Abstract
Paramutation is the directed, heritable alteration of the expression of one allele when heterozygous with another allele. Here, the isolation and characterization of a mutation affecting paramutation, mediator of paramutation1-1 (mop1-1), are described. Experiments demonstrate that the wild-type gene Mop1 is required for establishment and maintenance of the paramutant state. The mop1-1 mutation affects paramutation at the multiple loci tested but has no effect on alleles that do not participate in paramutation. The mutation does not alter the amounts of actin and ubiquitin transcripts, which suggests that the mop1 gene does not encode a global repressor. Maize plants homozygous for mop1-1 can have pleiotropic developmental defects, suggesting that mop1-1 may affect more genes than just the known paramutant ones. The mop1-1 mutation does not alter the extent of DNA methylation in rDNA and centromeric repeats. The observation that mop1 affects paramutation at multiple loci, despite major differences between these loci in their gene structure, correlations with DNA methylation, and stability of the paramutant state, suggests that a common mechanism underlies paramutation. A protein-based epigenetic model for paramutation is discussed.
INTRODUCTION
Recognition of the widespread nature and importance of epigenetic phenomena in many organisms has prompted extensive review (see Cell Vol. 93, No. 2; Trends in Genetics Vol. 13, No. 8; Plant Molecular Biology Vol. 43, No. 2/3). Epigenetics refers to altered gene expression associated with alternative chromatin or methylation states (or both) superimposed on an unchanged primary DNA sequence. Alternative epigenetic states are frequently heritable through mitosis (Holliday et al., 1996; Pirrotta, 1997; Sherman and Pillus, 1997) and sometimes through meiosis (Jorgensen, 1995; Grewal and Klar, 1996; Cavalli and Paro, 1998; Chandler et al., 2000). Epigenetic modifications are often attributed to trans-sensing or homology effects (Henikoff and Comai, 1998; Wu and Morris, 1999). Some of the earliest examples of epigenetic phenomena were identified by pigment variegation in maize and Drosophila (Muller, 1930; Hinton and Goodsmith, 1950; Baker, 1953, Brink, 1956; McClintock, 1957; Hessler, 1958; Coe, 1959).
Anthocyanin pigmentation is a valuable tool in plants for identifying and tracking genetic and epigenetic events (Dooner et al., 1991; Meyer, 1995). The amount of the nonessential anthocyanins present is very sensitive to subtle changes in expression of the regulatory genes, and these differences are readily visible. In maize, these genes were used by Barbara McClintock to identify and follow the behavior of transposable elements (reviewed in Fedoroff, 1983) and by Alexander Brink and several researchers since to identify and characterize a phenomenon Brink (1958)(1973) termed paramutation. More recently, the first examples of cosuppression—transgene silencing of a homologous endogenous gene—were identified by using anthocyanin genes in petunia (Napoli et al., 1990; van der Krol et al., 1990).
Paramutation has been extensively characterized at three maize loci: r1, b1, and pl1 (reviewed in Chandler et al., 2000). b1 and r1 (see Methods for a summary of maize nomenclature) encode functionally interchangeable basic helix-loop-helix (bHLH) factors (Styles et al., 1973; Ludwig et al., 1989; Goff et al., 1990; Radicella et al., 1991), whereas pl1 encodes a myb-related transcription factor (Cone et al., 1993). Activation of the anthocyanin biosynthesis pathway requires coexpression of a bHLH factor and a myb-related factor (Goff et al., 1992). In general, the pl1 locus is expressed in the vegetative body of the plant, whereas its functional equivalent, c1, is expressed in the embryo and the aleurone layer of the kernel endosperm. Therefore, when functional alleles of pl1 and c1 are present, the b1 and r1 alleles present are generally what determine the tissue-specific patterns of anthocyanin expression (Styles et al., 1973).
Paramutation is an interaction between two specific alleles that leads to a heritable alteration in one of the alleles at a very high frequency. Paramutation was first described at the r1 locus of maize (Brink, 1956, 1958). Brink observed that the amounts of aleurone pigment in genotypically identical kernels (triploid endosperm R-r/r-g/r-g) differed considerably, depending on whether R-r was previously homozygous or heterozygous with R-stippled (R-st). R-st (termed paramutagenic) induced a heritable decrease in the expression of standard R-r (termed paramutable); the altered R-r is designated R-r′ (reviewed in Kermicle, 1996). The r-g allele is null and does not participate in paramutation. The paramutable b1 and pl1 alleles are B-I and Pl-Rh, respectively, and when heterozygous with a paramutagenic B′ or Pl′ allele, the paramutable allele is heritably changed into a paramutagenic allele (Coe, 1966; Hollick et al., 1995). The paramutable B-I and Pl-Rh alleles are also unstable, spontaneously changing to B′ and Pl′ (Coe, 1966; Hollick et al., 1995). Most alleles of b1, r1, and pl1 do not participate in paramutation and are termed neutral. Paramutation-like phenomena are not restricted to pigment-regulatory genes; they have also been described in other plants (reviewed in Brink, 1973) and with transgenes in petunia and tobacco (Meyer et al., 1993; Matzke et al., 1994; reviewed in Hollick et al., 1997).
A variety of mechanisms have been proposed to explain paramutation (McClintock, 1963; Coe, 1966; Brink et al., 1968; Patterson et al., 1993; Chandler et al., 1996; Colot et al., 1996; Martienssen, 1996; Matzke et al., 1996; Hollick et al., 1997; Chandler et al., 2000). Given the numerous differences in paramutation phenomenology among b1, r1, and pl1 (Chandler et al., 2000), whether they share a common mechanism is not clear. An important approach to eventually understanding the underlying mechanism or mechanisms of paramutation is to isolate mutations in the genes that affect paramutation. Here, we describe a genetic screen and report the identification and characterization of a trans-acting gene that, when mutated, affects aspects of paramutation at b1, r1, and pl1.
RESULTS
Identification and Genetic Characterization of mop1
Paramutation always occurs when B′ and B-I are heterozygous, and B′ is extremely stable in subsequent generations (reviewed in Chandler et al., 2000). These characteristics enable a simple genetic screen to isolate mutations affecting paramutation. Furthermore, the phenotypes of B-I and B′ are readily distinguishable even in young seedlings; B-I seedlings have intense anthocyanin pigmentation, whereas B′ seedlings have little or no anthocyanin pigmentation. A screening population was generated by crossing B′ plants with B-I plants carrying active Mutator (Mu) transposable elements. F1 plants were screened for darkly pigmented (B-I–like) individuals, which could represent dominant mutations that prevented the establishment of paramutation. All F1 individuals (>3500) were lightly pigmented (B′), indicating that paramutation had occurred without exception. To identify recessive mutations that might represent failure to maintain the repressed B′ transcription state, F1 individuals were self-pollinated and F2 families were screened for darkly pigmented (B-I–like) seedlings segregating at 1/4 frequency. For brevity, in this article we refer to the darkly anthocyanin-pigmented B-I–like individuals as dark and the lightly anthocyanin-pigmented B′-like individuals as light. Several F2 families yielded dark seedlings (25 of 510 families screened). Segregation in subsequent crosses confirmed the presence of a recessive mutation at a single locus in each of these families (data not shown). The high frequency of families segregating dark seedlings was not observed in a similar screen resulting from the cross of a B′ Mu stock with B-I; none of the F2 families yielded dark seedlings (zero of 427 families screened). The high frequency in one set of stocks but not in the other suggested that a recessive mutation had been present in some of the B-I Mu stocks used in these experiments. Consistent with this hypothesis, genetic complementation tests among plants from the 25 families yielded dark seedlings, confirming that each of these families carried a mutation in the same gene (data not shown). Pedigree analyses demonstrated that this mutation was segregating in the B-I Mu stocks. On the basis of subsequent analyses described below, we designated the locus identified by this mutation as mediator of paramutation1 (mop1) and the mutant allele as mop1-1. Recessive mop1-1 likely represents a reduced or loss-of-function allele; individuals heterozygous for mop1-1 have the same amounts of pigment as the B′ seedlings, which is defined as wild type for this genetic background. Because these individuals possess the Mop1 allele normally present in most maize stocks, we refer to the Mop1 allele as wild type.
mop1-1 Alters the Phenotype of Both B′ and Pl′
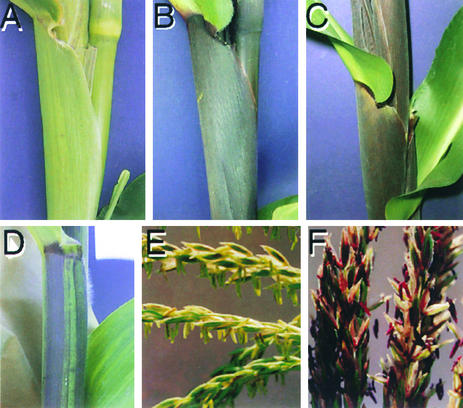
The fact that mop1-1 was isolated as dark B-I–resembling seedlings among light B′ siblings indicated that mop1-1 modified the phenotype of B′ seedlings to resemble a B-I phenotype. A similar modification of the B′ phenotype is observed in mature plants. Figures 1A and 1B show the adult phenotypes of B′ Mop1/mop1-1 and B′ mop1-1/mop1-1 plants. Figures 1B and 1C illustrate the very similar phenotype of B′ mop1-1/mop1-1 and B-I Mop1/Mop1 plants. These results are consistent with the mop1 gene having a role in the maintenance of the paramutant B′ state. Although B′ mop1-1 plants resemble B-I plants, they occasionally show somatic instability, manifest as sectors of B′-like pigmentation similar to that seen in Figure 1D. This observation suggests that the mop1-1 mutation is unstable and may be caused by a transposable element.
Figure 1.
Phenotypes Associated with the mop1-1 Mutation.
(A) B′ Mop1/mop1-1.
(B) B′ mop1-1/mop1-1.
(C) B-I Mop1/Mop1.
(D) B′ mop1-1/mop1-1 plant with B′-like sectors.
(E) Pl′ Mop1/- (either Mop1/Mop1 or Mop1/mop1-1).
(F) Pl′ mop1-1/mop1-1.
