stage
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1
|
Early S
|
Mid-S
|
Late S
|
G2
|
||||||
| Labeling Typea | % | n | % | n | % | n | % | n | % | n |
| Bright spots + nucleolus + | 71 | 267 | 63 | 236 | 66 | 259 | 67 | 262 | 74 | 298 |
| Bright spots + nucleolus − | 14 | 54 | 9 | 33 | 8 | 33 | 7 | 28 | 6 | 25 |
| Bright spots − nucleolus + | 8 | 28 | 14 | 52 | 15 | 59 | 11 | 43 | 10 | 38 |
| Bright spots − nucleolus − | 2 | 9 | 9 | 35 | 4 | 15 | 7 | 25 | 4 | 16 |
| Bright spots − uniform | 5 | 18 | 5 | 17 | 7 | 25 | 8 | 32 | 6 | 23 |
| Σ | 100 | 376 | 100 | 373 | 100 | 391 | 100 | 390 | 100 | 400 |
| Total nucleolus + | 79 | 295 | 77 | 288 | 81 | 318 | 78 | 305 | 84 | 336 |
| Total bright spots + | 85 | 321 | 72 | 269 | 75 | 292 | 74 | 290 | 80 | 323 |
Empty spots occur in most nuclei of all labeling types.
To determine whether the H3Ac14 labeling of metaphase chromosomes (with highly acetylated Fok element–free and nonacetylated Fok element–containing heterochromatin) persists or alters during interphase and whether Fok element–containing heterochromatin is subject also to histone H3 acetylation during interphase, we performed FISH with Fok elements after H3Ac14 immunolabeling (Figure 9E). Fok elements were mostly colocalizing with empty spots (lacking detectable H3Ac14), regardless of the presence or absence of bright H3Ac14 spots in the respective nuclei. Strongly immunolabeled spots did not contain Fok elements. Usually, Fok elements occurred in less polar positions than the brightest immunosignals. This is reasonable given that, in most cases, Fok elements are located more distantly from the centromeres than are the Fok element–free heterochromatic regions (Figures 1B, 1C, and 9E).
These studies gave the following results: (1) strong histone H3 acetylation is excluded from Fok element–containing heterochromatin, (2) Fok element–containing and Fok element–free heterochromatin occupy separate compartments within interphase nuclei, (3) only Fok element–free heterochromatin and nucleolus organizers are targets for very strong acetylation of histone H3, (4) H3 acetylation is not clearly related to the replication of euchromatin and heterochromatin domains, and (5) H3 acetylation shows no clear correlation with the transcriptional status of the investigated chromatin domains.
DISCUSSION
Overall Acetylation of Large Chromatin Domains Correlates with Replication Rather Than with Transcription
We have shown by FISH with Fok elements that the interstitial heterochromatin domains of individual field bean chromosomes form distinct compartments during interphase. Immunostaining of acetylated isoforms of histones H3 and H4 and subsequent identification of chromatin domains by FISH on meristematic nuclei, sorted according to their DNA content into five cell cycle fractions, allowed us for the first time to follow histone acetylation/deacetylation of defined chromatin domains of individual chromosomes through the cell cycle. BrdUrd pulse labeling allowed comparison of the acetylation intensity with replicational activity of the corresponding domains and showed that acetylation of H4 in euchromatin of the field bean is most pronounced during replication and is weaker from late S through M to G1. In contrast, H4 acetylation in early replicating rDNA is most intense during mitosis, decreases in G1 toward a minimum in early S (K12, K16) or mid-S (K5, K8), and increases again from late S onward (Figures 4A, 4B, and 5). The prominent interstitial heterochromatin domains replicate late, are transcriptionally silent, and represent hot spots of mutagen-induced chromosomal aberrations (reviewed in Fuchs et al., 1998). They become strongly acetylated at all acetylatable lysines of H4 (except K8) during late S phase, are deacetylated in G2 (>2 hr before mitosis; Belyaev et al., 1997), and remain deacetylated until the next replication. Only deacetylation of K16 of H4 is not always completed before mitosis, and acetylation of this residue may persist until the next G1 (for a summary, see Figure 10). A narrow time window of H4 acetylation at K5 and K12, correlating with replication, recently was reported also for mammalian heterochromatin (Taddei et al., 1999). In the field bean, H4 of euchromatin (as well as of centromeres and telomeres) and of all prominent interstitial heterochromatin thus shows most pronounced acetylation during and shortly after replication. This is in accordance with a phylogenetically conserved deposition-related acetylation at lysines 5 and 12 (Sobel et al., 1995). We predict that in plants lysine 16 also might be acetylated in a deposition-related manner.
