Abstract
The types of cells and tissues infected by a virus define its tissue tropism. Determinants of tissue tropism in animal-infecting viruses have been extensively investigated, but little is known about plant viruses in this regard. Some geminiviruses in the genus Begomovirus exhibit phloem limitation and are restricted to cells of the vascular system, whereas others can invade mesophyll tissue. To identify viral genetic determinants of tissue tropism, we established a model system using two begomoviruses and their common host plant, Nicotiana benthamiana. Analysis by DNA in situ hybridization confirmed that tomato golden mosaic virus invades mesophyll tissues in systemically infected leaves, whereas bean golden mosaic virus remains phloem limited. Through genetic complementation and analysis of recombinant hybrid viruses, we demonstrated that three genetic elements of tomato golden mosaic virus determine its mesophyll tissue tropism. A noncoding region of the viral genome is essential for the phenotype, but it must be accompanied by one of two different coding regions. To our knowledge, this is the first example documented in a plant virus of noncoding DNA sequences that determine tissue tropism.
INTRODUCTION
The ability to invade the tissues of a host organism is a fundamental requirement for a successful pathogen. Viruses are frequently able to disseminate systemically in their hosts to infect cells that are distal to the site of inoculation. The specific types of cells and tissues that become infected define a property of the virus referred to as its tissue tropism (Tyler and Fields, 1996). Because of the general importance of the disposition of systemically infected organs and tissues in the course of viral pathogenesis and disease, the molecular determinants of tissue tropism have been studied extensively in animal-infecting viruses. In such systems, whether the virus can infect a particular cell type is frequently determined by the presence or absence of a virus-specific cell-surface receptor (Tyler and Fields, 1996). Plant viruses are unlike their animal-infecting counterparts in that they mostly, if not exclusively, appear to spread symplastically through preexisting cytoplasmic connections formed by plasmodesmata (reviewed in Lucas and Gilbertson, 1994; Carrington et al., 1996). Molecular mechanisms that determine the tissue tropism of plant viruses are currently unknown.
To mount a systemic infection, a plant virus must be able to move into and through the vascular system of its host; such viruses thus may be considered to have a vascular tissue tropism. Some viruses remain confined to the vascular system and are said to be phloem limited. However, many plant viruses are also capable of moving out of the vascular system to infect mesophyll cells; these thus have a distinct mesophyll tissue tropism. The factors that govern whether a given plant virus can exit from the vascular system have not been extensively studied (reviewed in Nelson and van Bel, 1997). A phloem-limited infection might occur because the viral movement proteins involved are unable to function in cell types outside the vascular system. However, this relatively trivial mechanism is unlikely to be applicable to all phloem-limited plant viruses. In the case of bipartite geminiviruses in the genus Begomovirus, viruses that encode movement proteins with highly similar amino acid sequences nevertheless exhibit different tissue tropisms. Thus, we considered that these begomoviruses would provide an excellent system for investigating some of the viral and host cellular determinants that can affect the tissue tropism of plant viruses.
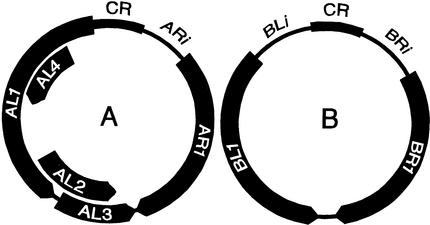
Several begomoviruses from the New World have been characterized extensively. In nature, these viruses are transmitted from plant to plant by whiteflies, but they can also be experimentally inoculated by various mechanical procedures, including microprojectile bombardment. The geminate virus particles contain circular single-stranded (ss) DNA. In the case of bean golden mosaic virus (BGMV), the type member of the genus, and its close relative, tomato golden mosaic virus (TGMV), the genome is divided between two DNA components, which are designated A and B. Pertinent features of the begomovirus genome are illustrated schematically in Figure 1. Each virus has six well-characterized genes, four located on DNA A and two on DNA B. An additional open reading frame (ORF), AL4, is either nonfunctional or encodes a product that is completely dispensable for the infection of plants by BGMV or TGMV (Elmer et al., 1988; Hoogstraten et al., 1996; Pooma and Petty, 1996). The two proteins encoded by DNA B, BL1 and BR1, interact to mediate viral intercellular movement (Sanderfoot and Lazarowitz, 1995; Schaffer et al., 1995). The BR1 protein can shuttle between the nucleus and the cytoplasm and is probably required for the intracellular trafficking of viral DNA (Sanderfoot et al., 1996; Ward and Lazarowitz, 1999), whereas the BL1 protein is probably responsible for the trafficking of viral DNA from cell to cell (Noueiry et al., 1994). However, other details of the cell-to-cell movement process remain controversial. The viral coat protein is encoded by the AR1 gene (Kallender et al., 1988; Azzam et al., 1994). The AL1 protein provides origin recognition and enzymatic functions that are essential for viral DNA replication (Fontes et al., 1994; Hanson et al., 1995; Hoogstraten et al., 1996; Orozco and Hanley-Bowdoin, 1998), whereas AL3 boosts the extent of viral DNA accumulation by an unknown mechanism (Sunter et al., 1990; Gladfelter et al., 1997). The transcription regulatory protein AL2 is required to activate expression of the AR1 (coat protein) and BR1 (movement protein) genes (Sunter and Bisaro, 1992). In addition to their protein-coding regions, geminivirus DNA components contain an intergenic region, which harbors viral promoters and the origin of DNA replication. The replication origin is contained in a sequence of ∼200 bp, which is nearly identical in both DNA components of a given virus (called the common region) (Fontes et al., 1994; Orozco et al., 1998). Unique, noncoding DNA sequences are located immediately upstream of the AR1, BR1, and BL1 ORFs and are referred to here as ARi, BRi, and BLi, respectively (Figure 1).
Figure 1.
Schematic Illustration of Begomovirus Genome Organization.
The location of coding and noncoding DNA sequences in the A and B genome components is shown. ORFs and their direction of transcription are depicted by arrows. Major noncoding regions on each DNA component contain a sequence of near-identity, termed the common region (CR), depicted by thick lines. In addition, unique noncoding sequences between the common region and the AR1, BL1, and BR1 ORFs, which are designated ARi, BLi, and BRi, respectively, are depicted by thin lines.
In the convenient model host Nicotiana benthamiana, TGMV infects most cell types of the leaf, including a large number of mesophyll cells (Rushing et al., 1987; Nagar et al., 1995). In beans, the tissue tropism of BGMV is phloem limited (Kim et al., 1978). To compare the determinants of tissue tropism directly, however, a host plant common to both viruses is required. Fortunately, BGMV is also able to infect N. benthamiana systemically, although it is less well adapted to this plant than is TGMV (Petty et al., 1995). BGMV replicates well in tobacco protoplasts (Fontes et al., 1994), but it accumulates poorly relative to TGMV in systemically infected leaves of N. benthamiana (Petty et al., 1995). These contrasting observations suggested that BGMV could not infect as many cells as TGMV in N. benthamiana and prompted us to investigate the tissue tropism of BGMV in this host. Not only did we find that BGMV infection is phloem limited in N. benthamiana, but we also confirmed the prediction of Rushing et al. (1987) that geminivirus tissue tropism has a viral genetic component. Regions of the TGMV genome that are required for mesophyll invasion in systemically infected leaves were identified, and potential mechanisms by which geminivirus tissue tropism may be determined are discussed.
