Abstract
The recessive nuclear vdl (for variegated and distorted leaf) mutant of tobacco was obtained by T-DNA insertion and characterized by variegated leaves and abnormal roots and flowers. Affected leaf tissues were white and distorted, lacked palisadic cells, and contained undifferentiated plastids. The variegation was due to phenotypic, rather than genetic, instability. Genomic and cDNA clones were obtained for both the mutant and wild-type VDL alleles. Three transcripts, resulting from alternate intron splicing or polyadenylation, were found for the wild type. The transcripts potentially encode a set of proteins (53, 19, and 15 kD) sharing the same N-terminal region that contains a chloroplast transit peptide capable of importing the green fluorescent protein into chloroplasts. The predicted 53-kD product belongs to the DEAD box RNA helicase family. In the homozygous vdl mutant, T-DNA insertion resulted in accumulation of the shortest transcript and the absence of the RNA helicase–encoding transcript. Genetic transformation of the homozygous mutant by the 53-kD product–encoding cDNA fully restored the wild-type phenotype. These data suggest that a plastid RNA helicase controls early plastid differentiation and plant morphogenesis.
INTRODUCTION
Compared with other eukaryotic cells, plant cells contain a unique class of organelles, plastids, that differentiate according to developmental stage and cell localization. The function of each type of plastid (chloroplasts, amyloplasts, and chromoplasts) is correlated with the particular physiological role of the cell type and is accompanied by modification of plastid morphology and enzymatic machinery. All plastids differentiate from the undifferentiated proplastids in meristems. In green tissues, chloroplasts are the major form of plastids; they perform not only photosynthesis but also other metabolic processes, such as synthesis of lipids, amino acids, and hormones. Chloroplast development from undifferentiated proplastids occurs by a series of biological processes involving a large number of proteins. Because of the limited coding capacity of the chloroplast genome, most chloroplast proteins are encoded in the nucleus. The biogenesis of photosynthetic complexes and thylakoid assembly during chloroplast development, therefore, are regulated by coordination between the chloroplast and nuclear genomes (Mullet, 1988; Taylor, 1989; Susek et al., 1993; Surpin and Chory, 1997; Somanchi and Mayfield, 1999).
The identification and characterization of nuclear mutants affecting chloroplasts or other plastids constitute an interesting approach to identifying the nuclear genes required for plastid biogenesis and function and to understanding nucleus–plastid interactions. Many genes involved in light perception and signaling have been identified (reviewed in Goldschmidt-Clermont, 1998; Khurana et al., 1998). If we consider genes that encode chloroplast proteins, several nuclear mutants that affect chloroplast development have been subjected to molecular analysis (see, e.g., Han et al., 1992; Keddie et al., 1996; Sundberg et al., 1997; Carol et al., 1999; Wu et al., 1999). However, most nuclear genes that encode chloroplast proteins, especially regulatory proteins essential for the early development of plastids, remain uncharacterized.
Plastid biogenesis is tightly coupled with temporal and spatial stages of plant development. Chloroplast development and leaf differentiation are both stimulated by light and are initiated in a coordinated fashion. However, certain mutants in which the light transduction pathway is blocked develop leaves in the dark in the absence of chloroplast development (Cabrera y Poch et al., 1993; Li et al., 1994), which suggests that these coordinated developmental processes can be separated. On the other hand, considerable evidence indicates that chloroplast development also regulates leaf differentiation, especially mesophyll morphogenesis. When chloroplast development is blocked at an early stage, formation of the palisade layer is also affected (Hudson et al., 1993; Reiter et al., 1994; Mandel et al., 1996), suggesting that a signal produced by the chloroplast during the early developmental stage affects leaf differentiation, particularly the full expansion of mesophyll cells (Surpin and Chory, 1997; Goldschmidt-Clermont, 1998). The isolation and characterization of mutations affecting chloroplast and leaf differentiation, therefore, constitute an interesting approach to understanding the early mechanisms involved in these developmental steps.
In this article, we report the identification of a tobacco T-DNA insertion mutant named vdl (for variegated and distorted leaf) that affects development of the leaves, flowers, and roots. The VDL locus encodes a putative chloroplast RNA helicase belonging to the DEAD box superfamily. These enzymes, which are present in both prokaryotes and eukaryotes, play important roles post-transcription, notably in cell growth and differentiation (Luking et al., 1998; De la Cruz et al., 1999). The possible role of this RNA helicase homolog in plastid differentiation and plant development is discussed.
RESULTS
Identification of the vdl Mutation
The vdl mutant was originally identified in distorted and variegated plants that appeared in the self-pollinated F1 progeny of a transgenic tobacco plant (E6A) obtained by transformation with an Agrobacterium T-DNA–derived vector containing the yeast nuclear gene MIP1, which encodes a mitochondrial DNA polymerase. Because E6A was the only transgenic plant to give rise to progeny with this phenotype, we suspected that the phenotype resulted from T-DNA insertion.
When germinated in vitro, the self-pollinated F1 progeny of E6A segregated into normal green seedlings and slow-growing, distorted seedlings characterized by albino or variegated cotyledons and leaves (Figure 1A). Later, the extent of distortion and variegation varied markedly (see Figures 1B to 1D for details). Green sectors or spots usually stood out, suggesting better cell proliferation in these areas. The root growth of the mutants was severely limited, even in plants with leaves that were almost completely green (Figure 1E).
Figure 1.
Phenotype of the vdl Mutant.
(A) A vdl seedling grown in vitro in Murashige and Skoog (MS) agar medium (see Methods).
(B) Four vdl plants grown in vitro and showing various phenotypes. The seedlings (5 weeks old) were grown in MS agar medium.
(C) and (D) Details of a vdl plant grown in vitro.
(E) Wild-type (left) and vdl (right) plants grown in vitro (4 weeks old).
(F) Leaves from various vdl plants grown in soil, showing various phenotypes.
(G) Wild-type (1 month old, at left) and vdl (2 months old, at right) plants grown in soil.
(H) Detail of a narrow leaf of a vdl plant similar to that shown in (G).
(I) Flower bud from wild-type (left) and vdl (right) plants.
(J) Flower from wild-type (left) and vdl (right) plants.
(K) Development of an axillary bud on green stem sections derived from wild-type (left) and vdl (right) plants and transferred for 2 weeks to MS agar medium.
(L) Leaf discs from wild-type (left) and vdl (right) green leaves transferred to an MS agar medium for shoot regeneration.
(M) A transgenic line (T6-5-8) resulting from the transformation of a vdl homozygous plant with a wild-type VDL-1 cDNA clone was grown until the flowering stage and then self-pollinated; seed was then sown on a growth medium without antibiotics. The photograph was taken 5 weeks after sowing.