Does mop1-1 also alter the phenotype of the paramutant Pl′ allele? To address this question, the pl1 expression phenotype must be assayed independent of changes in b1 expression; yet to activate the anthocyanin pathway, pl1 must be coexpressed with a bHLH factor (b1 or r1). The anthers represent a reliable tissue in which to assay pl1 expression because r1, but no b1 allele, is expressed in the anthers. Fortunately, the r1 gene expressed in the anthers does not undergo spontaneous paramutation; accordingly, changes in anther pigment reflect changes in pl1 expression. Because our homozygous B′ mop1-1 stocks lacked r1 expression in the anthers (homozygous for the null r-g allele), we introduced R-r, which is expressed in both seeds and anthers. In the presence of R-r, Pl′ gives variegated (light) anthers, whereas Pl-Rh gives fully pigmented (dark) anthers. Light-anthered B′/b-W23 Pl′/Pl′ Mop1/mop1-1 R-r/r-g individuals were self-pollinated, and purple kernels (inheriting dominant R-r) were planted. Recessive b-W23 produces green vegetative tissues when homozygous, but b1 segregation does not affect anther pigmentation. If mop1-1 intensifies Pl′, then one in four dark-anthered progeny are expected to segregate. Light-anthered individuals, as seen in Figure 1E, and dark-anthered individuals, as seen in Figure 1F, were observed among the F2 progeny at the expected frequency (nine of 47 dark;  ,
,  ). Three-quarters of these plants were B′/- (either B′/B′ or B′/b-W23), and intensification of the B′ phenotype in the dark-anthered individuals confirmed that they were mop1-1/mop1-1. This result demonstrated that mop1-1 is not specific to B′ but also affects the paramutant Pl′ allele.
). Three-quarters of these plants were B′/- (either B′/B′ or B′/b-W23), and intensification of the B′ phenotype in the dark-anthered individuals confirmed that they were mop1-1/mop1-1. This result demonstrated that mop1-1 is not specific to B′ but also affects the paramutant Pl′ allele.
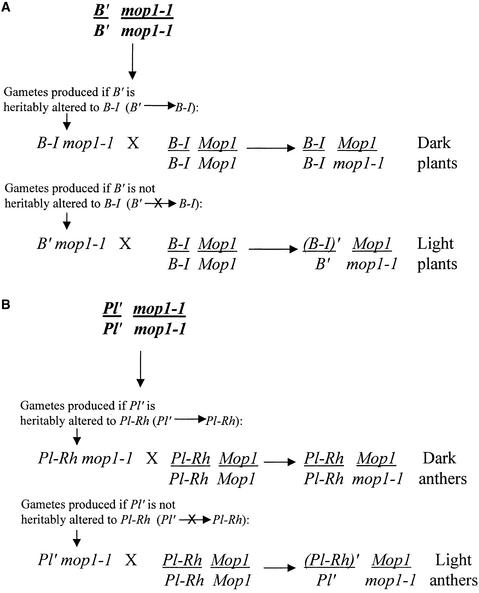
mop1-1 Does Not Heritably Alter B′ but Can Heritably Alter Pl′
Under some conditions, paramutant alleles can return to the higher expression state characteristic of the paramutable allele. This has been observed for paramutant alleles of pl1 and r1, although not for those of b1 (reviewed in Chandler et al., 2000). Genetic crosses were performed to test whether mop1-1 heritably alters B′ to B-I or Pl′ to Pl-Rh, as diagrammed in Figure 2. When B′ mop1-1 individuals are crossed with plants null for b1 and wild type for mop1 (Mop1), all progeny are light (B′/b Mop1/mop1-1; >400 plants from >20 families), thus demonstrating that the light pigment phenotype of B′ is not heritably altered by mop1-1. Furthermore, crosses between B′ mop1-1 and B-I Mop1 individuals, both of which are dark, also generate all light progeny (B′/[B-I]′, Mop1/mop1-1; >100 plants from six families), indicating that the B′ allele transmitted from homozygous mop1-1 plants is fully capable of causing paramutation (Figure 2A) in a Mop1/mop1-1 nucleus. Thus, mop1-1 does not disrupt the inheritance of the B′ state.
Figure 2.
Diagram Outlining Test for Heritability of B′ and Pl′ from Homozygous mop1-1 Individuals.
(A) If mop1-1 heritably alters B′ to B-I, then B′ mop1-1/mop1-1 individuals would generate B-I mop1-1 gametes and progeny would be dark. Alternately, if B′ is still B′, then all gametes would be B′ mop1-1, B′ would paramutate B-I (indicated by [B-I]′) in the next generation, and all progeny would be light.
(B) If mop1-1 heritably alters Pl′ to Pl-Rh, then Pl′ mop1-1/mop1-1 individuals would generate Pl-Rh mop1-1 gametes and progeny would have dark anthers. Alternately, if Pl′ is still Pl′, then all gametes would be Pl′ mop1-1, Pl′ would paramutate Pl-Rh (indicated by [Pl-Rh]′) in the next generation, and all progeny would have light anthers.
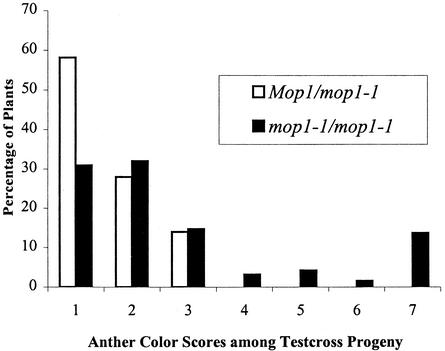
To test whether the Pl′ allele in homozygous Pl′ mop1-1 plants can be heritably altered to Pl-Rh, dark-anthered Pl′ mop1-1/mop1-1 individuals were crossed with Pl-Rh Mop1/Mop1 testers (Figure 2B). Progeny of this cross were scored with respect to anther pigmentation by using the anther color scale previously described (Hollick et al., 1995). In this scale, individuals with an Anther Color Score (ACS) of 7 are Pl-Rh. Most progeny (∼75%) had lightly pigmented anthers, indicating that Pl′ could be transmitted from homozygous mop1-1 plants. However, dark-anthered individuals (Pl-Rh–like, with ACS 7) were sometimes observed, as shown in Figure 3. In contrast, in control crosses, sibling Pl′ Mop1/mop1-1 plants were crossed with Pl-Rh testers, and all progeny had lightly pigmented anthers (Figure 3). Not all Pl′ mop1-1/mop1-1 individuals produced dark-anthered progeny. In a total of 12 progeny families, representing test crosses from 11 different individuals, only six families had some dark-anthered progeny. All six of these families were from homozygous Pl′/Pl′ mop1-1/mop1-1 individuals in which one of the Pl′ alleles had been exposed to homozygous mop1-1 for two consecutive generations. In contrast, no ACS 7 progeny were observed from any of the four Pl′ mop1-1/mop1-1 individuals in which the Pl′ alleles were exposed to homozygous mop1-1 for only one generation. Thus, mop1-1 can, but does not always, heritably alter Pl′ to Pl-Rh.
Figure 3.
ACSs of Pl′ mop1-1/mop1-1 Outcrosses versus Pl′ Mop1/mop1-1 Outcrosses.
Pl′ mop1-1/mop1-1 individuals versus Pl′ Mop1/mop1-1 individuals were outcrossed with Pl-Rh (Mop1/Mop1) testers. Individual progeny plants were scored for amount of anther pigment.
mop1-1 Increases Transcription of B′ and Amounts of Pl′ Transcripts
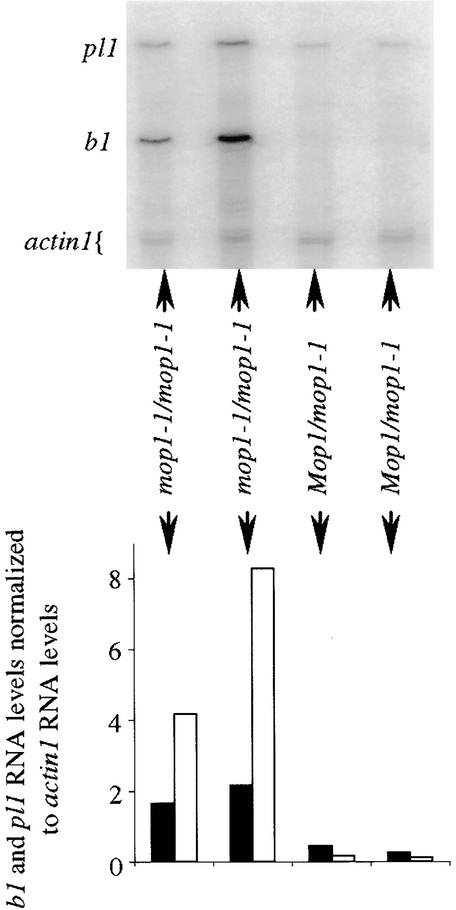
To test whether changes in pigment are the result of changes in transcript amounts and transcription, as observed with b1 and pl1 paramutation (Patterson et al., 1993; Hollick et al., 2000), RNA concentrations and transcription rate were determined. As depicted in Figure 4, analysis of RNA concentrations in husk tissue by using RNase protection assays showed a strong (46-fold) increase in b1 RNA in B′ mop1-1/mop1-1 relative to that in B′ Mop1/mop1-1 siblings. RNase protections also reveal a marked (5.6-fold) increase in pl1 RNA concentrations in Pl′ mop1-1/mop1-1 relative to those in Pl′ Mop1/mop1-1 siblings (Figure 4). These increases in b1 transcript values are consistent with (although greater than) the 10- to 20-fold differences in transcript values between B-I and B′ (Patterson et al., 1993). Increases in pl1 transcripts are also similar to (although less than) those differences seen between Pl′ and Pl-Rh (Hollick et al., 2000).
Figure 4.
Amounts of Transcripts in mop1-1 versus Wild-Type Siblings.
An example of RNase protections for pl1, b1, and actin1 on four sibling individuals. All individuals are homozygous B′ and Pl′ and segregate for mop1-1 as indicated. The bar graph shows the normalized amounts of b1 (open bars) and pl1 (closed bars) RNA from the RNase protection.