RESULTS
TGMV and BGMV Exhibit Differential Tissue Tropism in N. benthamiana
A DNA in situ hybridization procedure was used to detect geminivirus-infected cells in sections prepared from N. benthamiana leaves. Plants were inoculated by microprojectile bombardment with plasmids containing the A and B components of TGMV or BGMV, and systemic infections were allowed to develop. After 14 days, tissue samples were taken from leaves two nodes below the apex of the plant. These leaves represent the most highly symptomatic tissue produced during a systemic infection of N. benthamiana by wild-type TGMV. Virus-infected cells were detected by DNA in situ hybridization and visualized by light microscopy. As shown in Figure 2A, we found TGMV DNA localized to the nuclei in a variety of cell types in systemically infected N. benthamiana leaves, which is consistent with previous results obtained with other techniques (Rushing et al., 1987; Nagar et al., 1995). Tissue sections from uninfected control plants did not show nonspecific binding to the probe (Figure 2C). To quantify the ability of TGMV to invade mesophyll tissue, we identified and categorized infected cells as being either vascular associated (vascular parenchyma, companion cells, and bundle sheath cells) or mesophyll (palisade and spongy cells). The examination of only 10 tissue sections from each of two independently inoculated N. benthamiana plants allowed us to unambiguously identify 923 TGMV-infected cells, of which 279 (∼30%) were vascular associated and the remaining 644 (∼70%) were in the mesophyll.
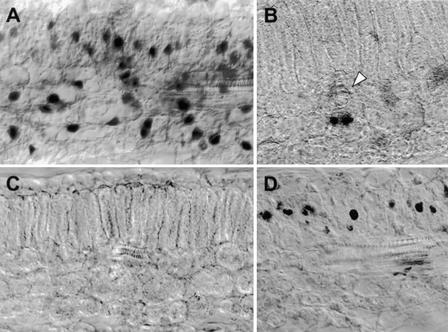
Figure 2.
In Situ Localization of Wild-Type Begomoviruses in Single and Double Infections.
All sections were made from systemically infected leaves of N. benthamiana, hybridized with digoxigenin (DIG)-labeled ssRNA probes, and viewed at ×400 magnification with differential interference contrast optics.
(A) Section from a plant infected with wild-type TGMV and hybridized with the TGMV-specific probe. Hybridization signals were detected in nuclei of spongy and palisade mesophyll cells as well as in those of vascular-associated cells.
(B) Section from a plant infected with wild-type BGMV and hybridized with the BGMV-specific probe. Hybridization signals were detected in nuclei of vascular-associated cells. A white arrowhead indicates the location of a vascular bundle passing perpendicular to the plane of the section.
(C) Section from an uninfected plant incubated with the TGMV-specific probe. No nonspecific hybridization was detected.
(D) Section from a plant infected with both wild-type BGMV and wild-type TGMV and hybridized with the BGMV-specific probe. The presence of hybridization signals in nuclei of palisade mesophyll cells indicates that BGMV can infect these cells in the double infection with TGMV. Infected spongy mesophyll cells were observed in other sections (not shown).
The lesser extent of DNA accumulation by BGMV compared with that of TGMV suggested that fewer cells were infected systemically by BGMV in N. benthamiana (Petty et al., 1995), but the types of cells infected had not previously been determined. Although BGMV produces an asymptomatic infection of N. benthamiana, tissue samples were taken from locations equivalent to those used for TGMV-infected plants. Examination of tissue sections from BGMV-infected plants by DNA in situ hybridization revealed that this virus was phloem limited in N. benthamiana (Figure 2B). Despite careful analysis of tissues surrounding the second- through fourth-order leaf veins, no substantial unloading of BGMV from the vascular system was observed. By combining the data from 38 tissue sections obtained from eight independently inoculated plants, we identified a total of 317 BGMV-infected cells. Of these BGMV-infected cells, 314 (∼99%) were vascular-associated cells and the remaining three (∼1%) were mesophyll cells. Transverse sections of minor veins were examined to determine which specific types of vascular-associated cells could be infected by BGMV. Among 32 BGMV-infected vascular cells that could be unambiguously identified, two were companion cells, 18 were phloem parenchyma cells, and 12 were bundle sheath cells. In this study, no viral DNA was detected in the phloem sieve elements.
These results show that TGMV and BGMV differ significantly in their ability to move into mesophyll tissue in systemically infected leaves of N. benthamiana (P < 0.001 by χ2 test). Consequently, the two viruses and their common host plant constitute an ideal experimental system for elucidating molecular mechanisms that can determine phloem limitation or mesophyll invasiveness in plant viruses.
Coinfection with TGMV Can Release the Phloem Limitation of BGMV in N. benthamiana
Coinoculation of BGMV with TGMV could enhance BGMV DNA accumulation in systemically infected leaves of N. benthamiana (Petty et al., 1995), although TGMV cannot replicate BGMV DNA, and both viruses could accumulate to similar extents in tobacco protoplasts (Fontes et al., 1994). Taken together, these results suggested that the enhanced BGMV DNA accumulation in coinfected plants is caused by an increase in the number of infected cells rather than a direct effect on DNA replication. To test this hypothesis, we coinfected plants with TGMV and BGMV and analyzed them using DNA in situ hybridization. Plants were inoculated with plasmids containing the TGMV A and B components, together with either plasmids containing the BGMV A and B components, or a plasmid containing the BGMV A component. The A component–specific probes for BGMV and TGMV do not cross-hybridize detectably during DNA in situ hybridization (data not shown). When coinoculated with TGMV, in either the presence or absence of BGMV DNA B, mesophyll cells were found to contain BGMV DNA A (Figure 2D). Thus, viral or host factors produced during a TGMV infection act in trans to allow BGMV DNA to move out of the vascular system and into mesophyll cells. The results of this experiment also demonstrate that the phloem limitation of BGMV, when inoculated alone, is not the result of an intrinsic inability of mesophyll cells to support the replication or spread of BGMV DNA.