In soil-grown plants, the phenotype varied according to the plant and the leaf (examples are shown in Figure 1F). Some albino leaves developed a main vein of normal length with very narrow and curled lamina (Figures 1G and 1H). Severely affected plants grew extremely slowly in soil, and most died before flowering; the survivors often had albino flower buds (Figure 1I) that fell off before opening. A few flowers developed variegated and distorted petals (Figure 1J) and had very poor male fertility. F1 seed could be obtained by pollinating with wild-type pollen, yielding germinated F1 seedlings that were 100% normal plants. In a few cases, self-fertilized seed was obtained from less-affected flowers; however, the germination rate was very low and the seedlings all displayed a mutant phenotype.
Cell Morphology
Closer inspection of the leaf morphology of the vdl mutant showed that the palisade layer was present in green sectors (Figure 2A) but absent from white sectors (Figure 2B). In narrow and curled leaves, spongy cells were irregularly shaped and the lamina thickness was variable (Figure 2C). The correlation between the absence of palisadic structure and albino phenotype was clear at junctions between green and albino sectors (Figure 2D) or at a white spot within a green sector (Figure 2E). Root structure, too, was disorganized (Fig-ures 2F and 2G).
Figure 2.
Light Microscopy Analysis of Wild-Type and Homozygous vdl Leaves and Roots.
(A) Cross-section from a green sector of a vdl leaf.
(B) Cross-section from a white sector of a vdl leaf.
(C) Cross-section from a curled vdl leaf similar to that shown in Figure 1H.
(D) Cross-section from a vdl leaf at a junction between a green (left) or white (right) sector.
(E) Cross-section from a white spot within a green sector of a vdl leaf.
(F) Cross-section from a vdl root. The epidermis and the endodermis are not well identified, and the vascular cylinder has a disorganized structure.
(G) Cross-section from a wild-type root.
 .
.
Electron microscopic analysis of cells from white leaf sectors (Figure 3 B) showed undeveloped plastids that lacked thylakoids and had vesiculated internal membranes. In green sectors, the chloroplasts were just as well developed as they were in the wild type (cf. Figures 3A and 3C). These data suggest that the vdl mutation affects both plastid differentiation and organ morphogenesis.
Figure 3.
Transmission Electron Micrographs of vdl Leaf Cells.
(A) Green sector of a vdl leaf. The plastid structure is similar to that found for the wild type shown in (C).
(B) White sector of a vdl leaf. Plastids show a vesicular instead of lamellar structure.
(C) Wild-type leaf. Chloroplasts show the typical stacking thylakoids.
 (A)
(A)  (A)
(A)  (C).
(C).
The vdl Mutation Is Genetically Stable
A somatic unstable phenotype is often associated with nonMendelian mutation or transposon-generated nuclear mutation. Although difficult to obtain, the self-fertilized progeny of vdl mutant plants were 100% mutant, whereas the progeny of vdl mutant plants crossed with wild-type tobacco were 100% normal (data not shown). In the self-fertilized offspring of the latter cross, mutant and normal plants typically were observed in a ratio of 214:750 (for example), which is close to the theoretical 1:3 ratio expected for a single recessive nuclear mutation. This was confirmed in subsequent crosses (data not shown).
Genetic instability of the mutation, for example, as induced by transposon excision, could be ruled out, because normal plants were never found in the self-fertilized progeny of a homozygous mutant when seed was obtained. In addition, when stem sections obtained from a phenotypically normal area of a mutant plant were grown in vitro, the new leaves that developed from the axillary bud adjacent to a normal leaf were always albino with green edges (Figure 1K). Finally, when placed on in vitro regeneration medium, leaf discs from mutant plants produced variegated or albino shootlets, regardless of whether the discs came from normal-looking green leaves (Figure 1L), green sectors of albino leaves, or albino leaves. Therefore, these data show that the vdl mutation is genetically stable even though its expression in the phenotype is variable.
Cloning and Characterization of the vdl Locus and Its Transcripts
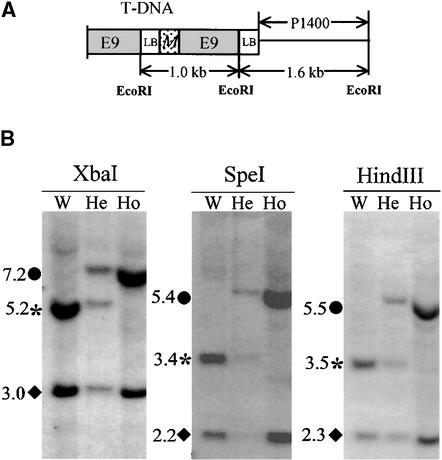
To demonstrate that the vdl mutation was induced by T-DNA insertion, DNA isolated from 36 plants produced by a cross between heterozygous vdl and wild-type (+) seedlings (18 +/+ and 18 +/vdl plants, as deduced from their self-crossed progeny) was digested with EcoRI or XbaI and subjected to DNA gel blotting with a T-DNA left border fragment as probe. One 7.2-kb XbaI fragment and two EcoRI fragments (1.6 and 1.0 kb) cosegregated with plants producing the mutant phenotype in their self-fertilized progeny (data not shown).
Specific primers corresponding to the T-DNA left border were designed to amplify the T-DNA flanking region by using inverse polymerase chain reaction (PCR). Cloning and sequencing showed that the 1.6- and 1.0-kb EcoRI fragments were adjacent to one another and were included within the 7.2-kb XbaI fragment (Figure 4A). The 1.0-kb fragment contained a partial T-DNA left border, the end of the MIP1 gene, and the rbcS E9 terminator sequence present in the original construct (Figure 4A). The 1.6-kb fragment contained a 0.2-kb T-DNA left border sequence and an unknown 1.4-kb sequence (P1400 in Figure 4A), presumably derived from the tobacco genome. DNA gel blotting of tobacco genomic DNA using this 1.4-kb sequence as the probe confirmed that the latter was included within the previously identified cosegregating 7.2-kb XbaI band and also within novel 5.2- and 3.0-kb XbaI (Figure 4B) fragments. Additional DNA gel blotting studies of heterozygous and homozygous vdl mutant and wild-type plants with SpeI and HindIII enzymes confirmed that wild-type tobacco, an amphitetraploid species, contained two VDL copies (Figure 4B). The VDL locus was designated as that disrupted by T-DNA, and VDL′ was the presumed locus from the other tobacco parental genome.
Figure 4.
A T-DNA−Flanking Genomic Fragment Cosegregates with the vdl Mutation.
(A) Structure of the cosegregating T-DNA insert. The recombined T-DNA insert consisted of two copies of the T-DNA left border (LB), part of the MIP gene (M), and the E9 terminator. The cloned 1.6- and 1.0-kb EcoRI T-DNA fragments and the 1.4-kb genomic fragment (P1400) are indicated.