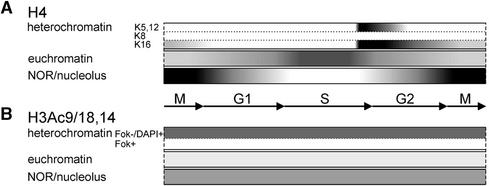
Figure 10.
Acetylation of Nucleosomal Histones at the NOR, Euchromatic, and Heterochromatic Domains of the Field Bean during the Cell Cycle.
(A) Histone H4. A strong cell cycle–dependent histone H4 acetylation occurs at the level of distinct chromatin domains. Heterochromatin contains acetylated H4 (except H4Ac8) during and (shortly) after replication; euchromatin, too, is most strongly acetylated during replication; the NOR contains acetylated H4 during mitosis, as do nucleoli in G1 and G2, but during S phase the histone H4 acetylation within nucleoli is considerably decreased.
(B) Histone H3. The intensity of H3 acetylation differs between Fok element–free heterochromatin, Fok element–containing heterochromatin, euchromatin, and the NOR/nucleolus but, unlike H4 acetylation, remains fairly constant throughout the cell cycle.
Still undetermined is the reason for the apparently futile strong postreplicative hyperacetylation within the heterochromatin. Because recombinative assembly of immunoglobulin genes in mammals has been found to be stimulated by histone acetylation (McBlane and Boyes, 2000; McMurry and Krangel, 2000), perhaps acetylation of histones (especially H4 in plants) is supportive also for recombination repair of DNA damage, which preferentially occurs during or shortly after replication. This might be particularly important with regard to heterochromatin, which is less acetylated in other cell cycle stages.
Although histone acetylation at the genic level may well be correlated with transcriptional activity (see Introduction), the overall acetylation of large chromatin domains is correlated with replication rather than with transcription (Figures 5 and 8B). However, there are two exceptions to this. One is the acetylation of H3, which is more or less persistent throughout the cell cycle, including the apparently sequence-specific above-average acetylation at lysines 9, 14, and 18 within Fok element–free heterochromatic regions. Perhaps H3 acetylation is required for maintenance of some heterochromatin domains, as is the case, for instance, in yeast (Thompson et al., 1994; Hecht et al., 1995; Braunstein et al., 1996; Grunstein, 1998). The other exception is the NOR.
H4 Is More Strongly Acetylated at the NOR during Mitosis, Whereas Nucleoli in S Phase Appear to Be Less Acetylated Than Euchromatin and May Be Transiently Free of Nucleosomes
The overall H4 acetylation patterns of field bean rDNA genes apparently are not strictly correlated with their transcriptional activity but are inversely correlated with the replication of rDNA, which takes place very early in S phase (Schubert and Rieger, 1979; Fuchs et al., 1998). This became evident from the finding that the strongest acetylation of H4 at the NOR occurred during mitosis, when rDNA is being neither replicated nor transcribed. The same was observed for barley (Idei et al., 1996) and onion (L. Malysheva and I. Schubert, unpublished findings) but not for mammals (Jeppesen and Turner, 1993) except for the NOR of the inactive X chromosome of female marsupials (Keohane et al., 1998). The presence of essential components of the rDNA transcriptional machinery at the NOR during mitosis (Scheer et al., 1993), which enables early, efficient initiation of transcription already in telophase/early G1 (Roussel et al., 1996; Gébrane-Younès et al., 1997; Klein and Grummt, 1999; Scheer and Hock, 1999), and our data on BrUTP incorporation indicate that rDNA is intensely transcribed during the entire interphase, regardless of H4 acetylation within the nucleolus. Strong H4 acetylation also appears frequently within nucleoli of G1 and G2 nuclei but only rarely during S phase, when replication occurs. Moreover, the results obtained after labeling of type II nuclei with antiserum R213, which recognizes histone H4 regardless of acetylation, show that in most nucleoli during S phase too little H4 is present for detection by immunolabeling. This indicates the absence of complete nucleosome core particles (the presence of acetylated H3 could be attributable to free histone molecules).
According to Lucchini and Sogo (1995), the coding regions of newly replicated rDNA genes usually are organized in nucleosomes, and transcriptional activation of rDNA requires disruption of preformed nucleosomes. This agrees with the apparent depletion of nucleosomes from field bean nucleoli shortly after rDNA replication in early S phase, which is not reversed until late S/G2, and invites the speculation that not only active rDNA genes but also the entire nucleolus is devoid of nucleosomes during most of S phase; consequently, histone acetylation cannot be detected.