BGMV-Based Hybrid Viruses Can Spread into the Mesophyll of Systemically Infected Leaves
To identify which viral genes determine the differential tissue tropism of BGMV and TGMV, we tested hybrid viruses for their ability to escape from the vascular system. We define a BGMV-based hybrid virus as one that encodes the BGMV replication initiator protein AL1 and has the BGMV replication origin on both the A and B components; however, it may contain any number of the remaining ORFs or cis-acting elements from TGMV. Conversely, a TGMV-based hybrid virus is one that contains the AL1 gene and common regions of TGMV. We were particularly interested in determining whether BGMV-based hybrid viruses could acquire the ability to escape from the vascular system, which would represent a gain-of-function phenotype in this system. Consequently, we first tested a BGMV-based hybrid virus that contained the ORFs for the TGMV coat protein (AR1), transcription regulator protein (AL2), replication booster protein (AL3), both movement proteins (BR1 and BL1), and the ARi and BRi noncoding regions. As illustrated in Figure 3A, DNA in situ hybridization showed that this hybrid virus, which contained much of the TGMV genome, was able to escape from the vascular system. Among a total of 729 infected cells that could be positively identified, 310 were mesophyll cells (∼41%). In addition to its TGMV-like tissue tropism, this BGMV-based hybrid virus exhibited TGMV-like DNA accumulation and elicited severe symptoms in systemically infected N. benthamiana (Petty et al., 2000).
Figure 3.
In Situ Localization of BGMV-Based and TGMV-Based Hybrid Viruses.
All sections were made from systemically infected leaves of N. benthamiana unless otherwise noted. Sections were hybridized with DIG-labeled ssRNA probes and viewed at ×400 magnification with differential interference contrast optics.
(A) Section from a plant infected with a BGMV-based hybrid virus that contains the TGMV AR1, AL23, BR1, and BL1 ORFs and the ARi and BRi noncoding regions. After hybridization with the BGMV-specific probe, signals were detected in nuclei of palisade mesophyll cells. Infected spongy mesophyll cells were observed in other sections (not shown).
(B) Section from a plant infected with a BGMV-based hybrid virus containing only the TGMV AL23 ORFs. Hybridization was with the BGMV-specific probe. Virus-infected cells were confined to the vascular system. A white arrowhead indicates the location of a vascular bundle passing perpendicular to the plane of the section.
(C) Section from a plant infected with a BGMV-based hybrid virus containing the AL23 ORFs and the BRi noncoding region from TGMV. Hybridization was with the BGMV-specific probe, and signals were detected in mesophyll cells.
(D) Section from a plant infected with a BGMV-based hybrid virus that contains the BRi noncoding region and the BL1/BR1 ORFs from TGMV. Hybridization was with the BGMV-specific probe, and signals were detected in mesophyll cells.
(E) Section from a plant infected with a TGMV al3 mutant A component and a BGMV-based hybrid virus containing only the TGMV BRi noncoding region (normally phloem limited). The presence of BGMV-specific hybridization signals in nuclei of mesophyll cells indicates that the coinoculated TGMV al3 mutant can complement the mesophyll invasion defect of a BGMV-based hybrid virus that contains only BRi from TGMV.
(F) Section from a plant infected with a TGMV-based hybrid virus containing the BRi noncoding region from BGMV. Hybridization was with the TGMV-specific probe. Virus-infected cells were confined to the vascular system. White arrowheads indicate the location of vascular bundles passing perpendicular to the plane of the section.
(G) Section from a directly inoculated leaf infected with a BGMV-based hybrid virus containing the AL23 ORFs from TGMV; this would be phloem limited in systemically infected leaves, as shown in (B). Hybridization was with the BGMV-specific probe, and signals were detected in mesophyll cells.
(H) Section from a directly inoculated leaf infected with a TGMV-based hybrid virus containing the BRi noncoding region from BGMV; this would be phloem limited in systemically infected leaves, as shown in (F). Hybridization was with the TGMV-specific probe, and signals were detected in mesophyll cells.
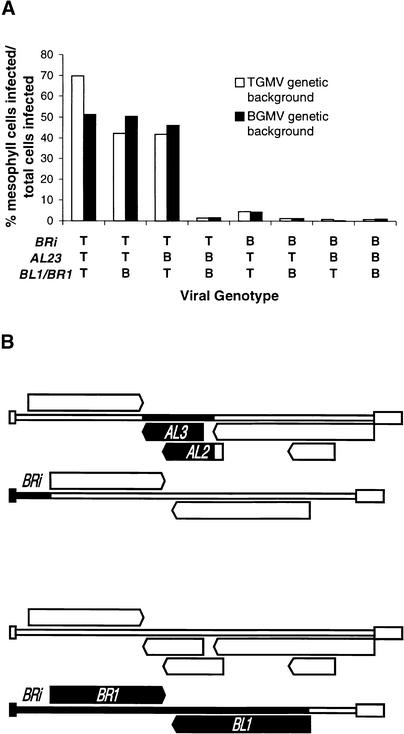
The preceding results allowed us to conclude that one or more of the TGMV AR1, AL2, AL3, BL1, and BR1 genes determined the ability of this virus to escape from the vascular system in systemically infected leaves of N. benthamiana. Previous studies have shown that TGMV ar1 (coat protein) gene null mutants can spread systemically in N. benthamiana (Gardiner et al., 1988; Jeffrey et al., 1996). To elucidate the potential role of the coat protein in determining tissue tropism, we analyzed a TGMV ar1 null mutant by DNA in situ hybridization and found that it could spread into the mesophyll of systemically infected leaves. Among 450 cells infected by the TGMV ar1 mutant that could be unambiguously identified, 174 were mesophyll cells (∼39%). This result indicated that expression of the coat protein gene was not essential for the escape of TGMV from the vascular system. Eliminating the coat protein ORF (AR1) and the corresponding upstream noncoding sequences (ARi) as potential determinants of mesophyll invasion allowed us to focus on three genetic elements: (1) AL23, the ORFs for the AL2 (transcription regulator) and AL3 (replication booster) proteins, which overlap extensively and cannot be physically separated in hybrid viruses (see Figure 1); (2) BL1/BR1, which were grouped together because the BL1 and BR1 proteins interact and cannot be functionally separated without compromising virus movement (Schaffer et al., 1995); and (3) BRi, the noncoding region upstream from the BR1 ORF. These three genetic elements were evaluated together, individually, and in all pairwise combinations to identify the TGMV components necessary for virus escape from the vascular system. The results of these experiments are presented in Table 1 and are summarized graphically in Figure 4A.
Table 1.