(B) Genomic DNA was extracted from wild-type (W), heterozygous (He), and homozygous (Ho) vdl plants and digested with the indicated restriction enzymes. After transfer, the DNA was hybridized with the primer-generated probe P1400. Circles indicate the vdl locus, asterisks indicate the wild-type VDL locus, and diamonds indicate the VDL′ locus.
A series of overlapping genomic fragments that included the VDL and VDL′ loci was cloned by successive inverse PCR amplifications and sequenced. Finally, the full genomic sequence was obtained by direct PCR for the VDL locus. The sequences of both loci were very similar, except for additional 1.3- and 2.6-kb fragments present in VDL introns 3 and 9, respectively, and complete divergence in the 5′ region (shown as hatched for VDL′ in Figure 5A). The T-DNA insert was incomplete as already suggested (Figure 4A).
Figure 5.
Genomic and cDNA Organization of the VDL and VDL′ Loci.
(A) Genomic map of the VDL and VDL′ loci showing exons (numbered black boxes), introns (open boxes), and the location of the T-DNA insert. EcoRI (E), HindIII (H), and XbaI (X) restriction sites are indicated. Because of the considerable difference in the 5′ untranslated region, the first exons of the VDL and VDL′ loci are named 1 and 1′. The hatched region in VDL′ represents an upstream sequence that is completely different from that in VDL. The size of VDL introns 3 and 9 is based on restriction mapping (some gaps in the sequence remain). The sequence downstream of VDL′ exon 10 has not been cloned.
(B) Alternative splicing products of the VDL and VDL′ loci. The filled boxes indicate exons. The asterisks in front of exons 5, 6, and 11 in VDL-6, VDL-7, and VDL-8, indicate that the 5′ or 3′ borders differed from those in the other cDNA clones, resulting in shorter transcripts; the exact sequence can be found in the databases.
A database search showed that two sequences had high homology with DEAD box RNA helicases (discussed in detail below). Corresponding cDNA clones were obtained from wild-type RNA by 5′ and 3′ rapid amplification of cDNA ends and direct reverse transcriptase (RT)–PCR with VDL- or VDL′-specific primers. cDNA clones corresponding to three partially overlapping transcripts were obtained for the VDL and VDL′ loci (Figure 5B), all of which contained the same 5′ region but differed in their 3′ region as a result of alternate polyadenylation or intron splicing. VDL-1 and VDL′-1 represented the longest transcripts and were assembled from 10 exons. In VDL-2 and VDL′-2, an alternative polyadenylation site terminated the transcript after exon 2 (2b in Figure 5B). In VDL-3 and VDL′-3, alternative splicing placed exon 3 (which terminates with a polyadenylation site), rather than exon 4, next to exon 2. For the VDL′ locus, cDNA clones for five additional transcripts (VDL′-4 to VDL′-8) were found. These corresponded to VDL′-1, except that some internal exons were absent, and in VDL′-6, VDL′-7, and VDL′-8, the splicing sites were slightly displaced for exons 5, 6, or 11, which were therefore shorter (Figure 5B). RT-PCR with primers corresponding to the untranslated 5′ region of exon 1 and the last exon (11) confirmed that a single band (corresponding to VDL-1) was found for VDL and that at least five bands were found for VDL′ (corresponding to VDL′-1 and VDL′-4 to VDL′-8; Figure 6A).
Figure 6.
RT-PCR Transcript Analysis of the VDL and VDL′ Loci.
(A) Poly(A)+ RNA from wild-type leaves was amplified by RT-PCR. The 5′ primers were located in the 5′ untranslated region and were specific to either VDL (pTA5*A) or VDL′ (pTABis5*A); the 3′ primers were common to VDL and VDL′ and were located in exon 11 (pTFL65B).
(B) Poly(A)+ RNA from wild-type (W), heterozygous (He), and homozygous (Ho) (white sections) mutant leaves was amplified by RT-PCR. The 5′ primers were as in (A); the 3′ primers were common to VDL and VDL′ and corresponded to exon 11 (pTFL57), exon 2b (pRH14), or exon 3 (pRH11).
(C) Poly(A)+ RNA from wild-type flower (F), leaf (L), or root (R) was amplified by nested RT-PCR with, for VDL, primers pTA5A and pTFL50 (5′) and pTFL57 (3′), and for VDL′, primers pTABis5A (5′) and pTFL65B and pTFL57 (3′). Note that, to simplify the pattern, the 3′ primer used here for exon 11 was chosen to cover a region not present in VDL′-6, VDL′-7, and VDL′-8, which have a shorter exon 11 because of their alternative splicing.
The numbers at left and right denote molecular mass markers (kilobases).
In the vdl Mutant, VDL-2 Transcript Accumulates, whereas VDL-1 and VDL-3 Transcripts Are Undetectable
In the vdl mutant, the T-DNA was inserted into intron 2. DNA gel blot analysis of genomic DNA cut independently with three restriction enzymes confirmed that the size of the restriction fragment corresponding to the VDL locus was increased by ∼2 kb, the size of the truncated T-DNA (Figure 4C). As expected, the heterozygous plant contained both the wild-type and T-DNA–inserted forms. In addition, a common band (e.g., 3 kb with XbaI) was found for the three plants and corresponded to the VDL′ locus.
Because the T-DNA sequence contains a copy of the rbcS E9 transcription terminator, the vdl transcript would be expected to terminate prematurely. To determine the effect of T-DNA insertion on the transcript profile, we performed RT-PCR analysis with a VDL-specific 5′ primer localized within the untranslated region of exon 1 and with 3′ primers corresponding to exons 11, 3, and 2b (VDL-1, VDL-3, and VDL-2, respectively). RT-PCR was chosen rather than RNA gel blotting because the strong similarity of the VDL and VDL′ sequences and the short untranslated sequences made it difficult to design probes specific enough for RNA gel blotting.
Using leaf RNA and the 3′ primer corresponding to exon 11, 3, or 2b, we obtained single bands for the wild type and the heterozygous vdl mutant (Figure 6B). These bands corresponded to VDL-1, VDL-3, and VDL-2, respectively. For the homozygous vdl mutant, no signal was found with the exon 11 (VDL-1) or exon 3 (VDL-3) primer, but a band was obtained with the exon 2b (VDL-2) primer. This band was stronger in the heterozygous and homozygous mutants but not for the VDL′ locus (Figure 6B). These data are in agreement with transcript termination within the T-DNA and polyadenylation at the VDL site downstream of exon 2b, leading to accumulation of the VDL-2 transcript.
