Abstract
To determine which components of the plant defense response make important contributions to limiting pathogen attack, an M2 mutagenized population of a transgenic Arabidopsis line was screened for mutants showing constitutive expression of β-glucuronidase activity driven by the promoter region of the CEVI-1 gene. The CEVI-1 gene originally was isolated from tomato plants and has been shown to be induced in susceptible varieties of tomato plants by virus infection in a salicylic acid–independent manner. We report here the characterization of a recessive mutant, detachment9 (dth9). This mutant is more susceptible to both virulent and avirulent forms of the oomycete Peronospora and also exhibits increased susceptibility to the moderately virulent bacterial pathogen Pseudomonas syringae pv maculicola ES4326. However, this mutant is not affected in salicylic acid metabolism and shows normal expression of pathogenesis-related (PR) genes after pathogen attack. Furthermore, after inoculation with avirulent pathogens, the dth9 mutant shows a compromised systemic acquired resistance response that cannot be complemented by exogenous application of salicylic acid, although this molecule is able to promote normal activation of PR genes. Therefore, the dth9 mutation defines a regulator of disease susceptibility that operates upstream or independently of salicylic acid. Pleiotropy is also evident in the dth9 mutant in the sense that the shoots of dth9 plants are insensitive to the exogenously applied auxin analog 2,4-dichlorophenoxyacetic acid.
INTRODUCTION
Plants have developed constitutive as well as inducible defense responses against pathogens. Systemic acquired resistance (SAR; Ross, 1961) is one such inducible defense response that is triggered in the plant by previous exposure to pathogens that cause cell death. SAR is long lasting and confers protection against a broad spectrum of pathogens (Ryals et al., 1996). A more rapid defense response that precedes the onset of SAR is the hypersensitive response (HR; Agrios, 1988), which is localized at the site of attempted pathogen entry. HR is characterized by programmed death of host cells and is a consequence of the interplay of the products of the pathogen avirulent genes (Avr) and the host disease resistance genes (R). Tightly correlated with the HR and the SAR is the production of antimicrobial compounds, the increased expression of a subset of the pathogenesis-related (PR) proteins, many of which possess antimicrobial activities, and the reinforcement of mechanical barriers such as cell walls (Sticher et al., 1997).
Salicylic acid (SA) is an important signal molecule in plant defense. SA accumulates in increased amounts during HR and SAR. Preventing the accumulation of SA compromises disease resistance and the accretion of PR proteins in infected plants (Gaffney et al., 1993; Delaney et al., 1994). Furthermore, exogenous application of SA induces disease resistance and the expression of PR genes (Ryals et al., 1996).
Studies on mutants altered in SAR responses suggest that plants have developed a complex network of interactions that ultimately orchestrate the activation of defense-related responses. In Arabidopsis, the NPR1/NIM1/SAI1 locus operates downstream from SA and has been shown to be a key component of SA-regulated PR gene expression and disease resistance, because npr1/nim1/sai1 mutants fail to express PR genes and display enhanced susceptibility to infection even after treatment with SA (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). Other mutants affected in plant resistance could be positioned upstream of the synthesis or action of SA. lsd (Dietrich et al., 1994) and acd2 (Greenberg et al., 1994) mutants have high amounts of SA, express lesions similar to that of an HR, and show enhanced resistance to infection. The cpr mutants (Bowling et al., 1994) also show constitutive expression of PR genes as well as constitutively high amounts of SA, suggesting that they act upstream of SA. However, although cpr1 is affected specifically in the SA pathway (Bowling et al., 1994), the cpr5 mutant (Bowling et al., 1997) shows constitutive expression of the PDF1.2 gene, which is independent of SA. Also, the cpr6 and ssi1 mutants are distinct in the sense that the expression of PR genes is independent of NPR1/NIM1/SAI1 (Clarke et al., 1998; Shah et al., 1999). In addition, dnd1, although exhibiting increased resistance and high constitutive expression of SA and PR genes, does not undergo an HR reaction when infected with avirulent pathogens (Yu et al., 1998).
In marked contrast, the pad4 mutant (Zhou et al., 1998) is impaired in the accumulation of SA and the phytoalexin camalexin as well as in expression of the PR1 gene after infection with a virulent pathogen. Likewise, the SA induction– deficient (sid) mutants (Nawrath and Métraux, 1999) are compromised in SAR and do not accumulate SA after pathogen infection, but only a subset of PR genes in these mutants is strongly reduced, and camalexin amounts are normal. The enhanced disease susceptibility (eds) mutants (Glazebrook et al., 1996; Parker et al., 1996; Rogers and Ausubel, 1997; Volko et al., 1998) represent another mutant type that is compromised in resistance, but the role of SA remains to be elucidated. Like sid mutants, the eds5 mutant (allelic to sid1) is defective only in the expression of PR1 but not in that of PR2 or PR5, suggesting that there is an SA-independent compensation pathway activated in the plant once the pathogen has been detected (Nawrath and Métraux, 1999).
We recently described the isolation of dth (detachment) mutants from Arabidopsis, which are defined as mutants displaying constitutive expression of the reporter β-glucuronidase (GUS) gene driven by the promoter of the tomato CEVI-1 gene (Mayda et al., 2000). In tomato plants, CEVI-1 expression was shown to be transcriptionally activated, along with other defense-related genes, during the course of compatible plant–virus interactions (e.g., tomato mosaic virus). Additionally, CEVI-1 showed rapid activation if leaf segments were detached from the plant. However, the expression of CEVI-1, either during disease or on detachment, was controlled by way of an SA-independent pathway, and neither ethylene nor jasmonic acid and wounding were able to induce expression of CEVI-1 in the plant (Mayda et al., 2000). These findings suggested that the expression of CEVI-1 could be controlled by a novel signaling pathway activated during disease in susceptible tomato plants. Additionally, the observation that the constitutive expression of CEVI-1::GUS in the Arabidopsis dth mutants concurs with an auxin-resistant phenotype in the plant prompted us to suggest that the mechanism upregulating the expression of CEVI-1 is related to an induced insensitivity to auxin that might be primed during disease (Mayda et al., 2000).