Thus, the question arises as to the degree of acetylated nucleosomal histones associated with transcriptionally active versus inactive rDNA during G1 and G2. Only a subset of rRNA genes is actively transcribed during interphase (Shaw et al., 1995), even in yeast with comparatively few rRNA genes (Dammann et al., 1993), and heavily transcribed cistrons are not organized in nucleosomal structures (e.g., Sogo et al., 1984; Conconi et al., 1992; Dammann et al., 1993). Apparently, transcriptional activity within nucleoli is upregulated by increased activity of already active cistrons that are free of nucleosomes (reflected by “Christmas tree”–like structures of nascent transcripts in electron microscopic images; Miller and Beatty, 1969) rather than by activation of (all) silent genes (Banditt et al., 1999). Transcriptionally silent rDNA copies forming condensed perinucleolar knobs are free of acetylated H4, as are some condensed rDNA blocks inside nucleoli during G1 and G2, whereas acetylation signals were found in nucleolar areas exhibiting diffuse or very faint signals after FISH with rDNA (Figure 8A). Arrays of inactive rDNA genes might become associated, forming intranucleolar condensed chromatin, whereas active rDNA genes loop out. These loops of less condensed rDNA might be still acetylated in G1 (type I and Ia nuclei) and already in G2 (type IIIa and I nuclei). When rDNA transcription increases during G1, at least some of these loops might become heavily covered by RNA polymerase I molecules, while at the same time being free of nucleosomes and histone H4 molecules (type II and III nuclei). Closely adjacent highly active and less active genes could explain the occasional proximity and partial overlap of acetylated H4 and condensed intranucleolar rDNA in G1 and G2 nuclei. Highly active rDNA genes are assumed to have a decondensed structure close to the limit of resolution of optical microscopy (Thompson et al., 1997). Such genes might be responsible for the faint diffuse signals observed after FISH with rDNA, as compared with the bright signals at condensed rDNA repeats. When the rate of rDNA transcription decreases in the course of G2, the fewer RNA polymerase I molecules per transcribed gene allow (acetylated) nucleosomes to bind to DNA, resulting in increased acetylation within the nucleoli. This acetylation pattern is then maintained at the NOR during mitosis and becomes reversed during the course of G1.
According to the hypothesis proposed by Jeppesen (1997), histone acetylation can provide a mechanism for propagating “cell memory.” He suggests that “genes in chromatin domains active before mitosis are marked by histone acetylation, and hence have the potential for being preferentially reactivated in the following G1 phase. Acetyl groups then serve as ‘tags’ for recognition by other proteins involved in regulating transcription.” This hypothesis could explain the temporal pattern of H4 acetylation at the NOR chromatin observed in the present study, which correlates with neither replication nor transcriptional activity.
Conclusions
The extent of H4 acetylation is greatest during or shortly after replication within eu- and heterochromatin of the field bean—except for rDNA chromatin, which was most highly acetylated at and around mitosis and was apparently free of H4 during S phase. Because heterochromatin is transcriptionally silent, but rDNA and large parts of euchromatin are transcribed throughout interphase, the overall H4 acetylation of large chromatin domains (except rDNA) apparently is linked to replication (and possibly postreplicative recombination repair) rather than to transcriptional activity.
The amount of H3 acetylation did not show a clear cell cycle dependence. Therefore, no clear correlation with replication or transcription could be stated with regard to the large chromatin domains in this species. Contrary to the situation in mammals, chromatin fractions of the field bean showed deviations between the acetylation patterns of H4 and H3; thus, the requirements for acetylation of these two histones may be different in plants.
The replication-associated stronger acetylation of K5 and K12 of H4 in late-replicating heterochromatin (Taddei et al., 1999) and the degree of acetylation of euchromatin (which is most intense during early to mid-S phase) probably are conserved for plants and animals, whereas the increased acetylation of K16 at heterochromatic domains—which occurs during late S and disappears between G2 and G1—might be plant specific.
Stronger acetylation of H4 at nucleolus organizers during mitosis (cf. with the situation for euchromatin) as well as the very high transcriptional activity within nucleoli (cf. with mammalian cells) (Jackson et al., 1993; Wansink et al., 1993; Sadoni et al., 1999) seems to be typical for several plant species.
Further investigation will show whether the apparently sequence-dependent strong acetylation of K9, K14, and K18 of H3 represents a particular feature of the DAPI-positive heterochromatin fraction of the field bean or is more widespread in other (plant) species.