Because roots or flowers were also affected in vdl plants, we examined whether transcripts corresponding to VDL-1 and VDL′-1 were expressed in those organs. A band corresponding to the full-length VDL transcript was indeed seen in all three organs, whereas signals for VDL′ were seen only in leaves (Figure 6C).
VDL and VDL′ Loci Encode a Group of Putative Proteins with the Same N-Terminal Region, the Longest Product Being a Putative RNA Helicase
The VDL-initiating translation codon was tentatively identified 86 (VDL) or 104 (VDL′) nucleotides downstream of the 5′ end of the longest cDNA clone. It is in a good initiating context (CCCATGG), is the beginning of a reading frame of 466 codons, and is preceded by an in-frame stop codon in VDL. Comparing VDL and VDL′ shows that the upstream sequence strongly diverges, whereas the downstream coding sequence is >98% identical.
The predicted VDL-1 or VDL′-1 protein (53 kD) is thought to be a putative DEAD box RNA helicase containing all of the conserved motifs (Figure 7). The proteins that show the greatest similarity with VDL-1 are DEAD box RNA helicases, which have been identified in several bacterial and animal species and are involved in various post-transcriptional steps in gene expression. The Arabidopsis genome project recently released a chromosome V contig that contains a sequence closely related to vdl-1 (67% identity between the predicted mature proteins).
Figure 7.
Alignment of the VDL-1/VDL′ -1 Protein Sequence with Sequences of Similar RNA Helicases.
Sequence alignment showing the conserved blocks and the consensus sequence of the aligned proteins. The seven motifs (I, Ia, II, III, IV, V, and VI) of DEAD box RNA helicases are indicated by asterisks. A conserved region of unknown function between motifs Ia and II is also indicated. Boldface letters indicate amino acids that are 100% conserved. Amino acid residues of the consensus sequence are in lowercase when conserved in at least 70% of the proteins displayed or in uppercase when they are 100% conserved. The number symbol (#) indicates the hydrophobic amino acid Met, Ile, Leu, or Val. aa, amino acids.
VDL-2/VDL′-2 and VDL-3/VDL′-3 consisted of 132 and 168 amino acids, respectively, including the same 112 N-terminal residues seen in VDL-1/VDL′-1. No homology with the C-terminal sequences specific to VDL2 or VDL3 was found in the databases.
VDL′-4 contained the seven RNA helicase motifs but lacked a stretch of nonconserved sequence between motifs Ia and II. VDL′-5, VDL′-6, and VDL′-8 contained some of the RNA helicase motifs, whereas VDL′-7 contained none because of a frameshift (data not shown).
The VDL N-Terminal Region Contains a Chloroplast Transit Peptide
The strong similarity between VDL-1 or VDL′-1 and RNA helicases starts at residue 74. The more upstream coding region, common to all predicted VDL or VDL′ products, has several of the features of plastid transit peptides (Von Heijne et al., 1989) because it is rich in Ser (13%) and Thr (16%) and has a very low content of Tyr, Asp, and Glu. This was further supported by a computer-based program (ChloroP; available at http://www.cbs.dtu.dk/services/), which predicted that the VDL N-terminal region contains a transit peptide with a cleavage site between residues 53 and 54 (with the typical Ala and Val at −1 and −3, respectively). To confirm this hypothesis experimentally, a translational fusion between the sequence encoding the first 73 N-terminal residues of VDL-1 and the green fluorescent protein (GFP) sequence was linked to the cauliflower mosaic virus (CaMV) 35S transcription promoter and electroporated into tobacco protoplasts for transient expression. The fusion protein clearly colocalized with the chloroplast chlorophyll, whereas a control GFP without transit peptide was detected in the cytosol (Figure 8).
Figure 8.
Predicted VDL Transit Peptide Imports the GFP into Chloroplasts.
The sequence coding for the first 73 amino acid residues of VDL-1 (VDL73) was inserted between the CaMV 35S promoter and the GFP sequence to give 35S-VDL73-GFP in a bacterial plasmid (see Methods). A 35S-GFP construct without the VDL sequence was used as a control. Both plasmids were electroporated into tobacco leaf protoplasts, and the fluorescence was analyzed after 24 hr with a confocal microscope.
(A) GFP fluorescence in a protoplast transformed with 35S-VDL73-GFP.
(B) Chlorophyll fluorescence of the protoplast shown in (A).
(C) Merging of (A) and (B).
(D) GFP fluorescence in a protoplast transformed with the control 35S-GFP.
Ectopic Expression of VDL-1 Restores a Wild-Type Phenotype
To prove that the defective phenotype is a direct consequence of the modified VDL-1 locus, we transformed homozygous vdl plant leaf discs with a cDNA clone corresponding to the VDL-1 transcript placed under the control of the CaMV 35S transcription promoter. A β-glucuronidase construct was used as a control. After transformation of the vdl mutant with the control construct and selection with hygromycin, only white and variegated calli and shoots developed, and those only slowly. However, in the VDL-1 construct, the majority of shoots were green and developed into normal plants (data not shown). When these plants were self-crossed, their progeny segregated into normal and variegated plants, showing Mendelian segregation (one, two, or more copies, depending on the transformant) of the complementing VDL-1 transgene. For example, plant T6-5-8 (Figure 1 M) gave 166 normal green plants and 65 small variegated plants, indicating a single locus for the ectopic VDL-1.
DISCUSSION
The tobacco vdl mutation has been identified within a gene that presumably encodes a chloroplast RNA helicase. The mutant leaves are variegated, the albino sectors are distorted and devoid of palisade parenchyma, and severely affected leaves are very narrow; however, the lamina is always present. The vdl mutant therefore differs from the phantastica mutant, which is characterized by a lack of lateral expansion (Waites and Hudson, 1995). Subcellular analysis showed that the plastids of albino cells do not develop stalked membranes. This correlation between defective chloroplast development and leaf morphogenesis supports a previous suggestion that a signal produced at the very early developmental stage of chloroplasts stimulates leaf palisade division and expansion (Reiter et al., 1994; Wetzel et al., 1994; Chatterjee et al., 1996; Keddie et al., 1996).
VDL-1, the product encoded by the largest VDL transcript identified, belongs to the DEAD box RNA helicase family. These ATP-dependent enzymes unwind RNA:RNA or RNA:DNA duplexes and are involved in several important biological processes in the nucleus, cytosol, and mitochondria, including RNA stability, processing, editing, export from the nucleus, and translation. Some of these enzymes are involved in the regulation of cell growth and cell differentiation (reviewed in Luking et al., 1998; De la Cruz et al., 1999). Because we also demonstrated that the N-terminal region of VDL-1 contains a functional chloroplast transit peptide, this enzyme is predicted to be a chloroplast DEAD box RNA helicase.