To study a causal link between the signal pathway mediating the activation of CEVI-1 and that mediating disease susceptibility in plants, we report the characterization of the dth9 mutant as a case study.
RESULTS
Identification of the dth9 Mutant
The Arabidopsis dth mutants were identified in a screen for constitutive expressers of a GUS gene driven by the promoter region of the CEVI-1 gene. The details of this screen have been described previously (Mayda et al., 2000). The CEVI-1 gene (which encodes an anionic peroxidase) originally was isolated from tomato plants and shown to be induced in susceptible tomato plants, but not in resistant plants, by viral infection (e.g., tomato mosaic virus; Mayda et al., 2000).
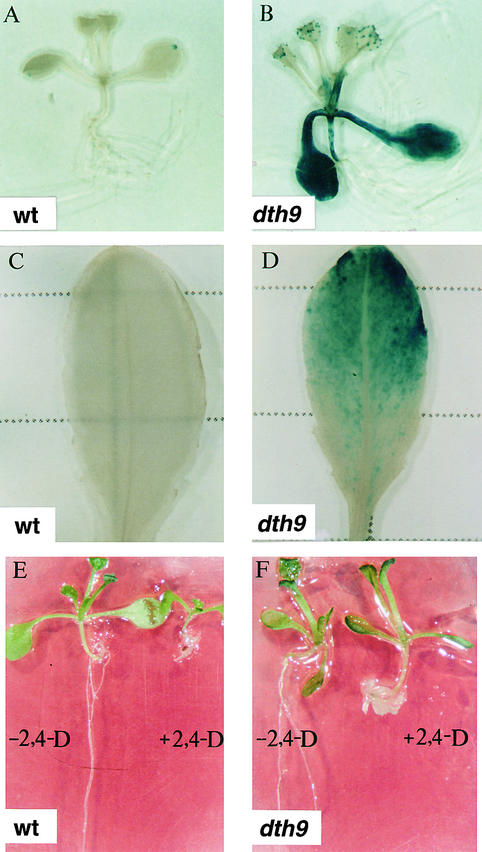
Here, we report the characterization of dth9. Figures 1A and 1B show the constitutive expression of the GUS reporter gene in the Arabidopsis dth9 mutant compared with the parental nonmutagenized transgenic CEVI-1::GUS line. The parental line showed no GUS activity except in a few cells located at the distal margin of the cotyledons, whereas the dth9 mutant showed intense GUS staining throughout the plant body except in the roots. In full-grown plants (Figures 1C and 1D), GUS staining was present along the rosette leaves in the dth9 plants, whereas expression of GUS activity in the parental line was again absent.
Figure 1.
Characterization of dth9 Plants and Comparison with Wild-Type Arabidopsis Plants Containing CEVI-1::GUS.
(A) Histochemical staining of GUS activity in a 12-day-old wild-type transgenic seedling grown on Murashige and Skoog (MS; see Methods) agar medium.
(B) GUS expression of a dth9 mutant plant grown as in (A).
(C) Fully expanded rosette leaf from a wild-type transgenic plant stained for GUS activity.
(D) Fully expanded rosette leaf from a dth9 plant stained for GUS activity.
(E) Twelve-day-old Arabidopsis wild-type seedlings grown on MS plates containing (right) or not containing (left) 0.2 μM 2,4-D for 12 days.
(F) Twelve-day-old Arabidopsis dth9 mutant seedlings grown on MS plates containing (right) or not containing (left) 0.2 μM 2,4-D for 12 days.
wt, wild type.
This tissue-specific distribution of GUS staining in dth9 plants was similar to that observed for the previously reported nonallelic dth2 and dth23 mutants isolated in the same screen (Mayda et al., 2000). In addition to their constitutive expression of GUS driven by the CEVI-1 promoter, dth2 and dth23 mutants also were characterized as auxin-insensitive mutants because they were resistant to the growth inhibition caused by (2,4-dichlorophenoxy)acetic acid (2,4-D; Mayda et al., 2000). To determine if the 2,4-D– resistant phenotype also concurred in the dth9 mutant, we germinated dth9 seed on agar plates containing 0.2 μM 2,4-D, and the extent of growth inhibition attributable to the perception of this hormone was compared with that observed in wild-type seed. Growth of wild-type seedlings was severely affected on this medium, whereas dth9 seedlings appeared to resist the inhibitory effect of 2,4-D, at least in the aerial part of the plant (Figures 1E and 1F). At variance with dth2 and dth23 mutants, which show the auxin-resistance phenotype in both aerial and root tissues (Mayda et al., 2000), roots of dth9 mutants did not elongate when grown in the presence of 2,4-D (Figure 1F).
Genetic Characterization of the dth9 Mutation
To determine the genetic basis of the phenotype described above in dth9 plants, we performed a backcross between dth9/dth9 plants and wild-type DTH9/DTH9 plants containing the CEVI-1::GUS transgene and then analyzed the progeny. In the F1 plants resulting from this cross, constitutive expression of GUS was absent in all 27 seedlings tested; in the F2 plants, expression was present in 19 of 90 seedlings. The F2 segregation ratio of the phenotype conferred by dth9 was 1:3.7 (constitutive expressers:nonexpressers), and the χ2 value calculated for goodness of fit to a single recessive nuclear mutation was 0.53 (0.1 > P > 0.5).