METHODS
Plant Material, Preparation of Slides, and Isolation and Sorting of Nuclei
Root tip meristems of the field bean (Vicia faba) karyotype ACB with individually distinguishable chromosome pairs (Fuchs et al., 1998) were used in all experiments. Suspensions of nuclei from unsynchronized root tip meristems and chromosomes from synchronized meristems (fixed in 4% formaldehyde, 10 mM Tris-HCl, 10 mM Na2 EDTA, 100 mM NaCl, and 0.1% Triton X-100, pH 7.5, for 20 min under vacuum) were prepared as described (Schubert et al., 1993). Isolated nuclei and chromosomes were centrifuged onto a microscopic slide by using a Cytospin3 (Shandon, Frankfurt, a.M., Germany) cytological centrifuge at 18g for 5 min; the loaded slides then were stored in glycerol at 4°C until use. Nuclei isolated from unsynchronized meristems (the first 2 mm of the root tips) and stained with 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) were sorted into G1, early S, mid-S, late S, and G2 fractions with a FACStarPlus (Becton Dickinson) flow cytometer. The gates for sorting were determined according to the histogram for nuclear suspensions (Figure 3). Approximately 1000 nuclei of each fraction were sorted onto a microscopic slide into a 15-μL drop of buffer consisting of 100 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2, 0.05% Tween 20, and 5% sucrose (Kubaláková et al., 1997). The drops with nuclei were nearly air-dried (sucrose prevents complete drying), and unless used immediately for immunolabeling or fluorescence in situ hybridization (FISH), the slides were stored at −20°C.
Indirect Immunodetection of Histone Isoforms
Polyclonal antisera against histones H3 and H4 acetylated at defined lysine residues were raised by immunization of rabbits with ovalbumin-conjugated synthetic peptides, as previously described (Turner and Fellows, 1989; White et al., 1999). The antisera used, and their specificities, were as follows: R41 (H4Ac5), R232 (H4Ac8), R101 (H4Ac12), R252 (H4Ac16), R243 (preferentially tri- and tetraacetylated H4), R213 (preferentially nonacetylated H4), R47 (H3Ac9, H3Ac18, or both), R224 (H3Ac14), and R222 (H3Ac23); see Turner et al. (1989), Belyaev et al. (1996), Stein et al. (1997), and White et al. (1999) for further details. The specificity of these sera to the same histone isoforms of plants was shown by protein gel blot analysis (Buzek et al., 1998). Preimmune sera reacted with neither the nuclear proteins (Buzek et al., 1998) nor chromosomes of the tested plants (Vyskot et al., 1999).
The nuclei were postfixed in 4% (w/v) paraformaldehyde in PBS for 20 min, washed three times in PBS, and blocked for 1 hr at 37°C in PBS containing 3% BSA and 10% horse serum. Slides then were incubated for 1 hr at room temperature (RT) in primary sera diluted 1:200 or 1:100 in AK (antibody) buffer (PBS containing 1% BSA, 10% horse serum, and 0.1% Tween 20; see ten Hoopen et al., 2000). After three washes in PBS, fluorescein isothiocyanate (FITC)–conjugated goat anti–rabbit secondary antibodies (Sigma) diluted 1:80 in AK buffer were applied for 1 hr at RT. The slides then were washed in PBS, and the DNA was counterstained with DAPI (1 μg/mL in mounting medium [Vectashield; Vector Labs, Burlingame, CA]). Secondary antibodies alone did not stain chromosomes or nuclei of the field bean.
Nascent RNA Labeling
5-Bromouridine-5′-triphosphate (BrUTP) was incorporated into isolated nuclei essentially as described (Thompson et al., 1997). In brief, unfixed nuclei from root tip meristems were released into MPB (modified physiological buffer: 100 mM potassium acetate, 20 mM KCl, 20 mM Hepes, 1 mM MgCl2, 1 mM ATP, and 1 mM DTT, pH 7.4) containing 1 M hexylene glycol (2-methyl-2,4-pentanediol), centrifuged onto a slide, washed in MPB, permeabilized in MPB plus 0.05% Tween 20 for 10 sec (Abranches et al., 1998), and incubated for 3 to 10 min at RT with the following transcription mix: 50 μM CTP, 50 μM GTP, and 25 μM BrUTP (all nucleotides purchased from Sigma), 0.5 mM phenylmethylsulfonyl fluoride in MPB, and 100 U/mL RNase inhibitor (RNA Guard; Pharmacia). After being washed in MPB, nuclei were fixed for 40 min in 4% paraformaldehyde in PBS, washed three times in PBS, and blocked for 1 hr at 37°C. Incorporation of BrUTP was detected by incubation for 1 hr at RT with mouse anti-BrdU monoclonal antibody (Becton Dickinson) diluted 1:10 in AK buffer, followed by three washes in PBS and incubation with the secondary FITC-conjugated sheep anti–mouse (Boehringer Mannheim) antibody diluted 1:30, or when combined with histone immunolabeling, in Alexa594-conjugated goat anti–mouse (Molecular Probes, Eugene, OR) antibody diluted 1:500 to 1:1000 in AK for 1 hr at RT.