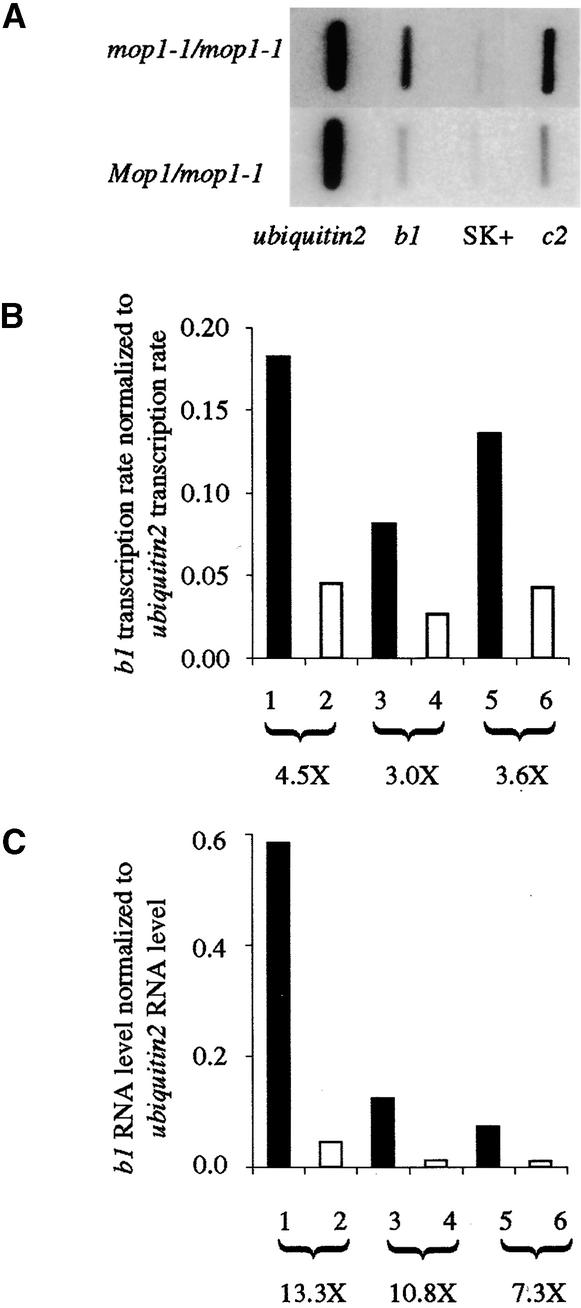
To determine whether the increases in transcripts of homozygous B′ mop1-1 plants are associated with increased transcription, in vitro transcription assays were performed on nuclei isolated from sheath tissue. As shown in Figure 5A, the transcription rate of b1 was increased in sheaths of B′ mop1-1/mop1-1 plants relative to that in Mop1/mop1-1 siblings. The results of three independent pairwise comparisons between sibling mop1-1/mop1-1 and Mop1/mop1-1 individuals are shown graphically in Figure 5B. RNase protection assays with the same samples were performed to determine the amounts of transcripts in the same sheath tissues. The results of pairwise comparisons of RNA concentrations for the same six individuals are summarized in Figure 5C. The in vitro transcription assays reveal a threefold to 4.5-fold increase in transcription rate, whereas RNase protection assays reveal a sevenfold to 13-fold increase in transcripts in the same tissues. Similar differences in RNA versus transcription rate increases (five- to eightfold versus 13- to 14-fold) are observed at c2, a gene regulated by b1 and pl1 (Figure 5A; data not shown). Overall, the marked pigmentation differences between B′ Mop1/mop1-1 and B′ mop1-1/mop1-1 siblings can be explained by increased b1 transcripts, which can be at least partially explained by an increase in b1 transcription rate.
Figure 5.
Transcription Rates in mop1-1 versus Wild-Type Siblings.
(A) An example of an in vitro transcription assay showing the SK+ plasmid negative control and the signal for b1, c2, and ubiquitin2 transcription in B′ mop1-1/mop1-1 versus B′ Mop1/mop1-1 individuals.
(B) Paired data for b1 transcription rate from several in vitro transcription assays for several mop1-1/mop1-1 (closed bars) versus Mop1/mop1-1 (open bars) individuals. The data represent three separate comparisons between a pair of sibling individuals. The n-fold (designated X) increase is given below each pairwise comparison.
(C) Paired data from RNase protection assays for b1 RNA quantities normalized to quantities of ubiquitin2 RNA for the same mop1-1/mop1-1 (closed bars) versus Mop1/mop1-1 (open bars) sibling individuals as examined in (B).
mop1 Does Not Affect Neutral Alleles
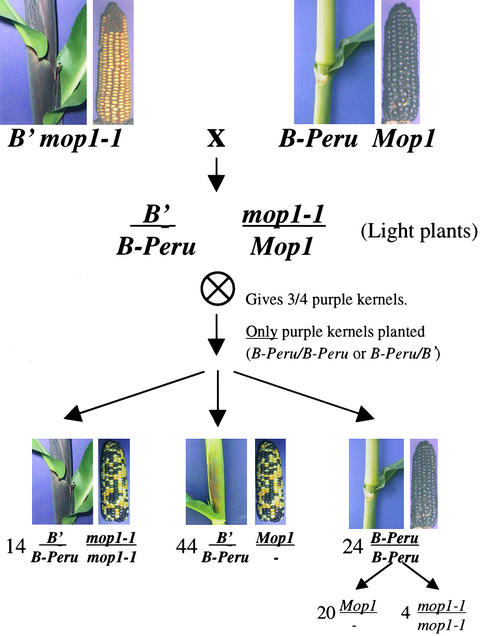
To determine whether mop1-1 is a general modifier of the anthocyanin regulators or is specific to those alleles that participate in paramutation, we tested the ability of mop1-1 to enhance the amount of pigment produced by b1 and pl1 alleles that do not participate in paramutation. The B-Peru allele, which does not participate in paramutation (Patterson et al., 1995), has dark aleurone pigment and extremely weak pigment in the vegetative body of the plant, which make it readily distinguishable from B′. As diagrammed in Figure 6, B′ Pl r-g mop1-1 plants were crossed with a B-Peru Pl r-g Mop1 stock, and four F1 individuals were self-pollinated. Only purple F2 kernels were planted to select for homozygous or heterozygous B-Peru. Among these progeny, three classes of plant color phenotypes were observed (Figure 6). Because intensification of B′ pigmentation would preclude any conclusions about mop1-1 intensification of the lighter B-Peru pigmentation in the vegetative body of the plant, it was essential to compare only homozygous B-Peru individuals, identified by the presence of 100% purple kernels on the ears produced by these individuals. All 24 individuals characterized as having typical B-Peru pigment in the vegetative body of the plant had ears with 100% purple kernels, confirming their b1 genotype as B-Peru/B-Peru. These plants were test-crossed with B′/B′ mop1-1/mop1-1 plants to determine their mop1 genotype. If any of these individuals were also homozygous for mop1-1, they would be expected to produce 100% dark progeny (vegetative plant phenotype) from the test crosses with B′ mop1-1. Four of the homozygous B-Peru individuals met this expectation and thus had been homozygous for mop1-1. The fact that their pigment phenotypes were indistinguishable from their B-Peru/B-Peru Mop1/mop1-1 or B-Peru/B-Peru Mop1/Mop1 siblings demonstrates that mop1-1 does not intensify B-Peru pigmentation. All plants that had the B′ mop1-1 or B′ Mop1/- phenotype (Figure 6) were confirmed to be heterozygous for B′.
Figure 6.
Diagram Showing Three Progeny Classes from B′/B-Peru Mop1/mop1-1 Self-Pollinations.
B′ mop1-1 plants were crossed with B-Peru Mop1 plants. The light F1 B′/B-Peru Mop1/mop1-1 plants were self-pollinated, and purple kernels (B-Peru/-) were planted. The progeny fell into three phenotypic classes; the numbers and genotypes of each phenotype are shown. The mop1 genotype of the 24 B-Peru/B-Peru plants was determined by test crosses with B′ mop1-1.
In the process of introducing mop1-1 into other genetic backgrounds, a stock was generated that contained B-bar and pl-W22, weakly pigmented neutral alleles of b1 and pl1, respectively. Three dark plants, subsequently shown to be B-bar/B′ mop1-1/mop1-1 pl-W22/pl-W22, were self-pollinated, producing 3:1 dark/light progeny (76 dark and 25 light). Polymerase chain reaction analyses confirmed that the light progeny were homozygous for B-bar. The fact that these light progeny resembled B-bar Mop1/- pl-W22 plants demonstrated that mop1-1 did not intensify B-bar. Because these plants were also homozygous for pl-W22, an allele of pl1 that does not participate in paramutation (Hollick et al., 1995), the effects of mop1-1 on a neutral pl1 allele could also be examined. All plants had weak pl-W22 anther color. Together with the results described above with B-Peru, these results suggest that the mop1-1 intensification of pigment is selective for paramutant alleles.
mop1-1 Inhibits Establishment of b1 Paramutation
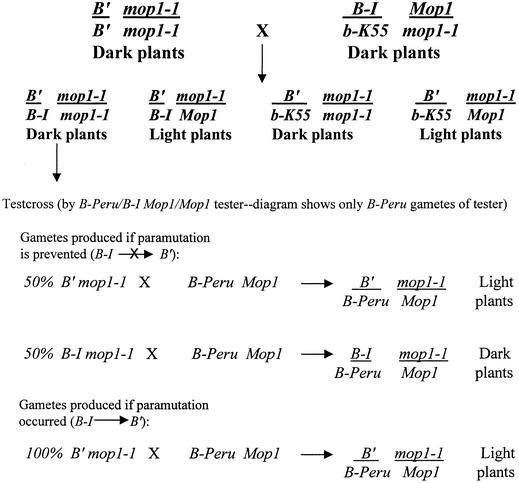
To determine whether mop1 is involved in the establishment of b1 paramutation, we asked whether B-I could be paramutated when encountering B′ in a homozygous mop1-1 nucleus. To address this question, homozygous B′ mop1-1 individuals were crossed, as diagrammed in Figure 7, with plants heterozygous for B-I/b-K55 and Mop1/mop1-1. The b-K55 allele is recessive, producing green vegetative tissues when homozygous. B′/B-I and B′/b-K55 were differentiated by restriction fragment length polymorphisms between the B-I and b-K55 alleles, and each class was crossed with both B-I/B-Peru testers (Figure 7) and with b1 testers (not diagrammed). A feature of the B-I/B-Peru cross is that the B-I allele is frequently more stable (less prone to spontaneous paramutation to B′) when heterozygous with a neutral allele (Chandler et al., 2000). Furthermore, the purple aleurone pigmentation of B-Peru kernels provides a useful marker for the allele, and the weak B-Peru pigmentation in the vegetative body of the plant is recessive to and readily distinguished from B-I or B′ pigmentation (Radicella et al., 1992; Patterson et al., 1995).