The primary structure of DEAD box RNA helicases is well conserved, especially within the seven motifs (Figure 7). Of these, motifs I, II, and VI are the most conserved and have been identified as ATP binding and nucleic acid binding sites. The function of the other four motifs remains unclear, although motif III might be involved in helicase activity. The lengths and sequences of the N- and C-terminal regions and the regions between the motifs differ between family members. These regions are believed either to have distinct biochemical activities that combine with the helicase activity to generate new enzyme activities or to be involved in RNA substrate specificity, which must be closely related to the biological function of the respective helicases (Bird et al., 1998; Eisen and Lucchesi, 1998). Thus, at this time, assigning a specific function to VDL-1 is difficult. An Arabidopsis open reading frame is very closely related to VDL-1. However, this putative gene was identified within sequences only recently generated by the Arabidopsis genome project and therefore does not give any clue about VDL-1 function.
The proteins with the greatest identity to VDL-1 were DEAD box proteins involved in post-transcription and translation regulation in other organisms (Figure 7). In Escherichia coli, the DEAD box protein can suppress a defective mutant in ribosomal S2 protein (Toone et al., 1991), SRMB can suppress a mutant defective in 50S ribosomal subunit assembly and may interact with 23S RNA (Nishi et al., 1988), DBPA can unwind both 23S and 16S RNA and may play a role in the assembly of the active center of 50S ribosomal subunits (Boddeker et al., 1997), and RHLB plays a role in postreplication repair, RNA processing, and mRNA degradation in the RNA degradosome (Py et al., 1996). In yeast, DED1 is involved in initiating translation (Iost et al., 1999). The mouse translation initiation factor, eIF4A, is involved in protein synthesis initiation and is required for mRNA binding to ribosomes and for cap recognition (Nielsen et al., 1985). The Drosophila vasa protein is important in oocyte formation and in specification of the posterior structures of the embryo (Hay et al., 1988; Styhler et al., 1998). The human growth-regulated nuclear 68 protein is involved in organ differentiation/maturation in the fetus (Stevenson et al., 1998). This survey clearly shows that DEAD box proteins are involved in various post-transcriptional steps and that some of them govern developmental processes.
Given the prokaryotic origin of plastids, we were not surprised to find substantial identity between VDL-1 and bacterial DEAD box proteins. However, VDL-1 is not closely related to a recently identified mitochondrial DEAD box RNA helicase (Gagliardi et al., 1999). The observation that VDL is expressed in nonphotosynthetic tissues and that its disruption also affects roots and flowers indicates that the encoded product is involved in the differentiation of all plastids. Transcriptional and post-transcriptional regulation of these organelles is complex (Rochaix, 1996; Sugita and Sugiura, 1996; Stern et al., 1997). During the conversion of proplastids into chloroplasts and other plastids, the gene expression machinery undergoes an important change, notably an increased synthesis of proteins involved in the transcription/translation apparatus (discussed in Goldschmidt-Clermont, 1998). For instance, chloroplasts contain a multisubunit complex that controls the amounts of specific RNAs at steady state (Hayes et al., 1999). A similar complex, called RNA degradosome, has been identified in E. coli and contains an RNA helicase presumably involved in unwinding structured RNA (Carpousis et al., 1999). We therefore propose that VDL-1 is involved in a general process of the plastid expression machinery. Clearly, a careful examination of the expression of several plastid genes and nuclear genes that encode plastid proteins in the vdl mutant is required to determine the stage at which VDL-1 plays a role.
The VDL gene is alternatively spliced. Two additional transcripts identified potentially encode proteins containing the same N-terminal chloroplast transit peptide as VDL-1. However, their predicted mature regions are short (13 and 8 kD, respectively), and the lack of homology with any database sequences makes it difficult to assign a possible function to these proteins. These same transcripts were found for VDL′, the VDL homolog presumably belonging to the second genome of tobacco (an amphitetraploid), but five additional alternative transcripts were also identified. These are thought to encode polypeptides containing some or all of the seven RNA helicase motifs; VDL′-4 contains all seven motifs, whereas VDL′-5 and VDL′-6 contain the most conserved motifs (motifs I, Ia, II, and VI), which provide the ATP binding and nucleic acid binding sites. Whether these all function as RNA helicases is difficult to predict at this time. The sequence polymorphism created by the alternative splicing might provide a means of altering substrate specificity or function. Oligomeric forms, generally dimers or hexamers, have been described for certain DNA helicases (Chao and Lohman, 1991; Amaratunga and Lohman, 1993) and are thought to provide the multiple nucleic acid binding sites necessary for helicase function. The crystal structure of RNA helicase from genotype 1b hepatitis C virus also suggests a putative functional dimer model (Cho et al., 1998). Some DEAD box proteins require accessory proteins to function in vivo (Rozen et al., 1990; Gibson and Thompson, 1994; Sommerville, 1999). A more exhaustive search for all alternative VDL′ transcripts at various developmental stages is required to gain more insight into the biological meaning of this finding.
Complementation of vdl disruption by ectopic expression of the longest VDL cDNA clearly indicated that the mutant phenotype results directly from the absence of the VDL-1 product rather than from the absence of VDL-3 or the increased expression of VDL-2. Although the VDL′ locus is also transcribed into a VDL-1–like transcript, this cannot fully substitute for VDL-1. Two differences between VDL and VDL′ might explain this observation. First, as mentioned above, perhaps the multiple VDL′ alternative splicing events undergo spatial or temporal regulation; therefore, the concentration of the longest transcript may be too low to sustain synthesis of the required amount of RNA helicase. Second, the region that starts 67 nucleotides upstream of the presumed translation initiation codon shows complete divergence, probably as a result of a recombination event; thus the two loci are expected to be transcribed differently, resulting in no overlap or only a partial overlap of expression. This is clearly the case in roots and flowers, in which no transcript corresponding to VDL′-1 was found by RT-PCR. In leaves, although VDL′-1 transcripts were found, expression of VDL and VDL′ does not necessarily overlap in all cell types and at all developmental stages. To determine whether this is the case would require careful examination of the expression of the two loci at the cellular level during development.