Replication Labeling
Main roots of 4-day-old seedlings were incubated in 5-bromo-2′-deoxyuridine (BrdUrd; 100 μM), fluorodeoxyuridine (0.1 μM), and uridine (5 μM), for 30 min in the dark. After a short rinse, the roots were immediately fixed in 4% formaldehyde/Tris-HCl buffer. After further washes in Tris-HCl buffer, nuclei were isolated and sorted as described above. Before immunodetection of BrdUrd, the nuclei were postfixed in 4% formaldehyde/PBS for 20 min and washed in PBS. DNA was denatured by treating the slides at 80°C for 1 min in 50% formamide/PBS. The slides then were immediately transferred into ice-cold PBS for 5 min and blocked. BrdUrd immunodetection was as described for BrUTP.
In Situ Hybridization
The following probes were used: Fok elements (59-bp tandem repeats, cleavable by the restriction endonuclease FokI [Kato et al., 1984], characteristic for ∼75% of the heterochromatic Giemsa bands of the field bean [Fuchs et al., 1994, 1998]) and pVER17 (with a 3.7-kb insert consisting of part of 18S, 5.8S, and most of the coding region of 25S rRNA genes of the field bean [Yakura and Tanifuji, 1983]). pVER17 was directly labeled with tetramethylrhodamine-5-dUTP (Boehringer Mannheim) by using a nick translation kit (Boehringer Mannheim) according to the manufacturer's instructions; FokI elements were amplified from genomic field bean DNA and labeled with tetramethylrhodamine-5-dUTP or digoxigenin-11-dUTP (Boehringer Mannheim) by polymerase chain reaction with sequence-specific primers.
When FISH was performed after immunolabeling, the slides first were evaluated for immunosignals, washed in 4 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) plus 0.1% Tween 20 to remove the cover slip, and then washed briefly in 2 × SSC. The nuclei were again postfixed in 4% paraformaldehyde/2 × SSC, washed in 2 × SSC, dehydrated in 70 and 96% ethanol, and air-dried. Before FISH with pVER17, slides were incubated with RNase (50 μg/mL) for 15 min at 37°C. The hybridization mixture containing probe, 50% formamide, 10% dextran sulfate, and 2 × SSC was denatured at 80°C for 10 min and cooled on ice. The target DNA was denatured together with the probe on slides at 80°C for 2 min. When using directly labeled probes (tetramethylrhodamine-5-dUTP), after posthybridization washes (3 × 5 min in 50% formamide in 2 × SSC at 42°C and 5 min in 2 × SSC at RT), the nuclei were counterstained with DAPI. Digoxigenin-11-dUTP– labeled probes were detected with FITC- or rhodamine-conjugated anti-digoxigenin antibodies (Boehringer Mannheim).
Microscopy, Image Processing, and Evaluation of Data
The preparations were inspected with an Axiophot 2 (Zeiss, Thornwood, NY) epifluorescence microscope equipped with a cooled charge-coupled device camera (Photometrics, Tucson, AZ). Images were taken with use of IPLab Spectrum software, pseudocolored, merged, and processed in Adobe Photoshop.
To determine the frequency of distinct immunolabeling patterns for each acetylated isoform of histone H3 and H4, we evaluated at least 100 nuclei in G1, early S, mid-S, late S, and G2 phases, respectively.
Confocal Microscopy
Confocal microscopy was performed with a Zeiss (Jena, Germany) LSM 410. FITC (H4Ac5 immunosignals) and rhodamine (Fok element FISH signals) signals were recorded separately with excitation wavelengths of 488 and 543 nm and bandpass filters at 510 to 525 nm and 575 to 640 nm, respectively. Optical sections of the whole nucleus were obtained at a step width of 500 nm with the pinhole adjusted to yield an axial resolution (full width at half maximum) of 3.1 μm. Image stacks of details in the rhodamine signal were recorded with a step width of 250 nm and an axial resolution of 1.1 μm. The lateral pixel size was 50 nm in all images.
Acknowledgments
We thank Joachim Bruder, Barbara Hildebrandt, and Martina Kühne for excellent technical assistance and Rigomar Rieger, Paul Fransz, and Rogier ten Hoopen for critical reading of the manuscript. This work has been supported by the Fonds der Chemischen Industrie (A.M., I.S.) and by The Wellcome Trust (B.M.T.).