Figure 7.
Diagram Outlining Test for Ability of mop1-1 to Prevent b1 Paramutation.
B′/B′ mop1-1/mop1-1 plants were crossed with B-I/b Mop1/mop1-1 plants, generating four types of segregating progeny. The B′/B-I and B′/b progeny were distinguished by restriction fragment length polymorphisms. The B′/b progeny were not analyzed further. Both classes of B′/B-I progeny (dark and light) were crossed with B-I/B-Peru Mop1/Mop1 testers (and with testers null for b1; not diagrammed) to test whether the B-I allele heterozygous with B′ had become B′ in the mop1-1/mop1-1 versus Mop1/mop1-1 plants. The expectations for crosses with B′/B-I mop1-1/mop1-1 with one type of tester (B-Peru Mop1, which gives purple kernels) are shown. The expectation for the B′/B-I Mop1/mop1-1 progeny is that all offspring will be light plants (not diagrammed).
Among offspring of B′/B-I mop1-1/mop1-1 plants crossed to B-I/B-Peru (Mop1/Mop1), purple kernels (inheriting the B-Peru allele of the tester) gave rise to plants segregating as 50% light (original B′) and 50% dark (unaltered B-I) individuals, as shown in Table 1. A similar result was obtained among offspring of B′/B-I mop1-1/mop1-1 plants crossed with b1 tester stocks (b-K55/b-K55 Mop1/Mop1 or b-K55/b-W23 Mop1/Mop1). Eighteen of 43 plants were dark or medium-dark, and the remaining 25 individuals were light (Table 1). Medium-dark individuals could be explained by spontaneous paramutation of B-I to B′, or Pl-Rh to Pl′, which is often observed with these alleles (Coe, 1966; Hollick et al., 1995). Some of these individuals were crossed with appropriate tester stocks, and the phenotypes of progeny were consistent with spontaneous paramutation of Pl-Rh causing the reduction in pigment. Control crosses of B′/B-I Mop1/mop1-1 plants with the same B-I/B-Peru and b1 tester stocks produced progeny that were all light colored. These results demonstrate that paramutation is not established in mop1-1/mop1-1 plants.
Table 1.
Results of B′/B-I mop1-1/mop1-1 Test Crosses with Several Tester Stocks
| Cross | No. of Dark Progeny |
No. of Light Progeny |
χ2 (P)a |
|---|---|---|---|
|
B′/B-I mop1-1/mop1-1 × b/b Mop1/Mop1 |
18b | 25 | 1.139 (0.29) |
|
B′/B-I mop1-1/mop1-1 × B-Peru Mop1c |
26 | 25 | 0.0196 (0.89) |
|
B′/B-I mop1-1/mop1-1 × B-I Mop1c |
3 | 46d | 37.7 (<0.0001) |
The hypothesis tested is 1:1 segregation.
Plants were scored as dark or medium-dark. Test crosses demonstrated that medium-darks were B-I Pl′.
B-Peru Mop1 and B-I Mop1 represent the different gametes produced by the B-I/B-Peru tester.
Plants were scored as medium (20) or light (26). Many mediums were darker at the base, consistent with spontaneous paramutation.
Surprisingly, colorless kernels from the same crosses with the B-I/B-Peru tester (inheriting the B-I allele of the tester) gave fewer than the expected 50% B-I plants (Table 1). Many individuals were dark toward the base of the plant but appeared to lighten progressively during development, so that at anthesis, 46 plants were scored as medium or light and only three as dark (B-I). Observation of B-I progeny at the anticipated 50% frequency requires three conditions: inheritance of an unaltered B-I allele from the B-I/B-Peru tester parent, inheritance of an unaltered B-I allele from B′/B-I mop1-1/mop1-1 individuals, and stability of both B-I alleles in the homozygous B-I progeny individuals. Homozygous B-I is generally less stable than is B-I/B-Peru (K.M. Kubo, G.I. Patterson, J.E. Dorweiler, and V.L. Chandler, unpublished data; discussed in Chandler et al., 2000). The developmentally progressive reduction in pigment of many (presumed B-I/B-I) progeny plants appeared similar to the progression sometimes observed during spontaneous paramutation of B-I to B′. This result could also be caused by a greater than usual rate of spontaneous paramutation of the B-I allele segregating from the B-I/B-Peru tester. Alternately, the B-I allele segregating from B′/B-I mop1-1/mop1-1 may be subtly destabilized. This subtle destabilization could be exacerbated in the next generation by homozygosity with a second B-I allele, whereas it could be overcome in the next generation by heterozygosity with a neutral allele such as B-Peru or b1. Nevertheless, mop1-1 is clearly able to inhibit the establishment of paramutation in B′/B-I mop1-1/mop1-1 plants, as shown by the dark offspring carrying B-I, which segregated from these individuals. This is in sharp contrast to wild-type stocks for mop1 in which paramutation always occurs (Coe, 1966; Patterson et al., 1993, 1995).
mop1-1 May Inhibit Establishment of pl1 Paramutation
We also examined whether mop1-1 inhibits the establishment of pl1 paramutation, although this experiment is complicated by the fact that Pl′ can occasionally be heritably altered to Pl-Rh in homozygous mop1-1 plants. To ensure that the allele entering the cross was the paramutagenic Pl′, light-anthered Pl′/Pl′ Mop1/mop1-1 plants (in which Pl′ is stable and does not change to Pl-Rh) were crossed with dark-anthered Pl-Rh/Pl-Rh Mop1/mop1-1 plants. Each of these stocks also carried the B′ allele, which enabled an independent assay of the mop1 genotype. Among the progeny, dark anther and vegetative tissue pigmentation perfectly cosegregated (mop1-1/mop1-1). Three dark-anthered individuals (Pl′/Pl-Rh) were crossed with a b-K55 Pl-Rh tester to determine whether paramutation occurred (all progeny should have light anthers) or whether the Pl-Rh allele could be inherited unaltered (50% dark- and 50% light-anthered plants). The resulting progeny segregated 39 purple-anthered Pl-Rh plants and 36 light-anthered Pl′ plants. The simplest explanation for this result is that mop1-1/mop1-1 prevented the establishment of paramutation and thus resulted in segregation of the original Pl-Rh and Pl′ alleles.
mop1-1 Inhibits Establishment of r1 Paramutation
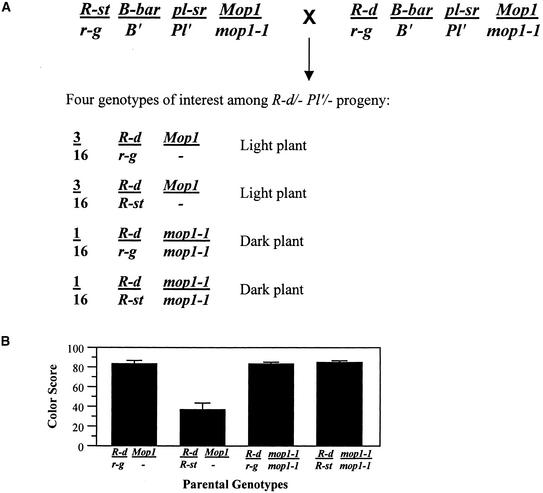
Because the phenomenology of r1 paramutation is very different from that of b1 (reviewed in Chandler et al., 2000), we asked whether mop1-1 is able to affect paramutation at r1. Our B′ mop1-1 stocks contained the r-g allele of r1, which does not participate in paramutation (Kermicle et al., 1995). These mop1-1 r-g stocks were crossed with stocks that were wild type for mop1 (Mop1) and contained r1 haplotypes that participate in paramutation. The term haplotype is used for complexes composed of multiple r1 genes. The r1 haplotypes used included the paramutagenic R-st haplotype and two paramutable haplotypes, standard R-r and R-d:Catspaw (R-d). The F1 progeny between R-st and mop1-1 (R-st/r-g; Mop1/mop1-1) were intercrossed with F1 progeny of the paramutable haplotypes (R-d/r-g or R-r/r-g; Mop1/mop1-1). Thus, the paramutagenic and paramutable haplotypes were combined in homozygous mop1-1 mutant progeny and in siblings that were wild type for mop1 (Mop1/-). Fifty percent of intercross progeny inherited the paramutable haplotype (R-d or R-r), and among these, four genotypes of interest segregated as shown for R-d in Figure 8A. The presence of B′ allowed the mop1 genotype to be easily monitored. Test crosses onto r-g/r-g females of Mop1/- (either Mop1/Mop1 or Mop1/mop1-1) individuals in which R-d was heterozygous with neutral r-g provided a baseline for wild-type kernel pigmentation. Test crosses of R-d/r-g plants homozygous for mop1-1 did not differ detectably from those of wild-type plants (Mop1/-), as shown in Figure 8B. Paramutation occurred as expected in the R-st/R-d Mop1/- individuals, manifested as less pigment in the R-d kernel progeny (Figure 8B). However, siblings homozygous for mop1-1 showed no decrease in R-d expression (Figure 8B). Thus, in individuals homozygous for the mop1-1 mutation, paramutable R-d exits the cross as if it had been heterozygous with a neutral allele rather than with paramutagenic R-st. Similar results were obtained with the paramutable standard R-r (data not shown). This demonstrates that mop1-1 is able to inhibit the establishment of r1 paramutation.
Figure 8.
Summary of mop1-1 Effects on r1 Paramutation.
(A) Parental genotypes and progeny classes used to evaluate the effect of mop1-1 on the establishment of r1 paramutation. mop1 genotypes were determined on the basis of the intensity of the vegetative tissue pigment within the Pl/- phenotypic class. Individuals belonging to the two R-d/- genotypic classes were identified by crossing with an r-g tester stock, and the R-d/r-g kernel progeny were then assayed for pigment intensity.
(B) The bar graph shows results of paramutation test with r1. Color scores for kernels inheriting the R-d allele are plotted from individuals having the parental genotype indicated along the x axis. The first two columns are Mop1/- genotype controls. The color score equals 100 minus the average reflectometer reading, as described by Alleman and Kermicle (1993). Error bars indicate sd.