To define the chromosomal map position of DTH9, we crossed the dth9 mutant, which is in a Columbia (Col-0) background, to wild-type Landsberg erecta (Ler), and F2 seedlings were phenotyped and examined using simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994). DNA was isolated from 31 dth9 homozygous plants, and segregation of SSLP markers indicated that dth9 showed linkage to the Nga1126 marker on chromosome 2. Of 62 chromosomes analyzed, 12 showed Ler alleles (12 Ler; 50 Col-0; data not shown).
dth9 Mutants Have Enhanced Susceptibility to Peronospora
The CEVI-1 gene has been shown to be induced in susceptible tomato plants infected with viral pathogens, and its expression correlated spatiotemporally with that of other defense-related genes (e.g., PR genes) in diseased plants (Mayda et al., 2000). However, the expression of CEVI-1 was not mediated by SA, ethylene, or jasmonate, thus favoring the interpretation that the signaling cascade controlling CEVI-1 expression runs parallel to that of other defense cascades activated during disease (Mayda et al., 2000).
To determine whether the increased amounts of GUS expression driven by the CEVI-1 promoter in dth9 plants correlated with alteration of the disease resistance state of the plant, we tested the response of dth9 plants to the obligate biotroph Peronospora (Figure 2).
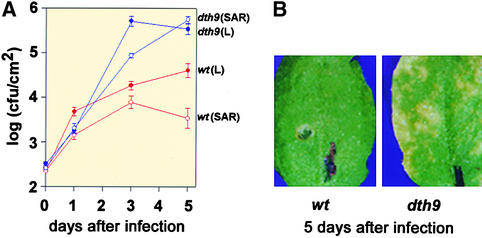
Figure 2.
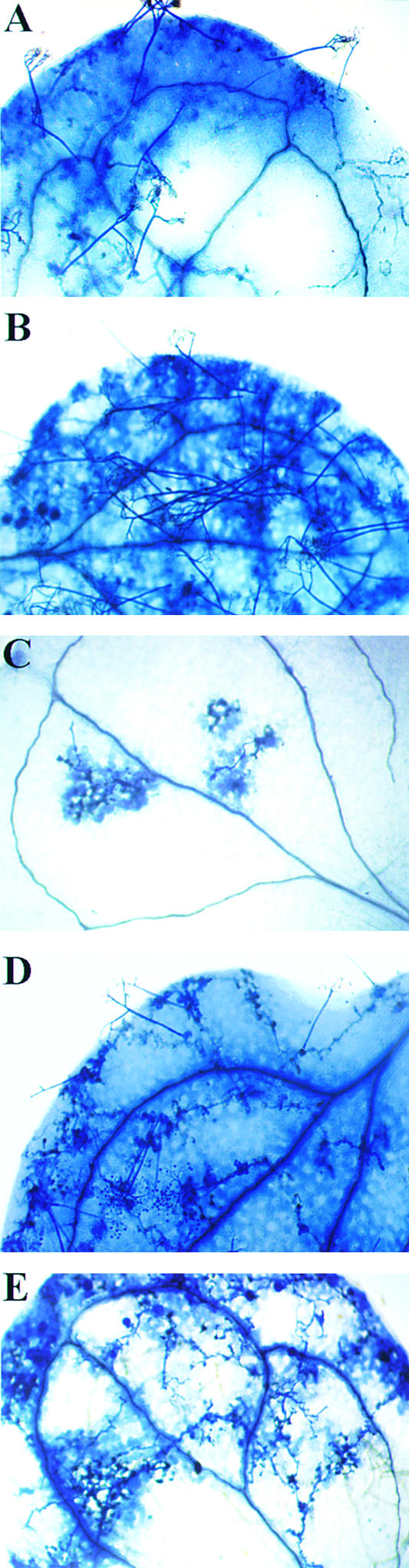
Resistance Response of Wild-Type and dth9 Mutant Arabidopsis Plants to Peronospora Isolates.
Seven days after spray inoculation of 2-week-old plants with 105 conidiospores per milliliter of Peronospora isolates, cotyledons were stained with lactophenol−trypan blue and viewed under a microscope.
(A) and (B) Plants inoculated with virulent isolate NOCO: wild-type plant with blue staining of hyphae and conidiospores (A) and dth9 mutant with blue staining of hyphae, conidiospores, and oospores (B).
(C) to (E) Plants inoculates with avirulent isolate EMWA: wild-type plant with blue staining of typical HR reactions (C) and dth9 mutant with blue staining of hyphae with trailing necrosis, conidiospores, and oospores ([D] and [E]).
We inoculated 25 dth9 plants as well as 25 wild-type parental plants with the Peronospora isolate NOCO (105 conidiospores per milliliter), which is virulent on Arabidopsis Col-0, and the appearance of conidiospores was scored for as long as 7 days later (Figures 2A and 2B). Whereas sporulation occurred on only 40% of the leaves from wild-type plants, 90 to 100% of the leaves from dth9 plants demonstrated its presence. Evaluation of the infection with a dissecting microscope revealed that the dth9 mutants became so heavily colonized that they consisted almost only of oomycetes. Therefore, even the susceptible Col-0 wild-type plants become even more susceptible when the dth9 mutation is present.
When the plants were similarly inoculated with the Peronospora isolate EMWA, which is avirulent on Arabidopsis Col-0, the wild-type plants were completely resistant, they reacted with an HR, and no sporulation was observed (Figure 2C). In contrast, the dth9 mutants showed a strong shift toward susceptibility, as demonstrated by extensive colonization of the plants (Figures 2D and 2E). On the plant side, cell death occurred in the form of trailing necrosis that followed the growing hyphae, as revealed by intense retention of trypan blue in the vicinity of the hyphae. However, these plant defense reactions seem to be insufficient, because Peronospora was able to complete its cycle, forming sexual oospores and asexual conidia on conidiophores. Because the growth of Peronospora EMWA in the dth9 plants always was associated with necrosis on the plant side, the resistance to EMWA was lost only in part.