References
- Abranches, R., Beven, A.F., Aragón-Alcaide, L., and Shaw, P.J. (1998). Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 143, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey, V., Faulkner, R.M., and Mirsky, A.E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banditt, M., Koller, T., and Sogo, J.M. (1999). Transcriptional activity and chromatin structure of enhancer-deleted rRNA genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4953–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaev, N.D., Keohane, A.M., and Turner, B.M. (1996). Differential underacetylation of histones H2A, H3 and H4 on the inactive X chromosome in human female cells. Hum. Genet. 97, 573–578. [DOI] [PubMed] [Google Scholar]
- Belyaev, N.D., Houben, A., Baranczewski, P., and Schubert, I. (1997). Histone H4 acetylation in plant heterochromatin is altered during the cell cycle. Chromosoma 106, 193–197. [DOI] [PubMed] [Google Scholar]
- Belyaev, N.D., Houben, A., Baranczewski, P., and Schubert, I. (1998). The acetylation patterns of histones H3 and H4 along Vicia faba chromosomes are different. Chromosome Res. 6, 59–63. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., Sobel, R.E., Allis, C.D., Turner, B.M., and Broach, J.R. (1996). Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzek, J., Riha, K., Siroky, J., Ebert, I., Greilhuber, J., and Vyskot, B. (1998). Histone H4 underacetylation in plant facultative heterochromatin. Biol. Chem. 379, 1235–1241. [DOI] [PubMed] [Google Scholar]
- Conconi, A., Widmer, R.M., Koller, T., and Sogo, J.M. (1989). Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57, 753–761. [DOI] [PubMed] [Google Scholar]
- Conconi, A., Sogo, J.M., and Ryan, C.A. (1992). Ribosomal gene clusters are uniquely proportioned between open and closed chromatin structures in both tomato leaf cells and exponentially growing suspension cultures. Proc. Natl. Acad. Sci. USA 89, 5256–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas, A. (1990). On the biological role of histone acetylation. Biochem. J. 265, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann, R., Lucchini, R., Koller, T., and Sogo, J.M. (1993). Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21, 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini, M., Pession, A., Licastro, F., and Novello, F. (1985). Electronmicroscopical evidence that ribosomal chromatin of human circulating lymphocytes is devoid of histones. Exp. Cell Res. 157, 50–62. [DOI] [PubMed] [Google Scholar]
- Döbel, P., Schubert, I., and Rieger, R. (1978). Distribution of heterochromatin in a reconstructed karyotype of Vicia faba as identified by banding- and DNA-late replication patterns. Chromosoma 69, 193–209. [Google Scholar]
- Fuchs, J., Pich, U., Meister, A., and Schubert, I. (1994). Differentiation of field bean heterochromatin by in situ hybridization with a repeated FokI sequence. Chromosome Res. 2, 25–28. [DOI] [PubMed] [Google Scholar]
- Fuchs, J., Strehl, S., Brandes, A., Schweizer, D., and Schubert, I. (1998). Molecular-cytogenetic characterization of the Vicia faba genome—Heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res. 6, 219–230. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez, M., Rocchini, C., and Ausio, J. (1995). Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 270, 17923–17928. [DOI] [PubMed] [Google Scholar]
- Gébrane-Younès, J., Fomproix, N., and Hernandez-Verdun, D. (1997). When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J. Cell Sci. 110, 2429–2440. [DOI] [PubMed] [Google Scholar]
- González-Melendi, P., Testillano, P.S., Mena, C.G., Muller, S., Raska, I., and Risueño, M.C. (1998). Histones and DNA ultrastructural distribution in plant cell nucleus: A combination of immunogold and cytochemical methods. Exp. Cell Res. 242, 45–59. [DOI] [PubMed] [Google Scholar]
- Grunstein, M. (1997). Histone acetylation in chromatin structure and transcription. Nature 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Grunstein, M. (1998). Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell 93, 325–328. [DOI] [PubMed] [Google Scholar]
- Hecht, A., Laroche, T., Strahl-Bolsinger, S., Gasser, S.M., and Grunstein, M. (1995). Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell 80, 583–592. [DOI] [PubMed] [Google Scholar]
- Hong, L., Schroth, G.P., Matthews, H.R., Yau, P., and Bradbury, E.M. (1993). Studies of the DNA binding properties of histone H4 amino terminus. J. Biol. Chem. 268, 305–314. [PubMed] [Google Scholar]