Although the variegation of several mutants with defective chloroplasts has been linked to either cytoplasmic mutation or transposon excision from a nuclear gene (Reidi, 1973; Martinez-Zapater et al., 1992; Martienssen and Baron, 1994; Chatterjee and Martin, 1997), we were able to exclude these possibilities in the case of vdl. The vdl mutation showed clear Mendelian inheritance, and plants regenerated from phenotypically reverted sectors always showed a mutant phenotype and gave mutant progeny. From the pattern of variegation, the albino phenotype apparently can appear at any developmental stage and at various places within a leaf. It affects all the cells across a section or only cells within some layers. Phenotypic reversion can also occur, as shown by tiny green spots appearing in large white areas. Other cases of variegated mutants unrelated to transposon activity have been described. This is the case, for instance, for the tomato ghost mutant (Scolnik et al., 1987) and the immutans mutant, a yellow variegated mutant in Arabidopsis (Carol et al., 1999; Chen et al., 1999; Wu et al., 1999). In no case has the variegation been explained in molecular terms, but a threshold hypothesis is often suggested. In accordance with the discussion above of the differences between VDL and VDL′, we suggest that VDL′ does not fully complement VDL, either because the exchanged transcription promoter region does not support correct expression or because possible regulation of the alternative splicing does not provide enough full-length transcripts. In those tissues or cells in which the required extent of VDL function is not reached, alteration of plastid gene expression might contribute, in a snowball effect, to further jamming of the expression machinery and finally lead to developmental arrest.
METHODS
Plant Materials
The vdl mutant was originally found in a population of transgenic tobacco (Nicotiana tabacum) plants expressing the yeast nuclear gene MIP1 encoding a mitochondrial DNA polymerase linked to the cauliflower mosaic virus (CaMV) 35S transcription promoter (Grec et al., 2000). The E6A transformant was chosen for this study because of the variegation phenotype in its F1 progeny. The mutant was propagated at the heterozygous stage in the greenhouse (16 hr of light at 25°C). The self-fertilized progeny of heterozygous plants were maintained in vitro (16 hr of light at 25°C) in Murashige and Skoog (MS) agar (Murashige and Skoog, 1962): MS salts, 3% sucrose, and 0.7% agar.
T-DNA Cosegregation Analysis
Total DNA was extracted as described by Rogers and Bendich (1994). Restricted DNA (10 μg) was electrophoresed on a 0.7% agarose gel and transferred to a nylon membrane (Hybond+; Amersham Corp.), which was prehybridized for 3 hr, then hybridized overnight at 42°C in 5 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate), 2 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 1% SDS, 330 μg/mL salmon sperm DNA, 0.5 mM EDTA, pH 8.0, and 50% formamide. 32P-labeled probes were prepared as follows. An 0.8-kb EcoRI/NruI fragment containing a 0.2-kb T-DNA left border region and a 0.6-kb flanking vector sequence was used for EcoRI-digested samples, whereas for XbaI-digested samples, a 1.4-kb XbaI/NruI fragment containing a 0.2-kb T-DNA left border region, a 0.6-kb E9 terminator, and a 0.6-kb flanking vector sequence was used. A 2.0-kb NheI fragment containing the nptII gene and the T-DNA right border was used for both EcoRI- and XbaI-digested samples. The blots were rinsed twice for 15 min at room temperature with 2 × SSC and 0.1% SDS, then twice for 15 min at 65°C with 0.1 × SSC and 0.1% SDS.
Inverse Polymerase Chain Reaction Amplification of T-DNA–Flanking Genomic Sequences
Genomic DNA was digested with the indicated restriction enzyme, then extracted with phenol/chloroform and precipitated with ethanol. Digested DNA (2 μg) was self-circularized overnight at 16°C in 1 mL of ligation buffer containing 20 units of T4 ligase. After inactivation for 10 min at 75°C and ethanol precipitation, standard conditions were used for polymerase chain reaction (PCR) amplification. The primers for amplification of the 1.0-kb T-DNA–containing fragment were pTLB1 (5′-CATGGATCCATTAAGTTGTCTAAGCGTC-3′) and pTLB2 (5′-GCCAAGCTTAACACATTGCGGACGTTTTT-3′). The primers for amplification of the 1.6-kb T-DNA–containing fragment were pTLB3 (5′-CGCACCGATCGCCCTTCCCAACAG-3′) and pTLB4 (5′-CGCTATTACGCCAGCTGGCGAAAG-3′). After cloning and sequencing, new pairs of primers were designed on the basis of the sequence information to obtain a series of overlapping genomic fragments.
3′ and 5′ Rapid Amplification of cDNA Ends
RNA was extracted from in vitro–grown wild-type seedlings by using the guanidinium method (Strommer et al., 1993) and poly(A)+ mRNA was purified with the PolyATtract mRNA Isolation System IV (Promega, Madison, WI). For 3′ rapid amplification of cDNA ends (RACE), the first cDNA strand was synthesized by using an oligo(dT) primer. PCR was performed with an oligo(dT) primer and either primer pRH1 (5′-GGAAGAGGTTGGATATGTGAT-3′) or primer pRH4 (5′-ACAGGTTCTGGAAAAACC-3′). For a second amplification, the oligo(dT) primer and either the nested primer pRH3 (5′-TCGGGACGTGATTGTGTGCTT-3′) or pRH5 (5′-CTTGGCATACCTGCTACAAATA-3′) were used.
For 5′ RACE, double-stranded cDNA was synthesized from purified mRNA by using the Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA) and ligated to adaptors. Specific 3′ primers were designed on the basis of the sequence obtained from the 3′ RACE clones: pRH8 (5′-AGTTCTGAAGGCTTTGCAGCGAGCA-3′), pRH11 (5′-GGTTTTGCTGGAGGCATTGTCAG-3′), and pRH14 (5′-GAAACA-ACACTCCTTACAGAAACC-3′). PCR was then performed with 3′-specific and adaptor primers.
PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced. Independent clones were sequenced to detect any mutations occurring during PCR.
The full-length cDNAs were obtained by PCR with specific 5′ and 3′ primers as follows: for VDL-1, pTA5*A (5′-GGATCCATCGTACTT-TTTTCCTATTGTGT-3′) and pTFL65B (5′-GGGGTACCCCTAAAGAAT-AGCTGTTCACAACA-3′); for VDL′-1, VDL′-4, VDL′-5, VDL′-6, VDL′-7, and VDL′-8, pTABis5*A (5′-GGATCCCTCGAAACGGCCTCTACGC-3′) and pTFL65B; for VDL-2, pTA5*A and pTFL60 (5′-GGGGTACCCCGAATAATGTTCTCCCTACCTAGAG-3′); for VDL′-2, pTABis5*A and pTFL60; for VDL-3, pTA5*A and pTB3*B (5′-GGCAGATGGAAAGTTAGTAATTC-3′); and for VDL′-3, pTABis5*A and pTB3*B.
The GenBank accession numbers for the cDNA and genomic sequences are AF261017 to AF261031.
RNA Extraction and Reverse Transcriptase–PCR
RNA was extracted from various tissues of wild-type and heterozygous and homozygous mutant plants by using the guanidinium method (defined above) and poly(A)+ mRNA was purified with the PolyATtract mRNA Isolation System IV (Promega). The first-strand cDNA was synthesized as for 3′ RACE. PCR was performed with the primers defined above and mentioned in the legends. Other primers were as follows: pRH16 (5′-GCTGGGGAATTGAAGCACTAGCAAA-3′), pTFL50 (5′-AGTCCCTACTGGCTACTTCCCC-3′), and pTFL57 (5′-AGATAACCTCCTCCTCCTCGAAGT-3′).