The dth9 Mutant Is More Susceptible to Pseudomonas syringae
Increased susceptibility of dth9 plants to pathogens was investigated further by using the virulent bacterial pathogen P. syringae pv maculicola ES4326 (see Methods). After infection of wild-type and dth9 plants with P. s. maculicola ES4326, the growth rate of these bacteria was monitored in extracts from infected leaves; the resulting growth curves are shown in Figure 3A. The number of P. s. maculicola ES4326 detected in dth9 plants was more than 10-fold greater than the number in wild-type plants after 3 to 5 days of growth (cf. wt[L] and dth9[L] growth curves in Figure 3A).
Figure 3.
Growth of P. s. maculicola ES4326 in Wild-Type and dth9 Plants during Local and SAR Responses.
(A) Bacterial growth curves. For local response studies (L), wild-type and dth9 plants were infected by infiltrating leaves with a P. s. maculicola ES4326 suspension at a 500-fold dilution (105 cfu/mL) at 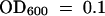 . Bacterial titer was determined at 0, 1, 3, and 5 days after infection. For SAR response studies (SAR), two leaves per plant first were infected by infiltration with a suspension of P. s. tomato DC3000 carrying avrRpm1 (
. Bacterial titer was determined at 0, 1, 3, and 5 days after infection. For SAR response studies (SAR), two leaves per plant first were infected by infiltration with a suspension of P. s. tomato DC3000 carrying avrRpm1 (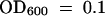 ); 3 days after this initial infection, the distal leaves were inoculated with a P. s. maculicola ES4326 suspension at a 500-fold dilution (105 cfu/mL) at
); 3 days after this initial infection, the distal leaves were inoculated with a P. s. maculicola ES4326 suspension at a 500-fold dilution (105 cfu/mL) at 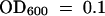 . Bacterial titer was determined at 0, 1, 3, and 5 days after the second inoculation. Error bars represent 95% confidence limits of log-transformed data. Eight samples were taken for each genotype at each time noted. The experiment was repeated three times with similar results. cfu, colony-forming unit.
. Bacterial titer was determined at 0, 1, 3, and 5 days after the second inoculation. Error bars represent 95% confidence limits of log-transformed data. Eight samples were taken for each genotype at each time noted. The experiment was repeated three times with similar results. cfu, colony-forming unit.
(B) Symptoms of bacterial infection in local leaves from wild-type (left) and dth9 (right) plants at 5 days after inoculation with P. s. maculicola ES4326.
wt, wild type.
To study this susceptibility of dth9 plants to the virulent bacteria in more detail, we addressed whether or not the dth9 mutant could be compromised in the SAR response. We inoculated wild-type and dth9 plants with the avirulent bacteria P. syringae pv tomato DC3000 AvrRpm1, which elicit an HR response in the inoculated leaf. Then, 3 days after this first inoculation, we challenged other leaves of the same plants with the virulent P. s. maculicola ES4326 strain. The plants were examined visually for disease symptoms as well as for growth of the newly inoculated P. s. maculicola ES4326.
The bacterial titer of P. s. maculicola ES4326 in the wild-type plants expressing SAR after the first inoculation with the incompatible bacteria was observed to decrease with time (Figure 3A: cf. wt[L] with wt[SAR] growth curves). This suggested that SAR developed appropriately in the wild-type plants. Conversely, the number of P. s. maculicola ES4326 in the dth9 mutants was 100-fold greater after 5 days of growth. Additionally, as shown for leaves photographed 5 days after the second inoculation with virulent bacteria (Figure 3B), severe chlorosis was observed in dth9 plants, whereas symptoms were absent in wild-type plants. These results suggested an effect of the dth9 mutation in interfering with the normal development of SAR as elicited by the avirulent P. s. tomato DC3000 AvrRpm1 pathogen.
Endogenous Amounts of SA Are Not Affected in dth9 Plants
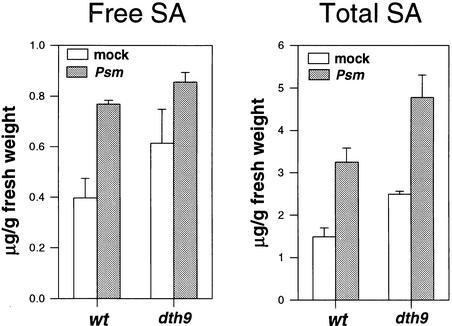
Because SA is the master regulatory molecule necessary for the normal activation of defenses and SAR response, measurements were made to determine whether the amount of endogenous SA was affected in dth9 plants. Free SA and conjugated salicylate glucoside (SAG) concentrations were examined in leaf tissues from dth9 and wild-type plants (Figure 4). In healthy noninoculated leaves of dth9 plants, the basal amount of SA was 1.7-fold greater than that observed in wild-type plants, the same difference observed when SAG contents were analyzed. In infected leaf tissue at 4 days after inoculation with P. s. maculicola ES4326, the amounts of free SA increased twofold in wild-type plants and 1.5-fold in dth9 plants (Figure 4). Similar increases in SAG were observed in both cases (Figure 4). This finding suggests that the increased susceptibility to pathogens observed in dth9 plants was not the result of major defects in SA production.
Figure 4.
SA and SAG Contents in Wild-Type and dth9 Plants.
Wild-type (wt) and dth9 plants were inoculated with P. s. maculicola ES4326 (Psm) or 10 mM MgCl2 (mock). Each bar represents the mean of five replicate samples. Error bars represent sd. SA and SAG were assayed in the same samples.