Tissue Tropism of Wild-Type and Hybrid Geminiviruses
| Cells Infecteda
|
|||||
|---|---|---|---|---|---|
| Viral Genotype | V | M | T | M/T % | χ2 |
| BGMV-based hybrid viruses | |||||
| Elements from TGMV | |||||
| None (wild-type BGMV) | 314 | 3 | 317 | 0.95 | a |
| AL23 | 308 | 4 | 312 | 1.28 | a |
| BL1/BR1 | 84 | 0 | 84 | 0.00 | a, b |
| BRi | 234 | 4 | 238 | 1.68 | a, b |
| AL23, BL1/BR1 | 395 | 18 | 413 | 4.36 | b |
| AL23, BRi | 383 | 387 | 770 | 50.26 | c |
| BRi, BL1/BR1 | 322 | 275 | 597 | 46.06 | c, d |
| AL23, BRi, BL1/BR1 | 293 | 307 | 600 | 51.17 | c |
| TGMV-based hybrid viruses | |||||
| Elements from BGMV | |||||
| None (wild-type TGMV) | 279 | 644 | 923 | 69.77 | e |
| AL23 | 458 | 327 | 785 | 41.66 | d |
| BL1/BR1 | 403 | 294 | 697 | 42.18 | d |
| BRi | 227 | 11 | 238 | 4.62 | b, f |
| AL23, BL1/BR1 | 135 | 2 | 137 | 1.46 | a, b |
| AL23, BRi | 130 | 1 | 131 | 0.76 | a |
| BRi, BL1/BR1 | 172 | 2 | 174 | 1.15 | a |
| AL23, BRi, BL1/BR1 | 233 | 2 | 235 | 0.85 | a |
Infected cells that could be unambiguously identified as vascular associated (V) or mesophyll (M) were scored. The total number (T) of infected cells (where T = V + M) and the percentage of the total that were mesophyll cells (M/T %) are also given. Mesophyll invasion efficiencies that were not significantly different (P > 0.05 by χ2 test) are indicated by like letters under χ2.
Figure 4.
Identification of the Minimal TGMV Sequences Required to Confer a Mesophyll-Invasive Phenotype.
(A) Mesophyll invasion efficiency of viruses with various combinations of the BRi noncoding region and AL23 or BL1/BR1 ORFs from either TGMV (T) or BGMV (B). Mesophyll invasion data for wild-type and hybrid viruses were taken from Table 1. All possible combinations of the three genetic elements were tested in TGMV-based hybrid viruses (TGMV genetic background, open bars) and in BGMV-based hybrid viruses (BGMV genetic background, solid bars). Wild-type TGMV and hybrid viruses that efficiently invade the mesophyll contain TGMV BRi paired with TGMV AL23 ORFs, TGMV BL1/BR1 ORFs, or both.
(B) Schematic illustration of the two BGMV-based hybrid viruses with the minimal TGMV sequences required for mesophyll invasion. The viral DNA components are shown as if linearized at the (+) strand origin of replication, with the A component on top and the B component beneath. The ORFs and noncoding regions are depicted with the symbols used in Figure 1. BGMV-specific sequences are shown with open symbols, and TGMV-specific sequences are shown with solid symbols. The mesophyll-invasive BGMV-based hybrid viruses contain either TGMV AL23 and TGMV BRi (top) or TGMV BRi and TGMV BL1/BR1 (bottom).
To be certain that these three sequence elements together contained all of the necessary components for spread into the vascular system and to confirm that the TGMV coat protein (AR1) gene was not required, we first tested a BGMV-based hybrid virus that contained the AL23 and BL1/BR1 ORFs and the BRi noncoding region from TGMV. As expected, this hybrid virus spread efficiently into mesophyll cells in systemically infected leaves (Table 1 and Figure 4A). Thus, in combination, the three sequence elements AL23, BL1/BR1, and BRi from TGMV are sufficient to confer on a BGMV-based hybrid virus the ability to move out of the vascular system. To determine the least amount of TGMV DNA that would allow a BGMV-based hybrid virus to escape from the vascular system, we analyzed hybrid viruses that contained each element individually. Interestingly, none of these hybrid viruses was able to escape efficiently from the vascular system; in each case, <2% of the infected cells were in the mesophyll (Figure 3B and Table 1). This result indicated that at least two of the three genetic elements from TGMV must be necessary for virus movement into mesophyll cells in systemically infected leaves.
To determine which of the three genetic elements from TGMV contributed to the phenotype, we analyzed BGMV-based hybrid viruses containing each pairwise combination by using DNA in situ hybridization. These experiments revealed that a combination of the TGMV BRi noncoding region with either the TGMV transcription regulator protein and replication booster protein ORFs (AL23) or the TGMV movement protein ORFs (BL1/BR1) allowed BGMV-based hybrid viruses to escape from the vascular system (Figures 3C and 3D). In contrast, a BGMV-based hybrid virus that contained the TGMV AL23 and TGMV BL1/BR1 ORFs was phloem limited. Thus, BRi, the noncoding region upstream from the TGMV BR1 ORF, is absolutely necessary for mesophyll invasion in systemically infected leaves, but either the TGMV AL23 ORFs or the TGMV movement protein ORFs (BL1/BR1) must also be present for the phenotype to be manifested (Figures 4A and 4B).
Reciprocal TGMV-Based Hybrid Viruses Are Phloem Limited
The analysis of BGMV-based hybrid viruses described so far had revealed that escape from the vascular system in systemically infected leaves of N. benthamiana was determined by the TGMV BRi noncoding region, in conjunction with either the TGMV movement protein ORFs (BL1/BR1) or the TGMV AL23 ORFs. To ensure that other features of the viral genetic background did not contribute significantly to the gain-of-function phenotype exhibited by BGMV-based hybrid viruses, we also tested an analogous series of TGMV-based hybrid viruses (Table 1 and Figure 4A). The results of DNA in situ hybridization showed that in contrast to wild-type TGMV, TGMV-based hybrid viruses were phloem limited when they contained either a BRi noncoding region derived from BGMV (Figure 3F) or a combination of the AL23 and BL1/BR1 ORFs from BGMV. Thus, the tissue tropism phenotypes of TGMV-based hybrid viruses were completely consistent with those of the BGMV-based hybrid viruses with equivalent genotypes (Figure 4A). Together, the experiments with hybrid viruses conclusively demonstrated that the differential tissue tropism exhibited by TGMV and BGMV in systemically infected leaves of N. benthamiana is genetically determined by three regions of the viral genome: the BRi noncoding region and the AL23 or BL1/BR1 ORFs.
Phloem-Limited Hybrid Viruses Can Infect Directly Inoculated Mesophyll Cells
The differential tissue tropism we observed in systemically infected N. benthamiana leaves could indicate either that mesophyll cells cannot support the replication or cell-to-cell movement of phloem-limited viruses or that such viruses are unable to efficiently gain access to the mesophyll. Because microprojectile bombardment introduces viral DNA directly into mesophyll cells in inoculated leaves, we could analyze such tissue by DNA in situ hybridization to distinguish between these possibilities. All of the phloem-limited hybrid viruses produced circular chlorotic lesions of various sizes on inoculated leaves, and in each case we examined, the virus had infected mesophyll cells within the chlorotic tissue. Specific examples are illustrated for a BGMV-based hybrid virus with the TGMV AL23 ORFs and for a TGMV-based hybrid virus with the BGMV BRi noncoding region (Figures 3G and 3H). These results indicated that hybrid viruses that remained phloem limited in systemically infected leaves were nevertheless capable of replicating within directly inoculated mesophyll cells. Because the formation of chlorotic lesions on inoculated leaves requires expression of the BL1 and BR1 movement proteins (Jeffrey et al., 1996), and because they contain many contiguous infected cells, the lesions probably represent radially spreading foci of virus infection. Thus, when introduced directly into mesophyll cells, phloem-limited hybrid viruses are not defective for either movement protein gene expression or cell-to-cell movement. Together, these data indicate that the phloem limitation observed in this system is not the result of an intrinsic inability of mesophyll tissues to support virus replication or movement; rather, it specifically reflects the failure of phloem-limited viruses to invade the mesophyll from the vascular system of systemically infected leaves.