Mutations in mop1 Correlate with Negative Pleiotropic Effects
As shown in Table 2 and Figures 9B, 9C, and 9D, plants homozygous for B′ mop1-1 can show deleterious pleiotropic phenotypes when compared with the wild type (Figure 9A). The range of effects seen include delayed flowering or shorter stature relative to wild-type siblings, spindly and sometimes barren stalks, and in some instances, aberrant development resulting in feminized tassels (such as that observed in tassel seed mutants [Irish et al., 1994]). We reproducibly see differences in flowering time, whereas other abnormalities appear stochastically. Perhaps, there is an environmental effect on the frequency with which these developmental abnormalities occur, because the frequency has been greatest in our Hawaii nursery during the past two winters. Two alternative hypotheses could explain the correlation of these deleterious effects with mop1-1. These effects could be caused either by other mutations that are linked to mop1-1 or by the mop1-1 mutation itself. Recently, a second allele of mop1 was isolated in a similar screening that was looking for modification of Pl′ instead of B′ and used ethyl methanesulfonate (EMS) as the mutagen (Hollick and Chandler, 2001). This independent allele (mop1-2EMS) provided independent data with which to judge the two alternative hypotheses. B′/- mop1-2EMS/mop1-2EMS and B′/- mop1-1/mop1-2EMS individuals showed the same suite of deleterious pleiotropic phenotypes as homozygous mop1-1 (Table 2). This strongly suggests that these negative pleiotropic effects are the result of the mutant mop1 locus rather than the result of linked mutations.
Table 2.
Developmental Phenotypes Associated with mop1 Mutants
| Phenotypesa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Segregation
|
Mop1
|
mop1
|
||||||||
| Family | Expected | Observed | χ2 (P) | Nb | O | N | STS | WTS | BT | Rc |
| mop1-1 segregating | 1:1 | 37:39 | 0.05 (0.82) | 36 | 1d | 13 | 15 | 3 | 3 | 5 |
| mop1-2EMS segregating | 3:1 | 75:15 | 3.33 (0.07) | 74 | 1e | 6 | 2 | 1f | 4 | 2 |
| mop1-1/mop1-2EMS segregating | 1:1 | 13:15 | 0.14 (0.71) | 13 | — | 6 | 3 | 1 | 2 | 3 |
Phenotypes are as follows: Mop1, lightly pigmented plant; mop1, darkly pigmented plant; N, normal; O, other; STS, strong tasselseed; WTS, weak tasselseed; BT, barrenized tassel; R, runty or scrawny plant.
The number of individuals represent progeny from four, five, and two plants for the mop1-1, mop1-2EMS, and mop1-1/mop1-2EMS segregating crosses, respectively.
One of five, one of two, and one of three plants also possessed a barrenized tassel, and one of five plants possessed a feminized tassel, which failed to emerge (see Figure 9C).
This plant was slightly deformed with a twisted stalk.
This plant was diseased.
This plant was shorter than were sibling plants and did not produce an ear.
Figure 9.
Phenotypes Characteristic of mop1 Mutations Relative to Wild-Type Siblings.
(A) A B′ Mop1/mop1-1 individual bearing a normal tassel.
(B) A B′ mop1-1/mop1-1 individual bearing a feminized tassel (strong tasselseed).
(C) A B′/- mop1-2EMS/mop1-2EMS individual bearing a severely barrenized tassel.
(D) A runty B′ mop1-1/mop1-1 individual in which the feminized terminal inflorescence failed to emerge.
mop1-1 Does Not Affect Global Methylation Levels
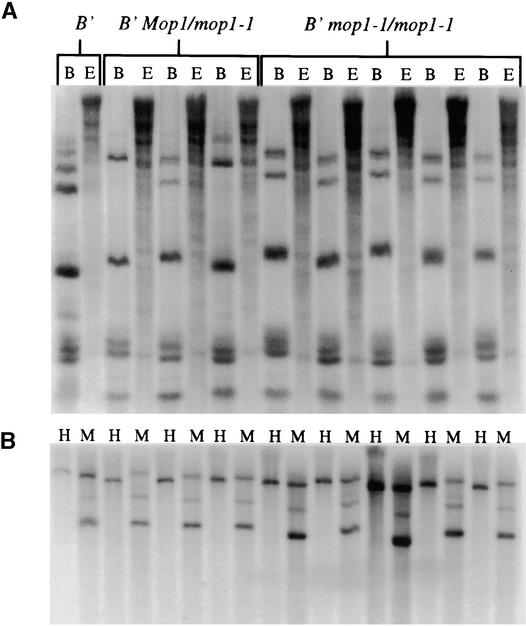
Although no differences in DNA methylation are observed in comparisons of the paramutagenic and paramutable alleles of b1 and pl1 (within the coding and ∼15 kb of the respective flanking regions) (Patterson et al., 1993; Hollick et al., 2000), paramutation of R-r correlates with DNA methylation in the transcribed region (Walker, 1998). In other systems, changes in DNA methylation often have correlated with differences in transcription; thus, in one model, mop1 encodes a protein that influences global DNA methylation levels such as a DNA methyltransferase (Finnegan et al., 1996) or DDM1 (for decrease in DNA methylation) (Vongs et al., 1993). Under this model, one could hypothesize that methylation differences occur at b1 and pl1, but the changes occur outside of the regions examined. We tested whether global DNA methylation levels were affected by mop1-1. DNA gel blots with methylation-sensitive restriction enzymes were prepared to compare homozygous B′ mop1-1 individuals, wild-type siblings (B′ Mop1/mop1-1), and B′ stocks (Mop1/Mop1). These blots were hybridized with repeated sequences of the 45S ribosomal region (McMullen et al., 1986) as well as a repeated sequence found at centromeres (Jiang et al., 1996). As shown in Figures 10A and 10B, no differences in DNA methylation were detected among the mop1 genotypes.
Figure 10.
DNA Gel Blots Assaying Methylation of Repeated Sequences.
Individual genotypes are indicated above the DNA gel blots (B' stands for B′/B′ Mop1/Mop1).
(A) Samples digested with the methylation-insensitive enzyme BstNI (B) and the methylation-sensitive enzyme EcoRII (E) were probed with the 45S ribosomal repeat.
(B) Samples digested with HpaII (H) or MspI (M) were probed with the centromere repeat.
DISCUSSION
Our results demonstrate that mop1 plays a central role in paramutation. The mop1-1 mutation disrupts the maintenance and establishment of paramutation at multiple maize loci but has no effect on alleles that do not participate in paramutation. The pleiotropic developmental phenotypes that stochastically occur in some families segregating mop1 mutations suggest that mop1 may play a broader role than simply regulating alleles that undergo paramutation. Experiments are in progress to determine whether developmental phenotypes are heritable independent of the mop1 mutation; if so, this would suggest that those phenotypes represent epimutations at other loci induced by the mop1 mutation. This has been observed with the ddm1 mutation, which encodes a putative chromatin remodeling protein. Plants that are homozygous for ddm1 for consecutive generations accumulate epimutations at other loci (Kakutani et al., 1996).
Paramutation at b1, pl1, and r1 Shares a Common Mechanism
The behavior of b1, pl1, and r1 alleles participating in paramutation has unique aspects (reviewed in Chandler et al., 2000). r1 paramutation involves structurally distinct haplotypes of a complex or repeated nature. In contrast, B′ and Pl′ are spontaneous derivatives of B-I and Pl-Rh, respectively. Furthermore, increased methylation is readily apparent in R-r′ relative to R-r (Walker, 1998), whereas such differences have not been observed for b1 and pl1 (Patterson et al., 1993; Hollick et al., 2000). We have demonstrated that mop1-1 disrupts the maintenance of B′ and Pl′ expression, the heritability of Pl′, and the establishment of b1 and r1 paramutation. These results provide compelling evidence that despite several phenomenological differences, all three loci use a common molecular factor for paramutation, strongly suggesting that a common mechanism underlies paramutation at these three loci.
A Role for MOP1 in Establishing and Maintaining the Paramutant State
Our results indicate that MOP1 function is clearly involved in all three phases of gene silencing: establishment, maintenance, and heritability (reviewed in Loo and Rine, 1995). When paramutable and paramutagenic alleles of b1, pl1, or r1 are heterozygous, paramutation always occurs (reviewed in Kermicle, 1996; Chandler et al., 2000), but in nuclei that were homozygous for mop1-1, paramutation was not established at b1, r1, or pl1. When Pl-Rh was introduced to Pl′ in homozygous mop1-1 plants, Pl-Rh appeared to be transmitted unaltered, because the offspring plants segregated as 50% dark-anthered (Pl-Rh) and 50% light-anthered (Pl′). This interpretation is complicated by the fact that homozygous Pl′ mop1-1 plants can sometimes produce dark-anthered Pl-Rh progeny. However, for the 50% dark-anthered plants to have resulted from full establishment of paramutation and subsequent reversion of half the Pl′ alleles to Pl-Rh seems quite unlikely. The establishment experiment used Pl′ alleles exposed to homozygous mop1-1 for a single generation, a condition in which in other experiments no Pl-Rh progeny were observed. Thus, the simplest explanation for the 50% segregation observed in this experiment is that mop1-1 inhibits the establishment of pl1 paramutation.
One possibility is that MOP1 function mediates or facilitates the interaction between the paramutagenic and paramutable alleles, such that loss of function disrupts that interaction. MOP1 function may be necessary for the paramutagenic properties of B′, Pl′, or R-st, such that loss of function makes these alleles more similar to neutral or paramutable alleles. For B′ and Pl′, the paramutagenic property may be the repressed state, and mop1-1 prevents the establishment of paramutation by derepressing these alleles. The appearance of phenotypically B′ sectors in B′ mop1-1 plants suggests that MOP1 can function throughout development to reduce the expression of B′ such that regaining MOP1 function in a somatic sector reestablishes the reduced expression state.