Defense-Related Genes Are Normally Induced in dth9 Plants
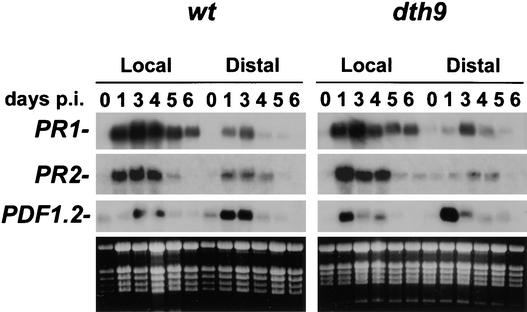
Common to the SAR response is the activation of defense-related genes (e.g., PR genes). Because no gross differences were observed in SA contents, we thought it possible that the SAR defect observed in dth9 plants was located downstream from SA, as observed in npr1/nim1 plants, which are also unusually susceptible to pathogen infection and do not express PR genes (Cao et al., 1994, 1997). To study this possibility, we inoculated wild-type and dth9 mutant plants with P. s. tomato DC3000 AvrRpm1 and determined by RNA gel blot analysis the extent and duration of the activation of the PR genes in both pathogen-treated leaves and nontreated systemic leaves. As shown in Figure 5, expression of PR1 and PR2 was highly induced in both pathogen-inoculated leaves and noninoculated systemic leaves. In wild-type plants and dth9 plants, the kinetics and intensity of PR induction were very similar. These results suggest that contrary to observations in other SAR-compromised mutants (e.g., npr1/nim1), dth9 mutant plants are not blocked in the induction of PR genes.
Figure 5.
Defense Gene Expression in Local (Inoculated) and Distal (Noninoculated) Leaves of Wild-Type and dth9 Plants at Different Times after Infection with P. s. tomato DC3000 Carrying avrRpm1.
Leaves were excised at 0, 1, 3, 4, 5, and 6 days after infection (p.i.), and the accumulation of PR1, PR2, and PDF1.2 mRNAs was tested. The experiment was repeated twice with similar results. wt, wild type.
Penninckx et al. (1998) reported an SA-independent resistance pathway in Arabidopsis that is characterized by the jasmonate-inducible PDF1.2 gene. To determine whether dth9 had a defect on the induction of this jasmonate-dependent pathway, we hybridized the RNAs mentioned above with the PDF1.2 probe. Figure 5 shows that wild-type and dth9 plants expressed PDF1.2 transiently when inoculated with P. s. tomato DC3000 AvrRpm1. This suggests that dth9 plants are not compromised in the signal pathway that activates PDF1.2.
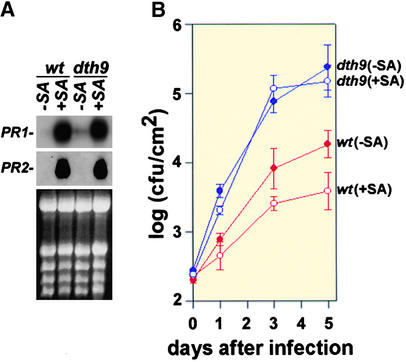
Exogenous SA Induces PR Gene Expression in dth9 but Does Not Complement the Compromised SAR Response
The fact that normal PR gene expression took place in dth9 plants after pathogen inoculation does not completely rule out the possibility that these genes could be activated in an SA-alternative or SA-independent pathway. To determine whether or not dth9 mutant plants can perceive SA, and also whether SA could restore the compromised SAR response, we treated wild-type and dth9 plants with 1 mM SA, and the induction of PR gene expression and the activation of SAR were studied. Figure 6A shows that after exposure of plants to SA, both wild-type and dth9 plants had high and comparable expressions of PR genes. However, when the growth curves of P. s. maculicola ES4326 were compared (Figure 6B), we observed that although SAR was induced effectively by SA in wild-type plants, the dth9 mutant still failed to develop the SAR phenotype. The growth of the bacteria in SA-treated dth9 plants was comparable to that observed in plants before treatment with SA.
Figure 6.
Exogenous SA Induced PR Gene Expression in Wild-Type and dth9 Plants but Enhanced Resistance to P. s. maculicola ES4326 in Wild-Type Plants Only.
Plants were sprayed with 1 mM SA or with buffer alone until uniformly wet.
(A) PR1 and PR2 gene expression in response to SA. Samples were taken 2 days after spraying with SA (+SA) or with buffer (−SA).
(B) Effect of spraying plants with SA or buffer alone on P. s. maculicola ES4326 growth. Two days after treatment, plants were infected with P. s. maculicola ES4326 at a 500-fold dilution (105 cfu/mL) at 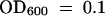 . Bacterial titer was determined at 0, 1, 3, and 5 days after infection. Each symbol represents the mean and sd of eight replicates. The experiments were repeated three times with similar results.
. Bacterial titer was determined at 0, 1, 3, and 5 days after infection. Each symbol represents the mean and sd of eight replicates. The experiments were repeated three times with similar results.
cfu, colony-forming unit. −SA, plants not treated with SA before infection; +SA, plants treated with SA before infection; wt, wild type.
The fact that the exogenous application of SA could promote the expression of PR genes but not complement the defect in SAR in dth9 plants suggests that the defective SAR response observed in this mutant was not caused by a lack of perception of SA.
The dth9 Mutant Accumulates Camalexin Normally
The accumulation of the phytoalexin camalexin is important for a plant's defense system, and some mutants impaired in camalexin production show marked increases in susceptibility to pathogens (Glazebrook and Ausubel, 1994; Glazebrook et al., 1996; Thomma et al., 1999). Furthermore, because camalexin is an indole-type compound with some similarities to auxin, we tested whether or not camalexin production could be altered in dth9 plants. Determination of camalexin content in mock-inoculated and P. s. tomato DC3000–inoculated plants revealed no marked differences between wild-type and dth9 plants with regard to camalexin accumulation (Table 1). Wild-type plants responded to the bacterial infection with a 148-fold induction of camalexin content; dth9 plants responded with a 113-fold induction. This finding suggests that the pathway controlling camalexin production is not affected in dth9 plants.
Table 1.