Effect of the AL23 ORFs on Mesophyll Invasion Is Mediated by AL2 Protein
Because the AL2 transcription regulator and AL3 replication booster proteins are involved in quite different aspects of the viral infection process, we wanted to determine if possible which of these proteins mediated the effect of the AL23 region on mesophyll invasion. Because the overlapping AL23 ORFs cannot be separated in hybrid viruses, we instead used a genetic complementation assay. As described above, a BGMV-based hybrid virus that contained only the TGMV BRi noncoding region was phloem limited (Table 1). However, when wild-type TGMV DNA A (which would produce TGMV AL2 and AL3 proteins in trans) was also included in the inoculum, the infection spread readily to the mesophyll. Among a total of 140 infected cells, 46 were mesophyll cells (∼25%). Importantly, when wild-type TGMV DNA A was replaced by a TGMV al3 null mutant A component (which could produce only TGMV AL2 in trans), the infection also spread into the mesophyll (Figure 3E). Among a total of 266 infected cells identified, 72 were mesophyll cells (∼21%). Thus, the contribution of the TGMV AL23 region to mesophyll invasion is apparently mediated by the AL2 protein. Unfortunately, we were unable to perform the complementary negative control experiment with a TGMV al2 null mutant A component because its systemic movement is inefficiently complemented by coinoculated BGMV (Gillette et al., 1998).
Mesophyll Invasion Requires Compatibility between the BL1 and BR1 Movement Proteins
Because our genetic data suggested that expression of the BR1 gene might be important for mesophyll invasion, we wished to determine whether TGMV BR1 alone was responsible for the contribution of the BL1/BR1 ORFs to this phenotype. Hybrid viruses containing the combination of BGMV BL1 and TGMV BR1 ORFs are viable, although imperfect compatibility between the heterologous movement proteins reduces viral fitness relative to that of otherwise similar hybrids in which both BL1 and BR1 are from the same virus (Schaffer et al., 1995). In contrast, hybrid viruses that encode TGMV BL1 and BGMV BR1 are not viable, so we could not evaluate the effect of this combination of movement proteins on tissue tropism. As shown in Table 2, when their tissue tropism was analyzed by DNA in situ hybridization, we found that hybrid viruses with the combination of BGMV BL1 and TGMV BR1 ORFs were all phloem limited, regardless of the source of BRi or AL2 or of other features of the viral genome. In particular, hybrid viruses that contained both TGMV AL23 and TGMV BRi were phloem limited when they encoded BGMV BL1 plus TGMV BR1 in either viral genetic background. In contrast, otherwise similar hybrids with both movement protein ORFs from the same virus, either from TGMV or BGMV, were able to invade the mesophyll in systemically infected leaves of N. benthamiana (Table 2). The exclusively phloem-limited phenotype of hybrid viruses with the mismatched BGMV BL1 and TGMV BR1 movement proteins did not allow us to distinguish the individual contributions of BR1 and BL1 to mesophyll invasion. However, given that the degree of compatibility between the BL1 and BR1 proteins is known to affect the efficiency of cell-to-cell movement (Schaffer et al., 1995), these results provide support for a mechanism in which a defect in virus movement leads to phloem limitation.
Table 2.
Tissue Tropism of Hybrid Geminiviruses with Mismatched versus Matched BL1 and BR1 Movement Proteins
| Genotypeb
|
Cells Infectedc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristicsa | AL23 | BRi | BL1 | BR1 | V | M | T | M/T % | χ2 |
| BGMV-based hybrid viruses | |||||||||
| Matched MPs (wild-type BGMV) | B | B | B | B | 314 | 3 | 317 | 0.95 | a |
| Mismatched MPs | B | T | B | T | 230 | 22 | 252 | 8.73 | b |
| Mismatched MPs | T | T | B | T | 260 | 11 | 271 | 4.06 | c |
| Matched BGMV MPs | T | T | B | B | 383 | 387 | 770 | 50.26 | d |
| Matched TGMV MPs | T | T | T | T | 293 | 307 | 600 | 51.17 | d |
| TGMV-based hybrid viruses | |||||||||
| Matched BGMV MPs | B | B | B | B | 233 | 2 | 235 | 0.85 | a |
| Mismatched MPs | B | T | B | T | 226 | 1 | 227 | 0.44 | a |
| Mismatched MPs | T | T | B | T | 211 | 22 | 233 | 9.44 | b |
| Matched BGMV MPs | T | T | B | B | 403 | 294 | 697 | 42.18 | e |
| Matched MPs (wild-type TGMV) | T | T | T | T | 279 | 644 | 923 | 69.77 | f |
Matched movement proteins (MPs) are defined as combinations of BL1 and BR1 from the same virus; mismatched combinations have BGMV BL1 paired with TGMV BR1. Controls were wild-type viruses.
Hybrid viruses contained the AL23 ORFs, BRi noncoding region, and BL1 and BR1 ORFs from either BGMV (B) or TGMV (T), as indicated.
Infected cells that could be unambiguously identified as vascular associated (V) or mesophyll (M) were scored. The total number (T) of infected cells (where T = V + M) and the percentage of the total that were mesophyll cells (M/T %) are also given. Mesophyll invasion efficiencies that were not significantly different (P > 0.05 by χ2 test) are indicated by like letters under χ2.
DISCUSSION
Our demonstration that the closely related begomoviruses TGMV and BGMV exhibit distinct mesophyll-invasive and phloem-limited phenotypes, respectively, in N. benthamiana confirmed that tissue tropism is at least in part genetically determined by the virus. However, because viruses are intimately associated with host cellular machinery during all phases of the infection, the interplay between host and viral factors is ultimately responsible for the manifestation of all viral phenotypes. For example, the developmental state of the host influences the tissue tropism of the begomovirus bean dwarf mosaic virus (Wang et al., 1996), and environmental factors such as temperature can affect the tissue tropism of RNA plant viruses (Ding et al., 1999). To simplify the genetic analysis, we kept environmental conditions constant and determined the viral tissue tropism only in systemically infected leaves that showed the most severe symptoms of infection (i.e., at a single host developmental stage).