MOP1 function also plays a role in maintenance of the b1 and pl1 paramutagenic states and affects the heritability of Pl′. Experiments are in progress to determine whether MOP1 functions similarly at r1. Inheritance of the Pl-Rh allele from Pl′ mop1-1 appears to be stochastic: only a few progeny plants are Pl-Rh; most are Pl′. This is true even among progeny derived from the outcross of a single Pl′/Pl′ mop1-1/mop1-1 tassel on a single day. Interestingly, only six of the 11 homozygous Pl′ mop1-1 individuals gave rise to Pl-Rh progeny. These were all derived from Pl′ Mop1/mop1-1 crossed with Pl′ mop1-1/mop1-1. Pl-Rh progeny were not observed from homozygous Pl′ mop1-1 individuals derived from self-pollinations of Pl′ Mop1/mop1-1 individuals. One interpretation is that Pl′ alleles exposed to mop1-1/mop1-1 for two consecutive generations are more susceptible to destabilization of the Pl′ state. The failure to detect purple anthers in progeny from some of the homozygous Pl′ mop1-1 individuals could also relate to the stability of Pl-Rh in the various tester stocks used, given that Pl-Rh can spontaneously change to Pl′. If such a change occurred, any Pl-Rh allele transmitted from Pl′ mop1-1 plants would be altered again to Pl′ and would not be detected. We suspect the latter explanation is unlikely, however, as it would require a high frequency of spontaneous paramutation. Because Pl′ has been observed to return to the more highly expressed state when heterozygous with a neutral allele or hemizygous (Hollick and Chandler, 1998), the varied effects of mop1-1 on the heritability of B′ versus Pl′ likely reflect the inherent differences in stability between B′ and Pl′. Thus, homozygous Pl′ mop1-1 represents another genotype in which Pl′ is capable of returning to the Pl-Rh state.
The effect of mop1-1 on B′ provides a very clear demarcation between two of the phases of gene silencing—maintenance and inheritance—as described by Loo and Rine (1995). Although functional MOP1 is required to maintain the reduced transcription state of B′, loss of MOP1 function is not sufficient to disrupt the inheritance of the B′ state; the latter is faithfully and completely restored when MOP1 function is recovered. In fact, we know of no condition in which B′ is heritably changed to B-I. We have speculated that this is because B′ represents the default transcription state specified by the DNA sequence (Patterson et al., 1993).
A Model for MOP1 Function and Paramutation
Numerous facets of b1, r1, and pl1 paramutation are consistent with chromatin structural changes, rather than DNA sequence changes, underlying paramutation. The absence of detectable DNA sequence differences between B′ and B-I, the multiple extents of Pl′ expression, and the instability of the paramutant R-r′ and Pl′ states fit well with a chromatin model (reviewed in Chandler et al., 2000). We propose that MOP1 functions as a chromatin-remodeling protein. As observed for some chromatin-related proteins, individuals mutant for mop1 can show developmental phenotypes (Kakutani et al., 1996; Grossniklaus et al., 1998; Eshed et al., 1999; Ogas et al., 1999). We hypothesize that MOP1 is involved in the assembly of a repressive chromatin structure at b1, pl1, and r1 during paramutation. This repressive structure could be analogous to polycomb group (PcG) complexes assembled at polycomb response elements (PRE) (Hollick et al., 1997; Pirrotta, 1998).
The distinct expression states of B-I versus B′ and Pl-Rh versus Pl′, as well as the spontaneous and directed alteration of the paramutable alleles, could be explained by this model. The alleles that participate in paramutation could possess PRE-like elements, but in the high-expression state (B-I or Pl-Rh), the elements are not efficiently recognized for PcG-like assembly. We postulate that the chromatin structure can spontaneously change to allow access of PcG-like proteins for assembly, which results in the B′ and Pl′ states. The fully assembled state of the B′ and Pl′ alleles, when together in a nucleus with the unassembled state, would induce assembly on the paramutable allele (Patterson et al., 1993; Hollick et al., 1997).
Variations on this model can explain differences in stability between the Pl′ and the B′ states and the different effect that mop1-1 has on the heritability of this state. Once assembled in B′, the PcG-like complex would remain quite stable because of multiple, strong interactions between members of the complex and DNA binding sites. When MOP1 function is lacking in B′ mop1-1 homozygotes, several remaining members of the complex would stay associated with the PRE-like binding sites, such that restoration of MOP1 function would quickly reestablish the repressive structure, just as some residual proteins are thought to mark PREs for rapid reassembly in Drosophila (Pirrotta, 1998). Divergence or fewer binding sites at pl1 could result in a less stable complex relative to b1. Loss of MOP1 function in Pl′ mop1-1 homozygotes, combined with fewer or compromised binding sites, could destabilize the complex such that residual association of the complex with the PRE-like element would be reduced in some nuclei, resulting in the Pl-Rh state.
Mutants Affecting Epigenetic Silencing or Allelic Interactions
Active research into many trans-sensing phenomena involves identifying mutations that disrupt the interaction. Because paramutation and other trans-sensing interactions share many features, these screens could identify mop1 homologs. Our results indicate that mop1-1 does not alter the extent of methylation of repetitive centromeric sequences or the 45S ribosomal repeated sequences. Thus, the mop1-1 mutation does not behave like mutations in either DNA methyltransferase or ddm1. The ddm1 locus was identified by its effect on methylation of repetitive sequences in Arabidopsis, such as the ribosomal genes and centromeric repeats (Vongs et al., 1993). The ddm1 locus has been cloned and shares similarity with SNF2, a yeast protein involved in chromatin remodeling (Jeddeloh et al., 1999). Interestingly, a related protein in humans has also been shown to decrease global DNA methylation when mutated (Gibbons et al., 2000). Similarly, decreased activity of the DNA methyltransferase of Arabidopsis, achieved by antisense expression of a MET1 transgene, reduces the extent of methylation of repeated sequences (Finnegan et al., 1996).
Several mutations altering the transcriptional silencing of plant transgenes have been identified (Furner et al., 1998; Mittelsten Scheid et al., 1998; Amedeo et al., 2000). Some of these mutations are allelic to ddm1 (Mittelsten Scheid et al., 1998; Jeddeloh et al., 1999), whereas one encodes a novel protein, MOM1 (Amedeo et al., 2000). Our mop1 alleles show pleiotropic developmental abnormalities, which is not seen with mom1 mutants. Several mutations have been identified that disrupt cosuppression or post-transcriptional gene silencing (Elmayan et al., 1998), but these are unlikely to represent mop1 homologs because post-transcriptional gene silencing appears to be mechanistically distinct from paramutation (reviewed in Meyer and Saedler, 1996; Chandler et al., 2000). A screen for mutations affecting transvection at the yellow locus of Drosophila identified exclusively cis-acting mutations at yellow (Morris et al., 1999). Numerous mutations in Drosophila have been isolated that affect position-effect variegation, identifying proteins associated with and involved in chromatin structure (Sass and Henikoff, 1998; Wakimoto, 1998; Wallrath, 1998; Cryderman et al., 1999).
Cloning of mop1, isolation and characterization of other loci required for paramutation, and characterization of the minimal cis-acting sequences required for paramutation will be critical for revealing mechanisms. A further understanding of proteins mediating paramutation in maize, transvection in Drosophila (Lewis, 1954), and many additional trans-sensing phenomena in a range of organisms (reviewed in Henikoff and Comai, 1998; Wu and Morris, 1999) is likely to reveal universal biological principles mediating cross-talk between alleles in the nucleus that influences their expression.
METHODS
Maize Nomenclature
In maize nomenclature, a gene is designated with lowercase italics (b1) (http://www.agron.missouri.edu/maize_nomenclature.html). Spe-cific alleles are indicated with an allele designation separated from the gene designation with a hyphen. Dominant alleles are indicated by an uppercase gene designation (B-Peru) and recessive alleles by a lowercase gene designation (b-K55). Gene products are indicated by capital letters and are not italicized (B).
Plant Stocks
All plant stocks contained dominant functional alleles for all the genes encoding the anthocyanin biosynthetic enzymes required in vegetative plant tissues. Because transcription of these genes in vegetative plant tissues is controlled by pl1 in combination with b1 or r1, the specific b1, pl1, and r1 alleles are indicated for relevant stocks. One exception is the distinction between Pl′-mahogany (Pl′) and Pl-Rhoades (Pl-Rh) (Hollick et al., 1995). Many stocks possess the r-g allele of r1 (no expression in the seed or vegetative body of the plant), which precludes reliable scoring of Pl′ versus Pl-Rh. In these stocks, we have used Pl to indicate the presence of either Pl-Rh or its spontaneous derivative Pl′.
Although stocks containing various b1 alleles have been maintained in the Chandler laboratory for several years, they were originally obtained from various sources: B-I Pl r-g (inbred W23 background), B′ Pl r-g (inbred K55 background), and b-K55 Pl r-g (inbred K55 background) stocks were from E.H. Coe, Jr. (University of Missouri, Columbia); B-bar was from E.D. Styles (University of Victoria); and B-Peru (inbred W22 background) was from M.G. Neuffer (University of Missouri). J.B.H. and V.L.C. have maintained Pl-Rh and Pl′-mahogany stocks originally obtained from E.H. Coe, Jr., and additional Pl-Rh stocks from the Maize Cooperative Stock Center. J.L.K. has maintained the R-stippled (R-st), R-r:standard (R-r:std, a specific accession of R-r, also known as standard R-r), and R-d:Catspaw (R-d) stocks (each containing B-bar and pl-W22, inbred W22 background) established by R.A. Brink and colleagues.
Genetic Screen
A B-I Pl r-g stock was used to generate a B-I Pl r-g Mu stock by sequential backcrosses into active Mutator stocks (Patterson et al., 1991). The B-I Pl r-g Mu stock, carrying functional alleles for all of the anthocyanin biosynthetic enzymes, was crossed with B′ Pl r-g (inbred W23 background). The B′ allele in this stock was a spontaneous derivative of the B-I allele obtained from E.H. Coe, Jr. F1 individuals between B′ and B-I Mu were self-pollinated to generate F2 families. F2 families were screened in sand benches for rare, darkly pigmented seedlings resembling B-I–like plants among siblings that were essentially green. The mop1-1 mutation was isolated from this screen.
Genetic Crosses
In all crosses, a single allele listing indicates homozygosity, whereas heterozygous individuals are indicated with alleles separated by a slash (/). In some instances, the identity of one (dominant) allele is known, but the second allele could be either of two possibilities (e.g., B′/-). The term family refers to plants grown from kernels on the same ear, all of which share a common tassel parent.