Accumulation of Camalexin in Leaves of Wild-Type and dth9 Plantsa
| Treatment | Camalexinb | Induction |
|---|---|---|
| dth9 + MgCl2 | 40.7 ng/g FW | |
| dth9 + P.s. DC3000 | 4587.7 ng/g FW | 113-fold |
| wt + MgCl2 | 24.6 ng/g FW | |
| wt + P.s. DC3000 | 3640.6 ng/g FW | 148-fold |
Four weeks after sowing, plants were vacuum-infiltrated (30 sec) with a suspension (in 10 mM MgCl2) of P. syringae pv tomato DC3000 at a concentration of 105 cfu/mL. Camalexin levels were determined 2 days after infection according to Nawrath and Métraux (1999). The experiment was repeated twice with similar results.
The camalexin content in each sample is expressed as nanograms per gram of fresh tissue. Each value is calculated from the peak height of each HPLC chromatogram and compared with o-anisic acid and SA standards.
FW, fresh weight; wt, wild type.
DISCUSSION
The constitutive expression of CEVI-1::GUS in the Arabidopsis dth9 mutant is similar to that observed in virally infected CEVI-1::GUS transgenic tomato plants (Mayda et al., 2000). This similarity indicates that the constitutive expression of GUS activity in intact dth9 plants probably results from a mutation in the same signaling pathway that controls its induction during disease in tomato. Furthermore, the absence of constitutive expression of PR genes in this mutant supports the notion that the dth9 mutation is distinct from mutations resulting in constitutive upregulation of SA-inducible genes (e.g., cpr [Bowling et al., 1994]).
To determine the effect of the dth9 mutation on susceptibility to pathogens, we examined the growth of two normally virulent pathogens. dth9 plants that had not been treated with any resistance inducers were challenged with either the fungal pathogen Peronospora NOCO or the bacterial pathogen P. s. maculicola ES4326. For both pathogens, dth9 plants showed greater susceptibility than did the parental wild-type plants, allowing the pathogens to grow massively (Figures 2A, 2B, and 3A).
To extend these studies in more detail, we determined whether or not activation of SAR also was affected in this mutant. After inoculating dth9 plants with the avirulent pathogen P. s. tomato DC3000 AvrRpm1 to elicit HR, we determined the extent of SAR responses by inoculating other leaves from the same plants with the virulent pathogen P. s. maculicola ES4326. These studies revealed that SAR was compromised in dth9 plants, which, unlike the wild-type plants, were unable to stop the growth of the bacteria (Figure 3). The inoculated leaves of dth9 plants, but not those of wild-type plants, very early developed the characteristic symptoms (e.g., pronounced chlorosis) of massive growth of the bacteria, further reinforcing the notion that the dth9 mutants failed to mount a proper SAR response.
SA is required for the induction of PR genes and for the activation of SAR responses (Gaffney et al., 1993). The pathway transducing the SA signal has been marked by different Arabidopsis mutants that represent only one genetic locus, designated NPR1/NIM1/SAI1. This type of mutant is nonresponsive to induction of SAR by avirulent pathogens, does not respond to SA, and is a nonexpresser of PR genes (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). Thus, the inability of dth9 plants to mount SAR could result initially from a defect in the downstream regulatory factor (e.g., NPR1/NIM1/SAI1) that transduces the SA signal to activate PR genes. However, our observation that in dth9 plants, the spatiotemporal expression of PR genes after infection with an avirulent pathogen (Figure 5) took place the same as in wild-type plants excludes this possibility. Furthermore, the exogenous application of SA also induced the expression of PR1 and PR2 in dth9 plants but was not able to correct the defective SAR response in this mutant (Figure 6). This finding favors the interpretation that SA functions normally for the activation of PR genes in dth9 plants and is consistent with the proposal that DTH9 operates upstream of SA or independently of NPR1/NIM1/SAI1 and PR gene expression or possibly both. Alternately, the inability of dth9 plants to mount SAR could reflect a defect in the synthesis and accumulation of SA without affecting the NPR1/NIM1/SAI1 locus. However, the observation that SA metabolism is not affected in dth9 plants (Figure 4) excludes the possibility that the observed compromised SAR response in dth9 plants is the result of a defect in SA metabolism.
Like dth9, the eds mutants (Glazebrook et al., 1996; Parker et al., 1996; Rogers and Ausubel, 1997; Volko et al., 1998) also show an altered susceptibility to virulent pathogens. However, at variance with dth9, some eds mutants are still capable of mounting a SAR response if they have been inoculated with an avirulent pathogen (Rogers and Ausubel, 1997). This finding suggests that dth9 and eds mutants represent different genes.
Another category of mutants with increased susceptibility to pathogens is represented by the phytoalexin-deficient (pad) mutants (Glazebrook and Ausubel, 1994). In particular, the pad4 mutant resembles the dth9 mutant in that SA can still induce defense gene expression (Glazebrook et al., 1996; Zhou et al., 1998). However, although dth9 plants accumulate normal amounts of SA (Figure 4) and camalexin (Table 1), the pad4 mutant is impaired in the synthesis of these two compounds (Zhou et al., 1998), suggesting that dth9 and pad4 mutants are different.
The sid mutants also have been described as more susceptible to pathogens (Nawrath and Métraux, 1999), but unlike dth9, the sid mutants do not accumulate SA and the expression of PR1 is strongly reduced.
Studies of the response of dth9 plants to the incompatible Peronospora isolate EMWA yielded an interesting observation. For this pathogen, dth9 plants show a reduction in resistance. However, this effect is only partial because the pathogen growth is followed by the development of massive cell death along hyphal tracks (trailing necrosis), suggesting that the R gene action in the mutant is only delayed. This phenotype is similar to that observed in nahG plants (Nawrath and Métraux, 1999) and also in the dominant phx3 mutant, which functions as a suppressor of the lsd5 cell death mutation (Morel and Dangl, 1999). However, dth9 plants allow the incompatible pathogen to complete its life cycle and sporulate, whereas the phx3 mutant does not. Interestingly, and although not as evident as for dth9, the phx3 mutant also shows some enhanced susceptibility to virulent P. syringae. Furthermore, because dth9 and phx3 retain the ability to express PR genes after treatment with SA, both mutants probably act upstream or independently of the point of action of SA. These observations indicate that the dominant phx3 mutant and the recessive dth9 mutant may participate in a similar signal transduction pathway evolved to limit pathogen growth.