Transformation and Complementation of vdl
The plasmid pBinhygTX (Gatz et al., 1992), which contained a hygromycin resistance selectable marker, a modified 35S promoter (Triple-op), and the octopin synthase terminator, was used for expression of the VDL-1 cDNA. A 1.5-kb SalI fragment containing the VDL-1 cDNA was subcloned into the SalI site of pBinhygTX to generate p35ShygVDL1. The plasmid BinhygTXGUS, containing the β-glucuronidase A (gusA) gene under the control of the Triple-op promoter (Gatz et al., 1992), was used as a control. Both plasmids were introduced into Agrobacterium tumefaciens. Leaf discs from a homozygous vdl plant were transformed as described by Rogers et al. (1986) and then selected in the presence of 50 μg/mL hygromycin B. Regenerated shoots were transferred to MS agar containing 50 μg/mL hygromycin B for root formation.
Expression of Green Fluorescent Protein Fusion Protein in Tobacco Protoplasts
We constructed a pUC-derived plasmid in which the green fluorescent protein gene (GFP) (Haseloff et al., 1997) was linked to the sequence encoding the 73 N-terminal amino acid residues of VDL-1. The junction read as follows: 5′- … CGGGAACTCTGTCATGGTCACCGGGGTACCATGAGTAAAGGAGAA … -3′ (VDL-1 and GFP nucleotides are shown in roman and italic type, respectively; the KpnI site is underlined). The chimeric gene was placed under the control of the CaMV 35S transcription promoter. This construct and a control containing the GFP gene without any additional coding sequence were introduced into tobacco protoplasts by electroporation (Lukaszewicz et al., 1998). GFP expression was analyzed by confocal microscopy (MRC-1024 laser scanning confocal microscope; Bio-Rad) 24 hr after transformation.
Light and Transmission Electron Microscopy of Leaf or Root Sections
Tissue processing, embedding, and sectioning were performed as described by Harris et al. (1994).
Acknowledgments
We thank Dr. Denyse Thinès for performing electron microscopy; Pierre Gosselin, Anne-Marie Faber, and Michèle Rochat for their excellent technical help; and Michel Hubermont and Pascal Veys for assistance with plant photography. This work was supported by grants from the Belgian National Fund for Scientific Research, the European Commission (BIOTECH program), and the Interuniversity Poles of Attraction program of the Belgian Government Office for Scientific, Technical, and Cultural Affairs.
References
- Amaratunga, M., and Lohman, T.M. (1993). Escherichia coli rep helicase unwinds DNA by an active mechanism. Biochemistry 32, 6815–6820. [DOI] [PubMed] [Google Scholar]
- Bird, L.E., Subramanya, H.S., and Wigley, D.B. (1998). Helicases: A unifying structural theme? Curr. Opin. Struct. Biol. 8, 14–18. [DOI] [PubMed] [Google Scholar]
- Boddeker, N., Stade, K., and Franceschi, F. (1997). Characterization of DbpA, an Escherichia coli DEAD box protein with ATP independent RNA unwinding activity. Nucleic Acids Res. 25, 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera y Poch, H.L., Peto, C.A., and Chory, J. (1993). A mutation in the Arabidopsis DET3 gene uncouples photoregulated leaf development from gene expression and chloroplast biogenesis. Plant J. 4, 671–682. [Google Scholar]
- Carol, P., Stevenson, D., Bisanz, C., Breitenbach, J., Sandmann, G., Mache, R., Coupland, G., and Kuntz, M. (1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis, A.J., Vanzo, N.F., and Raynal, L.C. (1999). mRNA degradation: A tale of poly(A) and multiprotein machines. Trends Genet. 15, 24–28. [DOI] [PubMed] [Google Scholar]
- Chao, K.L., and Lohman, T.M. (1991). DNA-induced dimerization of the Escherichia coli rep helicase. J. Mol. Biol. 221, 1165–1181. [DOI] [PubMed] [Google Scholar]
- Chatterjee, M., and Martin, C. (1997). Tam3 produces a suppressible allele of the DAG locus of Antirrhinum majus similar to Mu-suppressible alleles of maize. Plant J. 11, 759–771. [DOI] [PubMed] [Google Scholar]
- Chatterjee, M., Sparvoli, S., Edmunds, C., Garosi, P., Findlay, K., and Martin, C. (1996). DAGQ7, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 15, 4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Jensen, M., and Rodermel, S. (1999). The yellow variegated mutant of Arabidopsis is plastid autonomous and delayed in chloroplast biogenesis. J. Hered. 90, 207–214. [DOI] [PubMed] [Google Scholar]
- Cho, H.S., Ha, N.C., Kang, L.W., Chung, K.M., Back, S.H., Jang, S.K., and Oh, B.H. (1998). Crystal structure of RNA helicase from genotype 1b hepatitis C virus: A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 273, 15045–15052. [DOI] [PubMed] [Google Scholar]
- De la Cruz, J., Kressler, D., and Linder, P. (1999). Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24, 192–198. [DOI] [PubMed] [Google Scholar]
- Eisen, A., and Lucchesi, J.C. (1998). Unraveling the role of helicases in transcription. Bioessays 20, 634–641. [DOI] [PubMed] [Google Scholar]
- Gagliardi, D., Kuhn, J., Spadinger, U., Brennicke, A., Leaver, C.J., and Binder, S. (1999). An RNA helicase (AtSUV3) is present in Arabidopsis thaliana mitochondria. FEBS Lett. 458, 337–342. [DOI] [PubMed] [Google Scholar]
- Gatz, C., Frohberg, C., and Wendenburg, R. (1992). Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 2, 397–404. [DOI] [PubMed] [Google Scholar]
- Gibson, T.J., and Thompson, J.D. (1994). Detection of dsRNA-binding domains in RNA helicase A and Drosophila maleless: Implications for monomeric RNA helicases. Nucleic Acids Res. 22, 2552–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177, 115–180. [DOI] [PubMed] [Google Scholar]
- Grec, S., Wang, Y., LeGuen, L., Negrouk, V., and Boutry, M. (2000). Cryptic polyadenylation sites within the coding sequence of three yeast genes expressed in tobacco. Gene 242, 87–95. [DOI] [PubMed] [Google Scholar]
- Han, C.D., Coe, E.H., Jr., and Martienssen, R.A. (1992). Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. EMBO J. 11, 4037–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, N., Spence, J., and Oparka, K.J. (1994). General and enzyme histochemistry. In Plant Cell Biology: A Practical Approach, N. Harris and K.J. Oparka, eds (Oxford, UK: IRL Press), pp. 51–68.