Mixed infection experiments with TGMV and BGMV showed that trans-acting factors produced, or induced, by TGMV were able to alleviate the phloem limitation of coinoculated BGMV. Subsequently, substitution mapping, in which pieces of the BGMV genome were replaced with the homologous regions of TGMV, was used to precisely localize the viral genetic determinants responsible for mesophyll invasion in systemically infected leaves. Phloem-limited hybrid viruses could readily infect and spread through mesophyll tissue if they were introduced into it directly by microprojectile bombardment. Consequently, phloem limitation in this system must specifically result from the failure of such viruses to establish infection in the mesophyll of systemically infected leaves to which they are delivered through the vasculature. In N. benthamiana, escape of an RNA plant virus from the vascular system in a systemically infected leaf has been shown to occur only preceding the sink–source transition (Roberts et al., 1997). If this temporal window similarly constrains geminivirus egress from the vascular system, then two general types of model could explain the phenomenon of phloem limitation. In one, the movement of phloem-limited viruses may be compromised: either they reach upper leaves with delayed kinetics and simply miss the window of opportunity to transport into mesophyll cells, or they reach upper leaves efficiently but nonetheless fail to move across the bundle sheath–mesophyll cell boundary. Alternately, a systemically acquired host defense response of some kind may render mesophyll cells in upper leaves refractory to infection by phloem-limited viruses.
Comparison of BGMV-based and TGMV-based hybrid viruses revealed that the viral genetic background slightly affected the efficiency of mesophyll invasion in systemically infected leaves of N. benthamiana, but overall the results were consistent with the involvement of three genetic determinants. The primary determinant of the mesophyll-invasive phenotype was the noncoding sequence upstream from the TGMV BR1 ORF (i.e., TGMV BRi). Precedents among animal-infecting viruses illustrate the involvement of noncoding nucleotide sequences in determining tissue tropism (see Tyler and Fields, 1996), but the results of this study represent, to our knowledge, the first example documented in a plant virus. Although the TGMV BRi noncoding region was necessary for mesophyll invasion, it was not sufficient without the presence of either one of two accessory determinants from TGMV, namely, the AL23 or BL1/BR1 coding regions. Genetic complementation implicated the AL2 transcription regulator protein as the responsible factor contributed by the AL23 region, but because the BL1 and BR1 movement proteins are adapted to one another, we were unable to resolve their individual contributions to tissue tropism. Interestingly, however, hybrid viruses were phloem limited even when they contained TGMV BRi and TGMV AL23 if they also encoded BGMV BL1 and TGMV BR1, a combination of proteins that supports virus movement with reduced efficiency (Schaffer et al., 1995). This observation provides strong evidence that efficient cell-to-cell movement contributes directly to mesophyll invasion from the vascular system of systemically infected leaves.
The viral genetic determinants from TGMV that were necessary for mesophyll invasion included cis-acting (BRi) and trans-acting (AL2) factors required for BR1 movement protein gene expression (Sunter and Bisaro, 1992). Thus, effects on the extent or timing of BR1 gene expression are probably responsible for the mesophyll invasion phenotype, at least in part. Previous work has suggested that the BGMV AL2 protein and the BR1 and BL1 movement proteins are less well adapted to function in N. benthamiana than are their TGMV homologs (Schaffer et al., 1995; Gillette et al., 1998). Given these observations, the viral genetic data can be interpreted to indicate that a critical threshold of BR1 protein activity may be required for successful invasion of the mesophyll from the vascular system of systemically infected leaves. We propose that when the BR1 gene is under the control of the TGMV BR1 promoter, this threshold of activity can be attained either by enhanced BR1 expression mediated by the host-adapted TGMV AL2 protein or by the intrinsically greater activity of the host-adapted TGMV BR1 protein. A corollary of this interpretation is that the BGMV BR1 promoter apparently must provide insufficient BR1 gene expression to reach the putative threshold, regardless of the source of AL2 or BR1 proteins. Because BR1 gene expression is necessary for cell-to-cell movement and chlorotic lesion formation (Jeffrey et al., 1996), the BGMV BR1 promoter must be active in mesophyll cells of inoculated leaves. This conclusion is also supported by the observation that a fragment of BGMV DNA B that includes BRi can efficiently drive the AL2-dependent expression of a reporter gene in protoplasts prepared from N. benthamiana leaves (H.-C. Hung and I.T.D. Petty, unpublished results). However, attempts to establish a correlation between tissue tropism and relative BR1 promoter activity in leaf mesophyll protoplasts have been inconclusive (H.-C. Hung and I.T.D. Petty, unpublished results). This raises the possibility that biologically relevant differences in the extent of BR1 gene expression might occur not in the mesophyll cells themselves but within specific cell types of the vascular system from which the mesophyll invasion is launched in systemically infected leaves.
Although the viral genetic elements required for mesophyll invasion in systemically infected leaves were identified conclusively, and a model for their cooperative action could be proposed based on the extent of BR1 protein activity, still unresolved is whether the underlying mechanism involves enhanced viral cell-to-cell movement or suppression of a host defense response. Voinnet et al. (1999) suggested that plant virus tissue tropism could be determined by the interplay between the host response of post-transcriptional gene silencing (PTGS) and viral antisilencing factors. Indeed, some geminiviruses can suppress PTGS during infection of N. benthamiana, and both the AL2 and BL1 proteins have been implicated in this activity (Duan et al., 1997; Voinnet et al., 1999). Because invasion of mesophyll cells by hybrid geminiviruses required either AL2 or BL1 from TGMV, the ability of these proteins to counter PTGS might be responsible for their effect. However, the additional requirement for TGMV noncoding sequences (BRi) is not easily reconciled with a mechanism based on suppression of mesophyll-specific PTGS. As discussed above, TGMV BRi could affect the expression of BR1, and conceivably this protein could have an as yet undiscovered antisilencing activity. However, this model is inconsistent with the phloem-limited phenotype of a hybrid virus that contains all the elements of the TGMV genome except the BL1 ORF (Table 2). Instead, the phenotype of this hybrid virus suggests that cell-to-cell movement efficiency determines tissue tropism, an interpretation that would also be consistent with known activities of the BR1 protein. The threshold effect inferred for BR1 activity could then be explained by competition between the virus and normal cellular cargoes for access to endogenous intracellular or intercellular trafficking machinery (e.g., see Kragler et al., 1998). Thus, although suppression of PTGS may play an important role during systemic infection of N. benthamiana by geminiviruses, it appears unlikely that this alone determines their tissue tropism.
In conclusion, we have shown that the tissue tropism of begomoviruses is genetically determined and that, as in some animal-infecting viruses, a noncoding region of the viral genome contributes to the phenotype. Taken together, our results suggest that constraints on the efficiency of viral intercellular movement may lead to phloem limitation in the model system under study, although we cannot completely exclude a possible role for virus-mediated suppression of a host defense response. Further studies are required to distinguish between the various models with which our data are consistent and to determine whether similar mechanisms control the tissue tropism of other plant viruses.
