Abstract
Segregation analysis between Lysopersicon esculentum (cultivated tomato) and L. hirsutum (wild form) in conjunction with positional verification by using near-isogenic lines demonstrated that biosynthesis of two structurally different classes of sesquiterpenes in these species is controlled by loci on two different chromosomes. A locus on chromosome 6, Sesquiterpene synthase1 (Sst1), was identified for which the L. esculentum allele is associated with the biosynthesis of β-caryophyllene and α-humulene. At this same locus, the L. hirsutum allele is associated with biosynthesis of germacrene B, germacrene D, and an unidentified sesquiterpene. Genomic mapping, cDNA isolation, and heterologous expression of putative sesquiterpene synthases from both L. esculentum and L. hirsutum revealed that Sst1 is composed of two gene clusters 24 centimorgans apart, Sst1-A and Sst1-B, and that only the genes in the Sst1-A cluster are responsible for accumulation of chromosome 6–associated sesquiterpenes. At a second locus, Sst2, on chromosome 8, the L. hirsutum allele specified accumulation of α-santalene, α-bergamotene, and β-bergamotene. Surprisingly, the L. esculentum allele for Sst2 is not associated with the expression of any sesquiterpenes, which suggests that cultivated tomato may have a nonfunctional allele. Sesquiterpene synthase cDNA clones on chromosome 6 do not cross-hybridize on genomic DNA gel blots with putative sesquiterpene synthases on chromosome 8, an indication that the genes in Sst1 and Sst2 are highly diverged, each being responsible for the biosynthesis of structurally different sets of sesquiterpenes.
INTRODUCTION
Terpenoids are a structurally highly diverse and abundant family of natural compounds, of which only a limited number function in primary life processes, such as steroidal compounds as structural components of membranes, phytol and carotenoid pigments in photosynthesis, and the prenylated moieties ubiquinone and plastoquinone. Most terpenoids have been found to perform secondary functions, primarily in defense against insect herbivores, but also, for example, as components of the scent of flowers, such as limonene in citrus, geraniol in geranium and roses, and β-ionone in violets, clearly playing a role as attractants of pollinators.
Terpenoid biosynthesis begins with the commitment of three molecules of acetate to the formation of isopentenyl diphosphate (which contains five carbon atoms: C5). The subsequent key reaction in terpenoid biosynthesis is the isomerization of isopentenyl diphosphate to dimethylallyl diphosphate. Condensation of these C5 isomers by a prenyltransferase forms geranyl diphosphate (C10) as a precursor for monoterpenes. Subsequent additions of isopentenyl diphosphate result in the formation of farnesyl diphosphate (C15) as a precursor for sesquiterpenes, and geranylgeranyl diphosphate (C20) as a precursor for diterpenes. Terpene synthases transform these common linear prenyl diphosphate intermediates to a wide variety of mostly cyclic products. These compounds are then often modified further by oxidations, reductions, and isomerizations to form an even greater structural variety.
Several sesquiterpene synthases from various Solanaceous species have been cloned and heterologously expressed; elucidation of their three-dimensional protein structure has provided detailed insights into their reaction mechanisms (Lesburg et al., 1997; Starks et al., 1997). Sesquiterpene synthases accept the linear C15 precursor farnesyl diphosphate and generate a carbocation by releasing the negatively charged diphosphate group. The released negative charge of the diphosphate is offset through Mg2+ ions bound by a common DDXD sequence. Cyclization into a wide variety of cyclic products then occurs through an unusual carbocation mechanism to initiate the various hydrocarbon condensation and cyclization reactions.
Evolutionary studies of the chemical ecology of plants and insects suggest a series of reiterating reciprocal adaptation events between plants and herbivores, leading to a high diversity of defense compounds in plants and coevolving adaptation strategies in herbivores (Ehrlich and Raven, 1964; Gould, 1988; Harborne, 1993). Lycopersicon spp accumulate a wide array of secondary compounds, particularly in glandular trichomes on the leaf surface, where they form the first line of chemical defense against insect herbivores. Sesquiterpene hydrocarbons, sesquiterpene acids, and methyl ketones are found in the type VI glandular trichomes of L. hirsutum f typicum and L. hirsutum f glabratum and are implicated in resistance against spider mite, Colorado potato beetle, and beet armyworm (Sinha and Juvik, 1987; Carter et al., 1989a, 1989b; Weston et al., 1989; Synder et al., 1993; Eigenbrode et al., 1994). Several of the sesquiterpene acids accumulated by L. hirsutum LA1777, however, have been found to actually attract the moth Helicoverpa zea for oviposition (Coates et al., 1988), illustrating the mechanisms of plant–insect coevolution on a molecular level.