- Houben, A., Brandes, A., and Schubert, I. (1994). The distribution of cDNA sequences on field bean chromosomes. Genome 37, 1065–1067. [DOI] [PubMed] [Google Scholar]
- Houben, A., Belyaev, N.D., Turner, B.M., and Schubert, I. (1996). Differential immunostaining of plant chromosomes by antibodies recognizing acetylated histone H4 variants. Chromosome Res. 4, 191–194. [DOI] [PubMed] [Google Scholar]
- Houben, A., Belyaev, N.D., Leach, C.R., and Timmis, J.N. (1997). Differences of histone H4 acetylation and replication timing between A and B chromosomes of Brachycome dichromosomatica. Chromosome Res. 5, 233–237. [DOI] [PubMed] [Google Scholar]
- Idei, S., Kondo, K., Turner, B.M., and Fukui, K. (1996). Tomographic distribution of acetylated histone H4 in plant chromosomes, nuclei and nucleoli. Chromosoma 105, 293–302. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A., Hassan, A.B., Errington, R.J., and Cook, P.R. (1993). Visualization of focal sites of transcription within human nuclei. EMBO J. 12, 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, V., Shires, A., Tanphaichitr, N., and Chalkley, R. (1976). Modifications of histones immediately after synthesis. J. Mol. Biol. 104, 471–483. [DOI] [PubMed] [Google Scholar]
- Jeppesen, P. (1997). Histone acetylation: A possible mechanism for the inheritance of cell memory at mitosis. Bioessays 19, 67–74. [DOI] [PubMed] [Google Scholar]
- Jeppesen, P., and Turner, B.M. (1993). The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74, 281–289. [DOI] [PubMed] [Google Scholar]
- Kato, A., Yakura, K., and Tanifuji, S. (1984). Sequence analysis of Vicia faba repeated DNA, the FokI repeat element. Nucleic Acids Res. 12, 6415–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohane, A.M., Lavender, J.S., O'Neill, L.P., and Turner, B.M. (1998). Histone acetylation and X inactivation. Dev. Genet. 22, 65–73. [DOI] [PubMed] [Google Scholar]
- Klein, J., and Grummt, I. (1999). Cell cycle–dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA 96, 6096–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubaláková, M., Macas, J., and Dolezel, J. (1997). Mapping of repeated DNA sequences in plant chromosomes by PRINS and C-PRINS. Theor. Appl. Genet. 94, 758–763. [Google Scholar]
- Lee, D.Y., Hayes, J.J., Pruss, D., and Wolffe, A.P. (1993). A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72, 73–84. [DOI] [PubMed] [Google Scholar]
- Loidl, P. (1988). Towards an understanding of the biological function of histone acetylation. FEBS Lett. 227, 91–95. [DOI] [PubMed] [Google Scholar]
- Loidl, P. (1994). Histone acetylation: Facts and questions. Chromosoma 103, 441–449. [DOI] [PubMed] [Google Scholar]
- Lucchini, R., and Sogo, J.M. (1995). Replication of transcriptionally active chromatin. Nature 374, 276–280. [DOI] [PubMed] [Google Scholar]
- McBlane, F., and Boyes, J. (2000). Stimulation of V(D)J recombination by histone acetylation. Curr. Biol. 10, 483–486. [DOI] [PubMed] [Google Scholar]
- McMurry, M.T., and Krangel, M.S. (2000). A role for histone acetylation in the developmental regulation of V(D)J recombination. Science 287, 495–498.10642553 [Google Scholar]
- Miller, O.L., and Beatty, R.R. (1969). Visualization of nucleolar genes. Science 164, 955–957. [DOI] [PubMed] [Google Scholar]
- Rabl, C. (1885). Über Zelltheilung. Gegenbaurs Morphol. Jahrb. 10, 214–330. [Google Scholar]
- Roussel, P., André, C., Comai, L., and Hernandez-Verdun, D. (1996). The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 133, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Carrillo, A., Wangh, L., and Allfrey, V. (1975). Assembly of newly replicated chromatin. Science 190, 117–128. [DOI] [PubMed] [Google Scholar]
- Sadoni, N., Langer, S., Fauth, C., Bernardi, G., Cremer, T., Turner, B.M., and Zink, D. (1999). Nuclear organization of mammalian genomes: Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 146, 1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, U., and Hock, R. (1999). Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11, 385–390. [DOI] [PubMed] [Google Scholar]
- Scheer, U., Thiry, M., and Goessens, G. (1993). Structure, function and assembly of the nucleolus. Trends Cell Biol. 3, 236–241. [DOI] [PubMed] [Google Scholar]
- Schubert, I., and Rieger, R. (1979). Asymmetric banding of Vicia faba chromosomes after BrdU incorporation. Chromosoma 70, 385–391. [Google Scholar]