To test whether mop1-1 affects the paramutant Pl′ allele, B′ Pl r-g mop1-1 plants were crossed with a b-W23 Pl′ R-r stock, and F1 individuals were self-pollinated. Additional segregating families were generated by intercrossing light- and dark-anthered siblings. Inheritance of Pl′ was assayed from four mop1-1/mop1-1 individuals derived directly from the self-pollinated F1 individuals. Additional tests were conducted with individuals from segregating families derived from Pl′ Mop1/mop1-1 crossed with Pl′ mop1-1/mop1-1. Tests from these families totaled seven Pl′ mop1-1/mop1-1 and three Pl′ Mop1/mop1-1 individuals. The inheritance of Pl′ was tested by crosses with various Pl-Rh tester stocks. Specific stock constitutions are available upon request.
To test whether mop1-1 influences the expression of other b1 alleles, B′ Pl r-g mop1-1 plants were crossed with a B-Peru Pl r-g Mop1 stock (inbred W22 background), and the F1 plants were self-pollinated. Purple kernels were planted (B-Peru/-), and homozygous B-Peru individuals produced ears with 100% purple kernels. All F2 individuals were test-crossed with B′ Pl r-g mop1-1 to determine the mop1 genotype. B-bar stocks were derived as follows: B-bar pl-W22 R-r Mop1 (inbred W22 background) was crossed with B′ Pl r-g mop1-1; the F1 plants were self-pollinated; dark progeny (B′/- mop1-1/mop1-1) were backcrossed with B-bar pl-W22 R-r Mop1; progeny of the backcross were self-pollinated; and the dark progeny were self-pollinated again. The phenotypes of progeny from the last self-pollinations were used to discern whether mop1-1 intensifies B-bar.
To test the effects of mop1-1 on establishment of r1 paramutation, B′ Pl r-g mop1-1 plants were crossed with R-st and R-d. Intercrosses between the resulting R-st and R-d progenies produced paramutagenic R-d R-st and nonparamutagenic R-d r-g heterozygotes for comparison among mop1-1/mop1-1 and Mop1/- classes. Because mop1-1 does not intensify B-bar pigmentation, one in 13 plants classified as Mop1/- (light plant) is expected to be B-bar/B-bar mop1-1/mop1-1. Reduced paramutation in this genotype would underestimate paramutation in the control population and thus would underestimate the ability of mop1-1 to inhibit r1 paramutation. Test crosses onto W23 r-g (null for both aleurone and vegetative tissue expression of r1) served to distinguish parental R-d/r-g and R-d/R-st genotypes and provided R-d/r-g and R-d′/r-g kernels for evaluating paramutation. The amounts of kernel pigmentation were measured with a reflectometer as described previously (Alleman and Kermicle, 1993). A similar crossing scheme was used to test the effects of mop1-1 on the paramutable R-r:std haplotype. R-r:std, also known as standard R-r, indicates that this is the specific R-r accession originally tested by Brink (1956).
Analysis of RNA Concentrations and Transcription Rates
RNA concentrations were assayed by RNase protections, with actin or ubiquitin used as an internal control. RNA was isolated by using Trizol (Gibco BRL) according to manufacturer's directions, except that tissue was ground in liquid nitrogen with a mortar and pestle. RNase protections were performed as described in Selinger and Chandler (1999). Each hybridization used 5 μg of total RNA. The actin1 probe (5′ half of exon 2), the 315-bp cDNA probe from b1, and the 438-bp cDNA probe from c2 were as described previously (Selinger and Chandler, 1999). The ubiquitin probe is a 228-bp BglII fragment from ubi2 (Christensen et al., 1992), which encompasses one of the seven ubiquitin repeats. The pl1 probe is a 505-bp cDNA fragment that includes exons 1 and 2 and ends at the BglI site in exon 3 of the Pl-Rh allele (Hollick et al., 2000).
Transcription rates were determined by using in vitro transcription assays on isolated nuclei. Nuclei from 5 to 10 g of sheath and husk tissue from plants at anthesis were prepared by using a chromatin isolation protocol (Steinmuller and Apel, 1986) with the following changes. Ground material was suspended in 15 mL of isolation buffer (Steinmuller and Apel, 1986) and filtered through two layers of cheesecloth followed by filtration through a 53-μm (pore size) nylon sheet. Three centrifugations were performed (each for 15 min at −10°C and 6000g). After the first two centrifugations, crude nuclei pellets were resuspended in 15 mL of isolation buffer. After the last centrifugation, crude nuclei were resuspended in 2 mL of resuspension buffer modified to contain 20% glycerol (Hollick and Gordon, 1993). Nuclei (1 mL) were repelleted (for 30 sec at room temperature and 2040g in an Eppendorf microcentrifuge) and resuspended in 100 μL of resuspension buffer. Reactions and RNA isolations were performed as described in Hollick and Gordon (1993), except that 100 μCi of α-32P-CTP (800 Ci/mmol) was used and incubation was for 25 min at 30°C. Comparable amounts of labeled RNA for each genotype were used for filter hybridization.
Slot blots on nitrocellulose filter membrane were prepared with 100 ng of denatured purified gene fragments or the equivalent amount of denatured linearized plasmid per slot. The sequences used were ubiquitin, ∼975-bp PstI fragment of ubi2 from plasmid ca210 (Christensen et al., 1992); b1, ∼1970 bp from plasmid pBcDNA (Radicella et al., 1991); and c2, 1450 bp of cDNA from plasmid cLC46E (Wienand et al., 1986). Strips of nitrocellulose with the slot-blotted gene fragments were prehybridized for 3 hr at 42°C in 5 × SSPE (1 × SSPE is 0.15 M NaCl, 0.01 M sodium phosphate, and 0.001 M EDTA), 0.1% polyvinyl pyrrolidine, 0.1% Ficoll, 50% formamide, and 12.5 μg/mL tRNA. This was followed by hybridization for 60 to 72 hr with the radiolabeled heat-denatured RNA. Strips were briefly rinsed, washed twice for 15 min each at 42°C in 1 × SSPE containing 0.1% SDS, and washed once for 15 min at 42°C in 0.1 × SSPE containing 0.1% SDS.
The in vitro transcription assay filters and RNase protections were visualized by using a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, CA), and signals were quantified with ImageQuaNT (Molecular Dynamics) software. Background was subtracted from each signal before normalizing the b1 and pl1 signals to the signals for ubiquitin or actin.
Analysis of Global DNA Methylation
The extent of global DNA methylation was assayed using methylation-sensitive restriction enzymes and DNA gel blots. DNA was isolated from leaves (Dellaporta et al., 1983), and ∼4 μg of DNA was digested with restriction enzymes according to the manufacturers' (New England Biolabs, Beverly, MA; Gibco BRL; and Pharmacia Biotech.) specifications and size-fractionated by electrophoresis in 0.8% agarose gels with 45 mM Tris-borate buffer containing 1 mM EDTA. The DNA was transferred to Hybond N+ (Amersham) membrane with alkaline transfer buffer (0.4 M NaOH and 0.6 M NaCl). The 45S ribosomal repeat (McMullen et al., 1986, 1991) and the centromere repeat from sorghum (pSau3a9) (Jiang et al., 1996) were radioactively labeled using random hexamer priming (Feinberg and Vogelstein, 1983). Hybridization was performed overnight at 65°C in a reaction reagent of 5% SDS, 50 mM Pipes, pH 6.5, 50 mM NaHPO4, pH 7, 1 mM EDTA, 100 mM NaCl, and 100 μg/mL salmon sperm DNA. The membrane was washed for 2 min at 65°C with 1 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate)/0.1% SDS, for 30 min at 65°C with 0.5 × SSC/0.1% SDS, and for 10 min at room temperature with 0.1 × SSC/0.1% SDS. Blots were exposed to a phosphor-imaging screen and visualized with a Storm 860 PhosphorImager (Molecular Dynamics).
Acknowledgments
This work was supported by grants from the American Cancer Society (No. NP875) and the National Science Foundation (No. MCB-9603638) to V.L.C., from the Department of Energy (No. FG02-86ER13539) to J.L.K., and from the U.S. Department of Agriculture National Research Initiative (No. 97-35301-4430) to J.B.H.; and by National Science Foundation postdoctoral fellowships to J.E.D. (No. BIR-9626082), J.B.H. (No. BIR-9303601), and K.M.K. (No. BIR-9104373). The Molecular Dynamics Storm 860 System used in this work was purchased with Grant No. DAAG559710102 from the Department of Army Research. We thank Edward H. Coe, Jr., M. Gerald Neuffer, E. Derek Styles, and the Maize Cooperative Stock Center for providing maize stocks, and Shawn Kaeppler and Charles Papa for providing the 45S rDNA and centromeric probes and for recommendations regarding appropriate enzymes for use with these probes.
References
- Alleman, M., and Kermicle, J.L. (1993). Somatic variegation and germinal mutability reflect the position of transposable element Dissociation within the maize R gene. Genetics 135, 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedeo, P., Habu, Y., Afsar, K., Mittelsten Scheid, O., and Paszkowski, J. (2000). Disruption of the plant gene MOM re-leases transcriptional silencing of methylated genes. Nature 405, 203–206. [DOI] [PubMed] [Google Scholar]
- Baker, W.K. (1953). V-type position effects of a gene in Drosophila virilis normally located in heterochromatin. Genetics 38, 328–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R.A. (1956). A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41, 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R.A. (1958). Basis of a genetic change which invariably occurs in certain maize heterozygotes. Science 127, 1182–1183. [DOI] [PubMed] [Google Scholar]
- Brink, R.A. (1973). Paramutation. Annu. Rev. Genet. 7, 129–152. [DOI] [PubMed] [Google Scholar]
- Brink, R.A., Styles, E.D., and Axtell, J.D. (1968). Paramutation: Directed genetic change. Science 159, 161–170. [DOI] [PubMed] [Google Scholar]
- Cavalli, G., and Paro, R. (1998). The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93, 505–518. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., Kubo, K.M., and Hollick, J.B. (1996). b and pl paramutation in maize: Heritable transcription states programmed during development. In Epigenetic Mechanisms of Gene Regulation, V.E.A. Russo, R.A. Martienssen, and A.D. Riggs, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 289–304.