Despite the very recent divergence of the genus Lycopersicon from Solanum 12 million years ago, species and accessions within Lycopersion exhibit a remarkable chemical diversity of terpenes (Lundgren et al., 1985; Urbasch, 1985; Coates et al., 1988; Juvik et al., 1988; Rahimi and Carter, 1993; Moniz and Drouin, 1996). Although plant–insect coevolutionary mechanisms may account for the selective origin of this chemical diversity, the biochemical and genetic bases of terpene synthesis and diversity are largely unknown. The goal of the current study was to investigate aspects of these questions as they pertain to sesquiterpene biosynthesis in two Lycopersicon species, the wild tomato species L. hirsutum LA1777 and the tomato cultivar L. esculentum E6203. Specifically, we have investigated how the genetic organization of sesquiterpene synthases in the tomato genome, in conjunction with the biochemical characteristics of the individual sesquiterpene synthases, controls the observed diversity in sesquiterpenes. Another intriguing question we have addressed is whether any clear correspondence exists between the degree of sequence divergence between different sesquiterpene synthases and the structural diversity of their end products. An understanding of the biochemical and genetic intricacies of sesquiterpene biosynthesis eventually may allow evaluation of the current theories explaining the structural diversity and functions of secondary metabolites (reviewed in Gould, 1988; Harborne, 1993). To undertake this study of sesquiterpene biosynthesis in a combined biochemical, genetic, and evolutionary context in Lycopersicon, we found the availability of a well-developed genetic map and a host of genetic resources for Lycopersicon to be critical (Tanksley et al., 1992; Monforte and Tanksley, 2000).
RESULTS
Sesquiterpene Identification and Screening of Near-Isogenic Lines
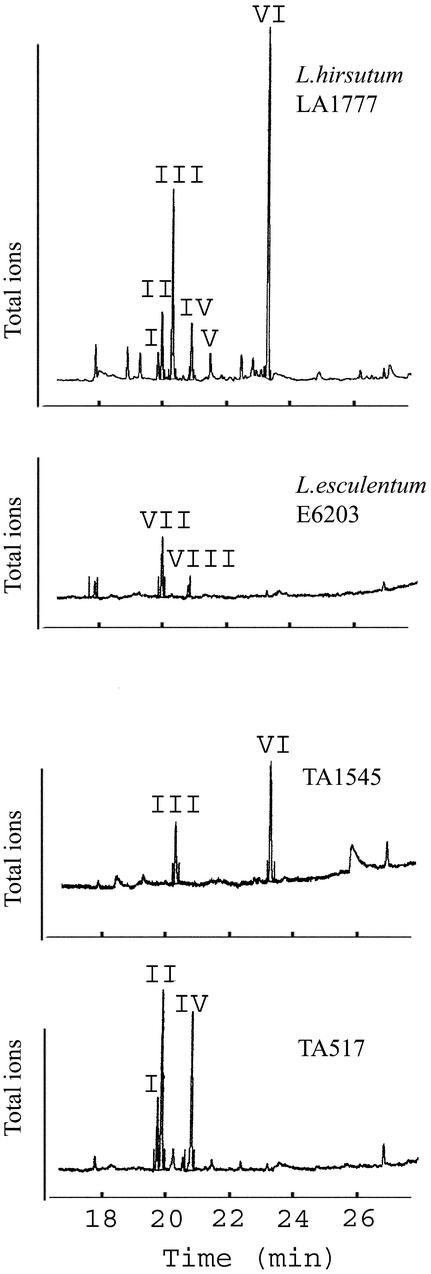
L. hirsutum LA1777 is resistant to a wide variety of insects and produces a sesquiterpene profile greatly different from that of the cultivated tomato, L. esculentum (Figures 1 and 2). To determine the genetic basis for the differences in sesquiterpene hydrocarbons between these two species, we examined a series of 38 near-isogenic lines (NILs) and L. esculentum E6203 and L. hirsutum LA1777 parents. The NILs carry defined introgressions of the L. hirsutum LA1777 genome in an otherwise L. esculentum background and were selected to cover most of the L. hirsutum genome (Bernacchi et al., 1998; Monforte and Tanksley, 2000). Only those NILs carrying introgressions encoding the L. hirsutum sesquiterpene synthases were expected to accumulate L. hirsutum sesquiterpenes, whereas all of the other NILs would display an L. esculentum sesquiterpene profile. This approach identified two NILs, TA1545 and TA517, each producing a subset of the L. hirsutum sesquiterpene spectrum and each representing two different structural classes (Figures 1 and 2).