- Schubert, I., Dolezel, J., Houben, A., Scherthan, H., and Wanner, G. (1993). Refined examination of plant metaphase chromosome structure at different levels made feasible by new isolation methods. Chromosoma 102, 96–101. [Google Scholar]
- Shaw, P.J., Highett, M.I., Beven, A.F., and Jordan, E.G. (1995). The nucleolar architecture of polymerase I transcription and processing. EMBO J. 14, 2896–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.G., Hill, R.S., and Baldwin, J.P. (1995). Plant chromatin structure and post-translational modifications. Crit. Rev. Plant Sci. 14, 299–328. [Google Scholar]
- Sobel, R.E., Cook, R.G., Perry, C.A., Annunziato, A.T., and Allis, C.D. (1995). Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 92, 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, J.M., Ness, P.J., Widmer, R.M., Parish, R.W., and Koller, T. (1984). Psoralen-crosslinking of DNA as a probe for the structure of active nucleolar chromatin. J. Mol. Biol. 178, 897–928. [DOI] [PubMed] [Google Scholar]
- Spencer, V.A., and Davie, J.R. (1999). Role of covalent modifications of histones on regulating gene expression. Gene 240, 1–12. [DOI] [PubMed] [Google Scholar]
- Stein, P., Worrad, D.M., Belyaev, N.D., Turner, B.M., and Schultz, R.M. (1997). Stage-dependent redistribution of acetylated histones in nuclei of the early preimplantation mouse embryo. Mol. Reprod. Dev. 47, 421–429. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 413, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl, K. (1998). Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Taddei, A., Roche, D., Sibarita, J.-B., Turner, B.M., and Almouzni, G. (1999). Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147, 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hoopen, R., Manteuffel, R., Dolezel, J., Malysheva, L., and Schubert, I. (2000). Evolutionary conservation of kinetochore protein sequences in plants. Chromosoma (in press). [DOI] [PubMed]
- Thiry, M., and Muller, S. (1989). Ultrastructural distribution of histones within Ehrlich tumour cell nucleoli: A cytochemical and immunocytochemical study. J. Histochem. Cytochem. 37, 853–862. [DOI] [PubMed] [Google Scholar]
- Thompson, J.S., Ling, X., and Grunstein, M. (1994). Histone H3 terminus is required for telomeric and silent mating locus repression in yeast. Nature 369, 245–247. [DOI] [PubMed] [Google Scholar]
- Thompson, W.F., Beven, A.F., Wells, B., and Shaw, P.J. (1997). Sites of rDNA transcription are widely dispersed through the nucleolus in Pisum sativum and can comprise single genes. Plant J. 12, 571–581. [DOI] [PubMed] [Google Scholar]
- Turner, B.M. (1991). Histone acetylation and control of gene expression. J. Cell Sci. 99, 13–20. [DOI] [PubMed] [Google Scholar]
- Turner, B.M. (1993). Decoding the nucleosome. Cell 75, 5–8. [PubMed] [Google Scholar]
- Turner, B.M. (2000). Histone acetylation and an epigenetic code. Bioessays 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Turner, B.M., and Fellows, G. (1989). Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur. J. Biochem. 179, 131–139. [DOI] [PubMed] [Google Scholar]
- Turner, B.M. O'Neill, L.P., and Allan, I.M. (1989). Histone H4 acetylation in human cells. Frequency of acetylation at different sites defined by immunolabeling with site-specific antibodies. FEBS Lett. 253, 141–145. [DOI] [PubMed] [Google Scholar]
- Vettese-Dadey, M., Grant, P.A., Hebbes, T.R., Crane-Robinson, C., Allis, C.D., and Workman, J.L. (1996). Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15, 2508–2518. [PMC free article] [PubMed] [Google Scholar]
- Vyskot, B., Siroky, J., Hladilova, R., Belyaev, N.D., and Turner, B.M. (1999). Euchromatic domains in plant chromosomes as revealed by H4 histone acetylation and early DNA replication. Genome 42, 343–350. [PubMed] [Google Scholar]
- Wansink, D.G., Schul, W., van der Kraan, I., van Steensel, B., van Driel, R., and de Jong, L. (1993). Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 122, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, D.A., Belyaev, N.D., and Turner, B.M. (1999). Preparation of site-specific antibodies to acetylated histones. Methods 19, 417–421. [DOI] [PubMed] [Google Scholar]
- Yakura, K., and Tanifuji, S. (1983). Molecular cloning and restriction analysis of EcoRI-fragments of Vicia faba rDNA. Plant Cell Physiol. 24, 1327–1330. [Google Scholar]
- Yoshida, M., Kijima, M., Akita, M., and Beppu, T. (1990). Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by Trichostatin A. J. Biol. Chem. 265, 17174–17179. [PubMed] [Google Scholar]