- Chandler, V.L., Eggleston, W.B., and Dorweiler, J.E. (2000). Paramutation in maize. Plant Mol. Biol. 43, 121–145. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H., Sharrock, R.A., and Quail, P.H. (1992). Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18, 675–689. [DOI] [PubMed] [Google Scholar]
- Coe, E.H., Jr. (1959). A regular and continuing conversion-type phenomenon at the B locus in maize. Proc. Natl. Acad. Sci. USA 45, 828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E.H., Jr. (1966). The properties, origin and mechanism of conversion-type inheritance at the b locus in maize. Genetics 53, 1035–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, V., Maloisel, L., and Rossignol, J.-L. (1996). Interchromosomal transfer of epigenetic states in Ascobolus: Transfer of DNA methylation is mechanistically related to homologous recombination. Cell 86, 855–864. [DOI] [PubMed] [Google Scholar]
- Cone, K.C., Cocciolone, S.M., Burr, F.A., and Burr, B. (1993). Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5, 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman, D.E., Morris, E.J., Biessmann, H., Elgin, S.C.R., and Wallrath, L. (1999). Silencing at Drosophila telomeres: Nuclear organization and chromatin structure play critical roles. EMBO J. 18, 3724–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA mini preparation. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Dooner, H., Robbins, T.P., and Jorgensen, R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 173–199. [DOI] [PubMed] [Google Scholar]
- Elmayan, T., Balzergue, S., Béon, F., Bourdon, V., Daubremet, J., Guénet, Y., Mourrain, P., Palauqui, J.-C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10, 1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V. (1983). Controlling elements in maize. In Mobile Genetic Elements, J.A. Shapiro, ed (New York, NY: Academic Press), pp. 1–63.
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner, I.J., Sheikh, M.A., and Collett, C.E. (1998). Gene silencing and homology-dependent gene silencing in Arabidopsis: Genetic modifiers and DNA methylation. Genetics 149, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, R.J., McDowell, T.L., Raman, S., O'Rourke, D.M., Garrick, D., Ayyub, H., and Higgs, D.R. (2000). Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24, 368–371. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Klein, T.M., Roth, B.A., Fromm, M.E., Cone, K.C., Radicella, J.P., and Chander, V.L. (1990). Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 9, 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., Cone, K.C., and Chandler, V.L. (1992). Functional analysis of the transcriptional activator encoded by the maize b gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Grewal, S.I.S., and Klar, A.J.S. (1996). Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86, 95–101. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and Comai, L. (1998). Trans-sensing effects: The ups and downs of being together. Cell 93, 329–332. [DOI] [PubMed] [Google Scholar]
- Hessler, A. (1958). V-type position effects at the light locus in Drosophila melanogaster. Genetics 43, 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton, T., and Goodsmith, W. (1950). An analysis of phenotypic reversions at the brown locus in Drosophila. J. Exp. Zool. 114, 103–114. [Google Scholar]
- Hollick, J.B., and Chandler, V.L. (1998). Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics 150, 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J.B., and Chandler, V.L. (2001). Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics, in press. [DOI] [PMC free article] [PubMed]
- Hollick, J.B., and Gordon, M.P. (1993). A poplar tree proteinase inhibitor-like gene promoter is responsive to wounding in transgenic tobacco. Plant Mol. Biol. 22, 561–572. [DOI] [PubMed] [Google Scholar]
- Hollick, J.B., Patterson, G.I., Coe, E.H., Jr., Cone, K.C., and Chandler, V.L. (1995). Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J.B., Dorweiler, J.E., and Chandler, V.L. (1997). Paramutation and related allelic interactions. Trends Genet. 13, 302–308. [DOI] [PubMed] [Google Scholar]
- Hollick, J.B., Patterson, G.I., Asmundsson, I.M., and Chandler, V.L. (2000). Paramutation alters regulatory control of the maize pl locus. Genetics 154, 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday, R., Ho, T., and Paulin, R. (1996). Gene silencing in mammalian cells. In Epigenetic Mechanisms of Gene Regulation, V.E.A. Russo, R.A. Martienssen, and A.D. Riggs, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 47–59.
- Irish, E.E., Langdale, J.A., and Nelson, T.M. (1994). Interactions between tassel seed genes and other sex determining genes in maize. Dev. Genet. 15, 155–171. [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jiang, J.M., Nasuda, S., Dong, F.G., Scherrer, C.W., Woo, S.S., Wing, R.A., Gill, B.S., and Ward, D.C. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93, 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A. (1995). Cosuppression, flower color patterns, and metastable gene expression states. Science 268, 686–691. [DOI] [PubMed] [Google Scholar]
- Kakutani, T., Jeddeloh, J.A., Flowers, S.K., Munakata, K., and Richards, E.J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L. (1996). Epigenetic silencing and activation of a maize r gene. In Epigenetic Mechanisms of Gene Regulation, V.E.A. Russo, R.A. Martienssen, and A.D. Riggs, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 267–287.
- Kermicle, J.L., Eggleston, W., and Alleman, M. (1995). Organization of paramutagenicity in R-stippled maize. Genetics 141, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, E.B. (1954). The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88, 225–239. [Google Scholar]
- Loo, S., and Rine, J. (1995). Silencing and heritable domains of gene expression. Annu. Rev. Cell Dev. Biol. 11, 519–548. [DOI] [PubMed] [Google Scholar]
- Ludwig, W.F., Habera, L.F., Dellaporta, S.L., and Wessler, S.R. (1989). Lc, a member of the maize r gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 86, 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A. (1996). Paramutation and gene silencing in plants. Curr. Biol. 6, 810–813. [DOI] [PubMed] [Google Scholar]
- Matzke, A.J.M., Neuhuber, F., Park, Y.-D., Ambros, D.F., and Matzke, M.A. (1994). Homology-dependent gene silencing in transgenic plants, epistatic silencing loci contain multiple copies of methylated transgenes. Mol. Gen. Genet. 244, 219–229. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Matzke, A.J.M., and Eggleston, W.B. (1996). Paramutation and transgene silencing: A common response to invasive DNA? Trends Plant Sci. 1, 382–388. [Google Scholar]
- McClintock, B. (1957). Genetic and cytological studies of maize. Carnegie Inst. Wash. Year Book 56, 393–401. [PubMed] [Google Scholar]
- McClintock, B. (1963). Further studies of gene-control systems in maize. Carnegie Inst. Wash. Year Book 62, 486–493. [Google Scholar]
- McMullen, M.D., Hunter, B., Phillips, R.L., and Rubenstein, I. (1986). The structure of the maize ribosomal DNA spacer region. Nucleic Acids Res. 14, 4953–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen, M.D., Phillips, R.L., and Rubenstein, I. (1991). Molecular analysis of the nucleolus organizer region in maize. In Chromosome Engineering in Plants: Genetics, Breeding and Evolution, P.K. Gupta and T. Tsuchiya, eds (New York: Elsevier Science), pp. 561–576.
- Meyer, P., ed (1995). Gene silencing in higher plants and related phenomena in other eukaryotes. Curr. Top. Microbiol. Immunol. 197, 1–232.7493486 [Google Scholar]
- Meyer, P., and Saedler, H. (1996). Homology-dependent gene silencing in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 23–48. [DOI] [PubMed] [Google Scholar]
- Meyer, P., Heidmann, I., and Niedenhoff, I. (1993). Differences in DNA methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 4, 89–100. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid, O., Afsar, K., and Paszkowski, J. (1998). Release of epigenetic gene silencing by trans-acting mutations in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.R., Chen, J.-l., Filandrinos, S.T., Dunn, R.C., Fisk, R., Geyer, P.K., and Wu, C.-t. (1999). An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics 151, 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H.J. (1930). Types of visible variations induced by x-rays in Drosophila. J. Genet. 22, 299–334. [Google Scholar]
- Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Kaurmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.I., Harris, L.J., Walbot, V., and Chandler, V.L. (1991). Genetic analysis of B-Peru, a regulatory gene in maize. Genetics 126, 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.I., Thorpe, C.J., and Chandler, V.L. (1993). Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics 135, 881–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.I., Kubo, K.M., Shroyer, T., and Chandler, V.L. (1995). Sequences required for paramutation of the maize b gene map to a region containing the promoter and upstream sequences. Genetics 140, 1389–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta, V. (1997). PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev. 7, 249–258. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V. (1998). Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 93, 333–336. [DOI] [PubMed] [Google Scholar]
- Radicella, J.P., Turks, D., and Chandler, V.L. (1991). Cloning and nucleotide sequence of a cDNA encoding B-Peru, a regulatory protein of the anthocyanin pathway in maize. Plant Mol. Biol. 17, 127–130. [DOI] [PubMed] [Google Scholar]
- Radicella, J.P., Brown, D., Tolar, L.A., and Chandler, V.L. (1992). Allelic diversity of the maize b regulatory gene: Different leader and promoter sequences of two b alleles determine distinct tissue specificities of anthocyanin production. Genes Dev. 6, 2152–2164. [DOI] [PubMed] [Google Scholar]
- Sass, G.L., and Henikoff, S. (1998). Comparative analysis of position-effect variegation mutations in Drosophila melanogaster delineates the targets of modifiers. Genetics 148, 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger, D.A., and Chandler, V.L. (1999). A mutation in the pale aleurone color1 gene identifies a novel regulator of the maize anthocyanin pathway. Plant Cell 11, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, J.M., and Pillus, L. (1997). An uncertain silence. Trends Genet. 13, 308–313. [DOI] [PubMed] [Google Scholar]
- Steinmuller, K., and Apel, K. (1986). A simple and efficient procedure for isolating plant chromatin which is suitable for studies of DNase I–sensitive domains and hypersensitive sites. Plant Mol. Biol. 7, 87–94. [DOI] [PubMed] [Google Scholar]
- Styles, E.D., Ceska, O., and Seah, K.-T. (1973). Developmental differences in action of r and b alleles in maize. Can. J. Genet. Cytol. 15, 59–72. [Google Scholar]
- van der Krol, A.R., Mur, L.A., Beld, M., Mol, J.N.M., and Stuitje, A.R. (1990). Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wakimoto, B.T. (1998). Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell 93, 321–324. [DOI] [PubMed] [Google Scholar]
- Walker, E. (1998). Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148, 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath, L. (1998). Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev. 8, 147–153. [DOI] [PubMed] [Google Scholar]
- Wienand, U., Weydemann, U., Niesbach-Klosgen, U., Peterson, P.A., and Saedler, H. (1986). Molecular-cloning of the c2 locus of Zea mays, the gene coding for chalcone synthase. Mol. Gen. Genet. 203, 202–207. [Google Scholar]
- Wu, C.-t., and Morris, J.R. (1999). Transvection and other homology effects. Curr. Opin. Genet. Dev. 9, 237–246. [DOI] [PubMed] [Google Scholar]