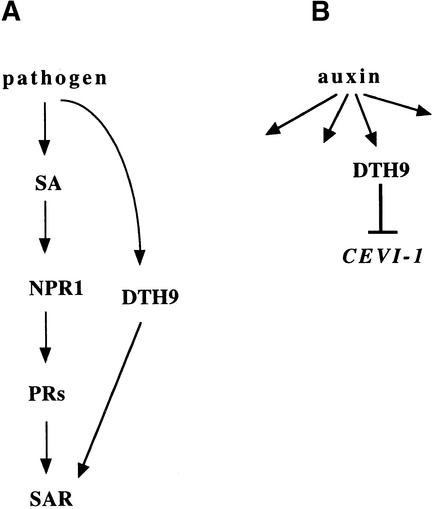
From the phenotype conferred by the dth9 mutation, we can speculate on the function of the wild-type DTH9 gene. DTH9 could function as a regulator of disease susceptibility operating upstream or independently of SA (Figure 7A). When this regulator is inactivated, the basic mechanism of resistance is blocked and, consequently, an increase in susceptibility to pathogens is primed. Such a factor could be inactivated permanently in dth9 plants, resulting in enhanced susceptibility to pathogens and the inability to mount an effective SAR response. A mutation of this type most likely would be recessive. Indeed, genetic analysis of progeny from a dth9/dth9 × DTH9/DTH9 backcross demonstrated that dth9 is a recessive mutation. Interestingly, pleiotropy is evident in the dth9 mutant, which is also insensitive to the auxin analog 2,4-D. Thus, the same DTH9 could be required for proper auxin sensitivity (Figure 7B). When this regulator is inactivated, a discrete auxin insensitivity character would be primed, leading to the expression of the CEVI-1 gene. Thus, we speculate that DTH9 could function as a regulator in both the pathogenic pathway and certain aspects of the auxin signaling pathway. This might indicate the existence of common components in certain aspects of the pathway that transduces auxin signals and that activates an effective resistance response, as suggested previously (Mayda et al., 2000).
Figure 7.
Model for Putative Roles of DTH9.
(A) DTH9 contributes in an SA-independent manner to promote an effective defense response when a pathogen is perceived.
(B) DTH9 mediates certain aspects of auxin perception.
One model does not necessarily exclude the other, and both pathways can operate in the same plant.
In conclusion, the identification of the dth9 mutant provides new information on the complex mechanism or mechanisms that control the susceptibility of plants to pathogens and adds support to the notion that a complex, interrelated network of signaling pathways has evolved in plants. Characterization of the DTH9 gene should provide further information on the elements involved in controlling the susceptibility to disease in plants.
METHODS
Plants and Growth Conditions
Arabidopsis thaliana plants were grown on soil or on plates containing Murashige and Skoog (1962) (MS) medium, as described previously (Mayda et al., 2000). The dth9 mutant was isolated in a screen for constitutive expressers of the CEVI-1::GUS reporter gene in transgenic Columbia (Col-0) plants mutagenized with ethyl methanesulfonate, as described by Mayda et al. (2000). The dth9 mutant line used in these experiments has been backcrossed twice to the wild-type parental line. Plants were grown in a growth chamber (20 to 22°C, 85% relative humidity (RH), 100 μE/m−2 sec−1 fluorescent illumination) on a 14-hr-light/10-hr-dark cycle. Unless indicated otherwise, fully expanded leaves of 4-week-old plants were used for all experiments.
To determine the response of dth9 and Col-0 parental line seedlings to 2,4-dichlorophenoxy acetic acid (2,4-D), surface-sterilized seed were germinated on MS plates supplemented with 0.2 μM 2,4-D, and the effects were monitored as described (Maher and Martindale, 1980).
Bacteria Inoculations and Quantification of Infection
Pseudomonas syringae pv maculicola ES4326 was grown at 28°C on King's B agar plates or in King's B liquid medium supplemented with 100 μg/mL streptomycin for selection. Bacteria then were collected by centrifugation, washed twice, and resuspended at 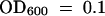 in a solution of 10 mM MgCl2. Wild-type and dth9 plants were grown on soil for 4 weeks and inoculated with the bacterial suspension at a 1:500 dilution by infiltration on the abaxial surface of leaves, using a 1-mL syringe without a needle. P. syringae pv tomato DC3000 carrying the avirulence gene AvrRpm1 was grown similarly, and selection was performed on medium containing 50 μg/mL kanamycin and 50 μg/mL rifampicin. Three leaf discs (∼100 mg) from infiltrated leaves and from 10 independent plants were excised from independent leaves at the indicated times after inoculation. The discs were divided randomly into sets of three and macerated in 10 mM MgCl2. The density of the bacterial populations was determined by plating serial dilutions on King's B medium supplemented with streptomycin (50 μg/mL) at 28°C and counting the colony-forming units (cfu). Data are reported as means and sd of the log (cfu/cm2) of at least six replicates.
in a solution of 10 mM MgCl2. Wild-type and dth9 plants were grown on soil for 4 weeks and inoculated with the bacterial suspension at a 1:500 dilution by infiltration on the abaxial surface of leaves, using a 1-mL syringe without a needle. P. syringae pv tomato DC3000 carrying the avirulence gene AvrRpm1 was grown similarly, and selection was performed on medium containing 50 μg/mL kanamycin and 50 μg/mL rifampicin. Three leaf discs (∼100 mg) from infiltrated leaves and from 10 independent plants were excised from independent leaves at the indicated times after inoculation. The discs were divided randomly into sets of three and macerated in 10 mM MgCl2. The density of the bacterial populations was determined by plating serial dilutions on King's B medium supplemented with streptomycin (50 μg/mL) at 28°C and counting the colony-forming units (cfu). Data are reported as means and sd of the log (cfu/cm2) of at least six replicates.