METHODS
Plant Infection with Recombinant Viral DNAs
Six-week-old Nicotiana benthamiana seedlings were inoculated by microprojectile bombardment with pUC-based plasmids containing partial tandem dimers of wild-type, mutant, or hybrid geminiviral DNA components, as described previously (Schaffer et al., 1995). Descriptions and sources of plasmids used in this study are presented in Table 3. All of the hybrid viral DNA components used have been previously described, except for pBTBRi1, which is based on the B component of bean golden mosaic virus (BGMV) and contains both the BRi noncoding region and BR1 open reading frame (ORF) from tomato golden mosaic virus (TGMV). This plasmid was constructed by isolating from pGTR (Schaffer et al., 1995) a 2292-bp SnaBI-BssHII fragment containing the TGMV BR1 ORF and the BGMV BL1 ORF, BLi and CR noncoding regions; this fragment was then used to replace the equivalent 2289-bp SnaBI-BssHII fragment of pGTBRSS (Table 3). Plant growth conditions were as described in Schaffer et al. (1995). At 14 days postinoculation, leaf tissue was harvested from the apex of the plant; the presence of viral DNA was confirmed, after nucleic acid extraction, by DNA gel blotting and analysis as described by Jeffrey et al. (1996).
Table 3.
Recombinant Viral DNA Components Used
| Description | Plasmid Designation | Reference |
|---|---|---|
| Wild-type and mutant viral DNA components | ||
| TGMV A, wild type | pTG1.3A | Fontes et al. (1994) |
| TGMV ar1 mutant | pTG1.3ARfsX | Pooma et al. (1996) |
| TGMV al3 mutant | pTGA45 | Sunter et al. (1990) |
| TGMV B, wild type | pTG1.4B | Fontes et al. (1994) |
| BGMV A, wild type | pGA1.2A | Fontes et al. (1994) |
| BGMV B, wild type | pGA1.2B | Fontes et al. (1994) |
| TGMV-based hybrid DNA components | ||
| Elements from BGMV | ||
| AL23 | pTBAL2SaBB | Gillette et al. (1998) |
| BL1 | pTBL3 | Schaffer et al. (1995) |
| BL1/BR1 | pTBLR2 | Schaffer et al. (1995) |
| BRi | pTG1.2B∇R | Petty et al. (2000) |
| BRi, BL1/BR1 | pGA1.2B-T3∇L | Petty et al. (2000) |
| BGMV-based hybrid DNA components | ||
| Elements from TGMV | ||
| AL23 | pGTAL2SaBB | Gillette et al. (1998) |
| ARi, AR1, AL23 | pGTARSSa | Petty et al. (2000) |
| BL1/BR1 | pGTLR2 | Schaffer et al. (1995) |
| BRi | pGTBRSS | Petty et al. (2000) |
| BRi, BL1/BR1 | pGTBSN | Petty et al. (2000) |
| BRi, BR1 | pBTBRi1 | This study |
DNA in Situ Hybridization
At 14 days postinoculation, pieces were cut from leaves two nodes below the plant apex. The leaf pieces were fixed with 4% paraformaldehyde and subsequently processed into ∼50-μm-thick tissue sections by using a Vibratome, as described by Nagar et al. (1995). Prehybridization, hybridization, and washing of tissue sections were performed as described for Drosophila embryo whole-embryo mounts by Tautz and Pfeiffle (1989). Plasmid clones containing a 182-bp NcoI-SalI fragment from the AL1 gene of either TGMV (pTNS) or BGMV (pBNS) in pBluescriptII KS+ (Stratagene) were used to generate probes for DNA in situ hybridization. Plasmids were linearized by digestion with EcoRI, and digoxigenin (DIG)-labeled in vitro transcripts were synthesized using bacteriophage T3 RNA polymerase and DIG-UTP RNA labeling mix (Boehringer Mannheim). The resulting DIG-labeled single-stranded (ss) RNAs were complementary to the viral ssDNA of the TGMV or BGMV A component. After the hybridized tissue sections were washed, the DIG-labeled probe was detected by incubation with horseradish peroxidase (HRP)–conjugated anti-DIG antiserum (Boehringer Mannheim), tyramide-mediated amplification of HRP (TSA Indirect kit; NEN Life Science Products), and visualization by reaction with the HRP substrate 3-amino-9-ethylcarbazole (Vector Laboratories). The tissue sections were mounted in 50% glycerol and observed by light microscopy with Hoffman modulation contrast optics, during which the identities and numbers of infected cells were recorded. Numerical data were recorded only for cells that could be unambiguously identified as vascular associated (companion cells, vascular parenchyma, and bundle sheath) or mesophyll (spongy cells or palisade cells) cells. We did not attempt to differentiate among the various types of vascular-associated cells except in transverse sections of minor veins, where they were identified according to the plan of N. benthamiana minor veins described in Roberts et al. (1997). Images of selected sections were captured by light microscopy with differential interference contrast optics by using charge-coupled device cameras and NIH-Image, Scion Image, or SPOT software. The contrast and gamma of digitized images were subsequently adjusted with Adobe Photoshop software (Adobe Systems, Inc., San Jose, CA).
Acknowledgments
We thank David Bisaro for kindly providing pTGA45, Tara Meade for making the pTNS and pBNS plasmids, and Arthur Weissinger for use of the gene gun. We are also grateful to Steve Nagar, Debra Sellon, and Yuri Yamamoto, who provided invaluable advice on in situ hybridization, and to Eric Miller for suggesting the quantitative analysis of mesophyll invasion. We thank Niki Robertson, Linda Hanley-Bowdoin, and Anton Callaway for their comments on the manuscript. Geminivirus-infected plants were maintained in the South Eastern Plant Environment Laboratory at North Carolina State University. This research was supported in part by the North Carolina Agricultural Research Service and by Public Health Service Grant No. GM-48067 from the National Institute of General Medical Sciences.