Figure 1.
Gas Chromatograms (Total Ion Chromatogram Mode) of L. hirsutum LA1777, L. esculentum E6203, and NILs TA517 and TA1545.
TA517 contains β-caryophyllene (VII) and α-humulene (VIII) in addition to the indicated compounds I, II, and IV, but these compounds are detected only as small shoulder peaks of α-santalene (II) and β-bergamotene (IV), respectively (not shown). Peak assignments correspond to those in Figure 2.
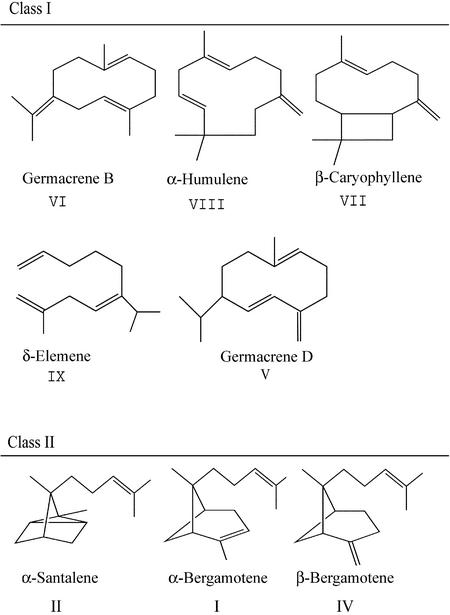
Figure 2.
Structures of Sesquiterpene Hydrocarbons Identified in the Hydrocarbon Fractions of Tomato Leaf Extracts and End Products of Heterologous Expression Studies.
Classification is based on structural similarities and genetic associations of sesquiterpenes. The molecular structure of compound III (in Figure 1) was not identified.
Class I compounds in L. hirsutum consisted of germacrene B (VI), trace amounts of germacrene D (V), and an unidentified sesquiterpene (unknown III) with a mass spectrum very similar to those for isocaryophyllene and β-caryophyllene but with a different retention time (Figures 1 and 2, and Table 1). Although in many L. hirsutum samples, unknown III appeared to be the second most abundant sesquiterpene hydrocarbon, insufficient amounts of this compound were found in the crude bulk preparations of L. hirsutum sesquiterpenes, precluding nuclear magnetic resonance (NMR) analysis. Plants for the crude bulk preparation were harvested relatively late in the growing season (mid-August 1998), and the near absence of unknown III might be the result of seasonal changes in accumulation of this compound. Spectral analysis and retention time comparison showed that unknown III did not represent one of the common thermal decomposition products of germacrene D (V) or germacrene B (VI), β-elemene (for V) and γ-elemene (for VI), respectively. In L. esculentum, class I compounds are represented by two sesquiterpenes, α-humulene (VIII) and β-caryophyllene (VII). The finding of these compounds is in agreement with other reports, although trace amounts of δ-elemene (IX) also have been reported in L. esculentum (Lundgren et al., 1985; Urbasch, 1985).
Table 1.
Quantification of Sesquiterpenes (Micrograms of Sesquiterpene/100 mg Fresh Weight Leaf Tissue)a
| Compound/Line | Peak No. | L. esculentum E6203 | L. hirsutum LA1777 | NIL TA517 | NIL TA1545 |
|---|---|---|---|---|---|
| Germacrene B | VI | 0 | 36.63 (8.57) | 0 | 5.02 (1.57) |
| Germacrene D | V | 0 | 2.17 (0.43) | 0 | NDb |
| Unknown III | III | 0 | 19.78 (5.93) | 0 | 2.71 (0.32) |
| β-Caryophyllene | VII | 2.33 (0.50) | 0 | ND | 0 |
| α-Humulene | VIII | 0.43 (0.32) | 0 | ND | 0 |
| α-Bergamotene | I | 0 | 2.17 (0.23) | 2.99 (0.32) | 0 |
| α-Santalene | II | 0 | 8.68 (1.15) | 8.51 (0.87) | 0 |
| β-Bergamotene | IV | 0 | 4.74 (0.89) | 7.22 (0.71) | 0 |
Average values and standard deviation (in parentheses) of three plants per line are given.