For chemical treatments, plants were sprayed with 1 mM salicylic acid (SA) in phosphate buffer, pH 7.1, or with buffer alone.
Inoculation with Peronospora parasitica
Peronospora isolate NOCO was transferred every week on 2- to 3-week-old Arabidopsis Col-0 plants by spray inoculation with a spore suspension. Peronospora isolate EMWA was cultivated every week on Arabidopsis Wassilewskija plants. Plants inoculated with Peronospora were kept in a 12-hr-light/12-h-dark cycle with a night temperature of 19°C for the first day and the last day of the growth cycle; plants were kept in 100% RH to ensure infection and sporulation, respectively.
For resistance experiments, wild-type parental plants and dth9 mutant plants were sprayed with a suspension of 105 conidiospores per milliliter of Peronospora isolate NOCO or EMWA and incubated as described above. Plants were examined for sporulation, or leaf samples were stained with lactophenol–trypan blue at different intervals (days) after inoculation and examined under a microscope (Koch and Slusarenko, 1990).
RNA Gel Blot Analysis
Tissue samples of four to five leaves were collected, frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted, and 5 μg of total RNA per sample was separated on a formaldehyde–agarose gel (Mayda et al., 2000). Ethidium bromide was added to each sample to allow visualization of RNA under UV light for confirmation of equal sample loading. 32P-labeled DNA probes for PR1, PR2, and PDF1.2 were prepared, and filters were hybridized as described (Mayda et al., 2000).
Genetic Analysis
Crosses were performed by dissecting and emasculating unopened buds and then using the pistils as recipients for pollen from five opened flowers. Backcrosses with the parental CEVI-1::GUS transgenic line were performed by using CEVI-1::GUS plants as the pollen donor. The reciprocal crosses also were performed. F1 and F2 plants were grown on MS plates and tested for β-glucuronidase (GUS) activity as described previously (Mayda et al., 2000). Segregation of constitutive GUS activity in the F2 generation was analyzed with the χ2 test for goodness of fit.
Polymerase Chain Reaction–Based Mapping
A dth9 plant (in the Col-0 background) was crossed with a Landsberg erecta (Ler) plant, and the progeny that segregated dth9 homozygous mutants when selfed were used for mapping. Thirty-five seedlings in the F2 population were selected for DNA extraction, and recombinant seedlings were identified by using simple sequence length polymorphism (SSLP) markers according to the protocol described by Bell and Ecker (1994) as well as new markers reported on the Arabidopsis database World Wide Web site (http://genome-www.stanford.edu).
Determination of Endogenous Amounts of SA and Camalexin
To determine how much free and conjugated SA was present, we infected leaves of 4-week-old wild-type plants and dth9 plants with P. s. maculicola ES4326 or mock-inoculated them with 10 mM MgCl2 as described above. Four days after inoculation, samples were collected (1 g of tissue per sample from six plants) and frozen in liquid nitrogen. SA and salicylate glucoside (SAG) were determined as described previously (Mayda et al., 1999).
For analysis of camalexin, leaves of 4-week-old wild-type plants and dth9 plants were vacuum-infiltrated for 30 sec with a suspension (in 10 mM MgCl2 ) of P. s. tomato DC3000 at a concentration of 105 cfu/mL. To achieve better penetration, we added 10 μL/100 mL Silwet L-77. The control plants were infiltrated only with MgCl2 containing Silwet L-77. Camalexin amounts were determined 2 days after infection according to Nawrath and Métraux (1999).
Acknowledgments
We thank M.D. Comín for taking care of the plants and C. Marques for creating some of the figures. We also thank Dr. J. Dangl for providing the bacterial strains used in these experiments and Dr. P. Tornero for helpful discussions. We acknowledge the financial support of the Spanish Ministry of Science and Education to P.V.
References
- Agrios, G.N. (1988). Plant Pathology. (London: Academic Press).]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6–1 mutant. Plant Cell 10, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Delaney, T., Friedrich, L., and Ryals, J. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced systemic acquired resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance responses. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negretto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., and Ausubel, F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91, 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 7, 551–563. [DOI] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher, E.P., and Martindale, S.J.B. (1980). Mutants of Arabidopsis thaliana with altered responses to auxin and gravity. Biochem. Genet. 18, 1041–1053. [DOI] [PubMed] [Google Scholar]
- Mayda, E., Tornero, P., Conejero, V., and Vera, P. (1999). A tomato homeobox gene (HD-Zip) is involved in limiting the spread of programmed cell death. Plant J. 20, 591–600. [DOI] [PubMed] [Google Scholar]
- Mayda, E., Marqués, C., Conejero, V., and Vera, P. (2000). Expression of a pathogen-induced gene can be mimicked by auxin insensitivity. Mol. Plant-Microbe Interact. 13, 23–31. [DOI] [PubMed] [Google Scholar]
- Morel, J.-B., and Dangl, J.L. (1999). Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics 151, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nawrath, C., and Métraux, J.-P. (1999). Salicylic acid induction–deficient mutants of Arabidopsis express PR2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Métraux, J.-P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathway is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, E.E., and Ausubel, F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR1 gene expression. Plant Cell 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A.F. (1961). Systemic acquired resistance induced by localized virus infections in plants. Virology 14, 340–358. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid–insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ss1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher, L., Mauch-Mani, B., and Métraux, J.-P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35, 235–270. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Nelissen, I., Eggermont, K., and Broekaert, W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19, 163–171. [DOI] [PubMed] [Google Scholar]
- Volko, S.M., Boller, T., and Ausubel, F.M. (1998). Isolation of new Arabidopsis mutants with enhanced disease susceptibility to Pseudomonas syringae by direct screening. Genetics 149, 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, I.-C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease response resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]