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and sub-cellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, B., Jan, L.Y., and Jan, Y.N. (1988). A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55, 577–587. [DOI] [PubMed] [Google Scholar]
- Hayes, R., Kudla, J., and Gruissem, W. (1999). Degradating chloroplast mRNA: The role of polyadenylation. Trends Biol. Sci. 24, 199–202. [DOI] [PubMed] [Google Scholar]
- Hudson, A., Carpenter, R., Doyle, S., and Coen, E.S. (1993). Olive: A key gene required for chlorophyll biosynthesis in Antirrhinum majus. EMBO J. 12, 3711–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost, I., Dreyfus, M., and Linder, P. (1999). Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274, 17677–17683. [DOI] [PubMed] [Google Scholar]
- Keddie, J.S., Carroll, B., Jones, J.D., and Gruissem, W. (1996). The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 15, 4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Khurana, J.P., Kochhar, A., and Tyagi, A.K. (1998). Photosensory perception and signal transduction in higher plants—Molecular genetic analysis. Crit. Rev. Plant Sci. 17, 465–539. [Google Scholar]
- Li, H.M., Altschmied, L., and Chory, J. (1994). Arabidopsis mutants define downstream branches in the phototransduction pathway. Genes Dev. 8, 339–349. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz, M., Jerouvile, B., and Boutry, M. (1998). Signs of translational regulation within the transcript leader of a plant plasma membrane H+-ATPase gene. Plant J. 14, 413–423. [DOI] [PubMed] [Google Scholar]
- Luking, A., Stahl, U., and Schmidt, U. (1998). The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33, 259–296. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., Feldmann, K.A., Herrera-Estrella, L., Rocha-Sosa, M., and Leon, P. (1996). CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 9, 649–658. [DOI] [PubMed] [Google Scholar]
- Martienssen, R., and Baron, A. (1994). Coordinate suppression of mutations caused by Robertson's mutator transposons in maize. Genetics 136, 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Gil, P., Capel, J., and Somerville, C.R. (1992). Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell 4, 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet, J.E. (1988). Chloroplast development and gene expression. Annu. Rev. Plant Physiol. 39, 475–502. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nielsen, P.J., McMaster, G.K., and Trachsel, H. (1985). Cloning of eukaryotic protein synthesis initiation factor genes: Isolation and characterization of cDNA clones encoding factor eIF-4A. Nucleic Acids Res. 13, 6867–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, K., Morel-Deville, F., Hershey, J.W., Leighton, T., and Schnier, J. (1988). An eIF-4A–like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature 336, 496–498. [DOI] [PubMed] [Google Scholar]
- Py, B., Higgins, C.F., Krisch, H.M., and Carpousis, A.J. (1996). A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381, 169–172. [DOI] [PubMed] [Google Scholar]
- Reidi, G.P. (1973). Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat. Res. 118, 149–162. [Google Scholar]
- Reiter, R.S., Coomber, S.A., Bourett, T.M., Bartley, G.E., and Scolnik, P.A. (1994). Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell 6, 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.D. (1996). Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol. Biol. 32, 327–341. [DOI] [PubMed] [Google Scholar]
- Rogers, S.G., Horsch, R.B., and Fraley, R.T. (1986). Gene transfer in plants: Production of transformed plants using Ti plasmid vectors. Methods Enzymol. 118, 627–640. [Google Scholar]
- Rogers, S.O., and Bendich, A.J. (1994). Extraction of total cellular DNA from plants, algae and fungi. In Plant Molecular Biology Manual (Dordrecht, The Netherlands: Kluwer Academic Publishers), D1, pp. 1–8.
- Rozen, F., Meerovitch, K., Dever, T.E., Merrick, W.C., and Sonenberg, N. (1990). Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik, P.A., Hinton, P., Greenblatt, I.M., Giuliano, G., Delanoy, M.R., Spector, D.L., and Pollock, D. (1987). Somatic instability of carotenoid biosynthesis in the tomato ghost mutant and its effect on plastid development. Planta 171, 11–18. [DOI] [PubMed] [Google Scholar]
- Somanchi, A., and Mayfield, S.P. (1999). Nuclear–chloroplast signalling. Curr. Opin. Plant Biol. 2, 404–409. [DOI] [PubMed] [Google Scholar]
- Sommerville, J. (1999). Activities of cold-shock domain proteins in translation control. Bioessays 21, 319–325. [DOI] [PubMed] [Google Scholar]
- Stern, D., Higgs, D., and Yang, J. (1997). Transcription and translation in chloroplasts. Trends Plant Sci. 2, 308–315. [Google Scholar]
- Stevenson, R.J., Hamilton, S.J., MacCallum, D.E., Hall, P.A., and Fuller-Pace, F.V. (1998). Expression of the ‘dead box’ RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J. Pathol. 184, 351–359. [DOI] [PubMed] [Google Scholar]
- Strommer, J., Gregerson, R., and Vayda, M. (1993). Isolation and characterization of plant mRNA. In Methods in Plant Molecular Biology and Biotechnology (London: CRC Press), pp. 51–53.
- Styhler, S., Nakamura, A., Swan, A., Suter, B., and Lasko, P. (1998). vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125, 1569–1578. [DOI] [PubMed] [Google Scholar]
- Sugita, M., and Sugiura, M. (1996). Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 32, 315–326. [DOI] [PubMed] [Google Scholar]
- Sundberg, E., Slagter, J.G., Fridborg, I., Cleary, S.P., Robinson, C., and Coupland, G. (1997). ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin, M., and Chory, J. (1997). The co-ordination of nuclear and organellar genome expression in eukaryotic cells. Essays Biochem. 32, 113–125. [PubMed] [Google Scholar]
- Susek, R.E., Ausubel, F.M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. [DOI] [PubMed] [Google Scholar]
- Taylor, W.C. (1989). Regulatory interaction between nuclear and plastid genomes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 211–233. [Google Scholar]
- Toone, W.M., Rudd, K.E., and Friesen, J.D. (1991). deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 173, 3291–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne, G., Steppuhn, J., and Herrmann, R.G. (1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180, 535–545. [DOI] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). PHANTASTICA—A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154. [Google Scholar]
- Wetzel, C.M., Jiang, C.-Z., Meehan, L.J., Voytas, D.F., and Rodermel, S.R. (1994). Nuclear–organelle interactions: The immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 6, 161–175. [DOI] [PubMed] [Google Scholar]
- Wu, D., Wright, D.A., Wetzel, C., Voytas, D.F., and Rodermel, S. (1999). The immutans variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]