References
- Azzam, O., Frazer, J., de la Rosa, D., Beaver, J.S., Ahlquist, P., and Maxwell, D.P. (1994). Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology 204, 289–296. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X.S., Flasinski, S., and Nelson, R.S. (1999). Infection of barley by brome mosaic virus is restricted predominantly to cells in and associated with veins through a temperature-dependent mechanism. Mol. Plant-Microbe Interact. 12, 615–623. [Google Scholar]
- Duan, Y.-P., Powell, C.A., Purcifull, D.E., Broglio, P., and Hiebert, E. (1997). Phenotype variation in transgenic tobacco expressing mutated geminivirus movement/pathogenicity (BC1) proteins. Mol. Plant-Microbe Interact. 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Elmer, J.S., Brand, L., Sunter, G., Gardiner, W.E., Bisaro, D.M., and Rogers, S.G. (1988). Genetic analysis of the tomato golden mosaic virus II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 14, 7043–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes, E.P.B., Gladfelter, H.J., Schaffer, R.L., Petty, I.T.D., and Hanley-Bowdoin, L. (1994). Geminivirus replication origins have a modular organization. Plant Cell 6, 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, W.E., Sunter, G., Brand, L., Elmer, J.S., Rogers, S.G., and Bisaro, D.M. (1988). Genetic analysis of tomato golden mosaic virus: The coat protein is not required for systemic spread or symptom development. EMBO J. 7, 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette, W.K., Meade, T.J., Jeffrey, J.L., and Petty, I.T.D. (1998). Genetic determinants of host-specificity in bipartite geminivirus DNA A components. Virology 251, 361–369. [DOI] [PubMed] [Google Scholar]
- Gladfelter, H.J., Eagle, P.A., Fontes, E.P.B., Batts, L., and Hanley-Bowdoin, L. (1997). Two domains of the AL1 protein mediate geminivirus origin recognition. Virology 239, 186–197. [DOI] [PubMed] [Google Scholar]
- Hanson, S.F., Hoogstraten, R.A., Ahlquist, P., Gilbertson, R.L., Russell, D.R., and Maxwell, D.P. (1995). Mutational analysis of a putative NTP-binding domain in the replication-associated protein (AC1) of bean golden mosaic geminivirus. Virology 211, 1–9. [DOI] [PubMed] [Google Scholar]
- Hoogstraten, R.A., Hanson, S.F., and Maxwell, D.P. (1996). Mutational analysis of the putative nicking motif in the replication-associated protein (AC1) of bean golden mosaic geminivirus. Mol. Plant-Microbe Interact. 9, 594–599. [DOI] [PubMed] [Google Scholar]
- Jeffrey, J.L., Pooma, W., and Petty, I.T.D. (1996). Genetic requirements for local and systemic movement of tomato golden mosaic virus in infected plants. Virology 223, 208–218. [DOI] [PubMed] [Google Scholar]
- Kallender, H., Petty, I.T.D., Stein, V.E., Panico, M., Blench, I.P., Etienne, A.T., Morris, H.R., Coutts, R.H.A., and Buck, K.W. (1988). Identification of the coat protein gene of tomato golden mosaic virus. J. Gen. Virol. 69, 1351–1357. [Google Scholar]
- Kim, K.S., Shock, T.L., and Goodman, R.M. (1978). Infection of Phaseolus vulgaris by bean golden mosaic virus: Ultrastructural aspects. Virology 89, 22–33. [DOI] [PubMed] [Google Scholar]
- Kragler, F., Monzer, J., Shash, K., Xoconostle-Cázares, B., and Lucas, W.J. (1998). Cell-to-cell transport of proteins: Requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J. 15, 367–381. [Google Scholar]
- Lucas, W.J., and Gilbertson, R.L. (1994). Plasmodesmata in relation to viral movement within leaf tissues. Annu. Rev. Phytopathol. 32, 387–411. [Google Scholar]
- Nagar, S., Pedersen, T.J., Carrick, K.M., Hanley-Bowdoin, L., and Robertson, D. (1995). A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, R.S., and van Bel, A.J.E. (1997). The mystery of virus trafficking into, through and out of vascular tissue. Prog. Bot. 59, 477–533. [Google Scholar]
- Noueiry, A.O., Lucas, W.J., and Gilbertson, R.L. (1994). Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76, 925–932. [DOI] [PubMed] [Google Scholar]
- Orozco, B.M., and Hanley-Bowdoin, L. (1998). Conserved se-quence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273, 24448–24456. [DOI] [PubMed] [Google Scholar]
- Orozco, B.M., Gladfelter, H.J., Settlage, S.B., Eagle, P.A., Gentry, R.N., and Hanley-Bowdoin, L. (1998). Multiple cis elements contribute to geminivirus origin function. Virology 242, 346–356. [DOI] [PubMed] [Google Scholar]
- Petty, I.T.D., Miller, C.G., Meade-Hash, T.J., and Schaffer, R.L. (1995). Complementable and noncomplementable host adaptation defects in bipartite geminiviruses. Virology 212, 263–267. [DOI] [PubMed] [Google Scholar]
- Petty, I.T.D., Carter, S.C., Morra, M.R., Jeffrey, J.L., and Olivey, H.E. (2000). Bipartite geminivirus host-adaptation determined cooperatively by coding and noncoding sequences of the genome. Virology, 277, 429–438. [DOI] [PubMed] [Google Scholar]
- Pooma, W., and Petty, I.T.D. (1996). Tomato golden mosaic virus open reading frame AL4 is genetically distinct from its C4 analogue in monopartite geminiviruses. J. Gen. Virol. 77, 1947–1951. [DOI] [PubMed] [Google Scholar]
- Pooma, W., Gillette, W.K., Jeffrey, J.L., and Petty, I.T.D. (1996). Host and viral factors determine the dispensability of coat protein for bipartite geminivirus systemic movement. Virology 218, 264–268. [DOI] [PubMed] [Google Scholar]
- Roberts, A.G., Santa Cruz, S., Roberts, I.M., Prior, D.A.M., Turgeon, R., and Oparka, K.J. (1997). Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushing, A.E., Sunter, G., Gardiner, W.E., Dute, R.R., and Bisaro, D.M. (1987). Ultrastructural aspects of tomato golden mosaic virus infection in tobacco. Phytopathology 77, 1231–1236. [Google Scholar]
- Sanderfoot, A.A., and Lazarowitz, S.G. (1995). Cooperation in viral movement: The geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Ingham, D.J., and Lazarowitz, S.G. (1996). A viral movement protein as a nuclear shuttle. Plant Physiol. 110, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R.L., Miller, C.G., and Petty, I.T.D. (1995). Virus and host-specific adaptations in the BL1 and BR1 genes of bipartite geminiviruses. Virology 214, 330–338. [DOI] [PubMed] [Google Scholar]
- Sunter, G., and Bisaro, D.M. (1992). Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4, 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter, G., Hartitz, M.D., Hormuzdi, S.G., Brough, C.L., and Bisaro, D.M. (1990). Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179, 69–77. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and Pfeiffle, C. (1989). A nonradioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals a translational control of the segmentation gene hunchback. Chromosoma 98, 81–85. [DOI] [PubMed] [Google Scholar]
- Tyler, K.L., and Fields, B.N. (1996). Pathogenesis of viral infections. In Virology, B.N. Fields, D.M. Knipe, P.M. Howley, R.M. Chanock, J.L. Melnick, T.P. Monath, B. Roizman, and S.E. Straus, eds (Philadelphia: Lippincott-Raven Publishers), pp. 173–218.
- Voinnet, O., Pinto, Y.M., and Baulcombe, D.C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.L., Gilbertson, R.L., and Lucas, W.J. (1996). Spatial and temporal distribution of bean dwarf mosaic geminivirus in Phaseolus vulgaris and Nicotiana benthamiana. Phytopathology 86, 1204–1214. [Google Scholar]
- Ward, B.M., and Lazarowitz, S.G. (1999). Nuclear export in plants: Use of geminivirus movement proteins for a cell-based export assay. Plant Cell 11, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]