ND, not defined. Concentrations could not be quantified due to the low abundance of these compounds and coelution of β-caryophyllene and α-humulene with α-santalene and β-bergamotene, respectively.
NIL TA1545, containing a large L. hirsutum introgression on chromosome 6, accumulated the class I hirsutum sesquiterpenes germacrene B, unknown III, and trace amounts of germacrene D. However, L. esculentum class I β-caryophyllene and α-humulene were lacking in this NIL. Substitution of the germacrenoid L. hirsutum sesquiterpenes for the L. esculentum caryophyllenes (β-caryophyllene and α-humulene) strongly suggests that the responsible synthases for these class I sesquiterpenes are encoded by different alleles at one or more loci on chromosome 6.
Class II sesquiterpenes of L. hirsutum are α-santalene (II), α-bergamotene (I), and β-bergamotene (IV) (Figures 1 and 2, and Table 1). Interestingly, L. esculentum had no sesquiterpenes structurally related to the class II sesquiterpenes, which share a common unsaturated methyl-isobranched C6 chain that suggests their differential biogenic origin from class I sesquiterpenes (Figure 2). Previously, the presence of α-santalenoic, α-bergamotenoic, and β-bergamotenoic acid was reported in L. hirsutum LA1777 (Coates et al., 1988). Those authors anticipated the presence of the corresponding sesquiterpene hydrocarbons, as is shown here, in L. hirsutum LA1777 as substrates for oxidation to their cognate sesquiterpene acids.
The second NIL, TA517, with a large introgression on chromosome 4 (and a previously undetected small introgression on chromosome 8, as discussed in the next section), accumulated the class II L. hirsutum sesquiterpenes α-santalene, α-bergamotene, and β-bergamotene, while still producing the class I L. esculentum sesquiterpenes α-humulene and β-caryophyllene. However, the L. esculentum sesquiterpenes were difficult to observe in TA517 because their lower abundance and their retention times were almost identical with those for α-santalene and β-bergamotene, respectively (Figure 1 and Table 1). The occurrence of both class I and class II sesquiterpenes in TA517 suggests that the synthases for the two structural classes are nonallelic and can be genetically separated, as demonstrated by sesquiterpene profiles of NILs TA1545 and TA517.
In our opinion, the TA1545 and TA517 introgressed regions can safely be assumed to somewhere contain the class I and class II L. hirsutum sesquiterpene synthases, respectively. However, we cannot at this point exclude the possibility that other regulatory genes are involved in the observed separation of class I and class II sesquiterpene biosynthesis in the introgression lines. Nonetheless, we think it highly unlikely that a regulatory locus alone, without additionally introgressed L. hirsutum sesquiterpene synthases, could have resulted in the observed separation of L. hirsutum sesquiterpene classes, because any hypothetical L. hirsutum regulatory locus, in an otherwise L. esculentum background, would have to interact with one of the L. hirsutum sesquiterpene synthases to cause accumulation of L. hirsutum sesquiterpenes. The only other scenario in which this could happen is in the unlikely situation in which an L. hirsutum regulatory locus somehow would modify the end product specificity of an L. esculentum sesquiterpene synthase (or synthases) to yield a specific subset of L. hirsutum sesquiterpenes. In addition, the involvement of any regulatory locus in the NILs would predict the presence of two controlling loci (the regulatory locus and the actual sesquiterpene synthase) within the boundaries of the L. hirsutum introgressed regions. Therefore, to confirm these notions and to examine further the genetic basis of sesquiterpene biosynthesis, populations that segregated for classes I and II sesquiterpenes were subjected to segregation analysis.
Molecular Mapping of Class I Sesquiterpene Synthases
Because the chromosome 6 NIL (TA1545) displayed an altered class I sesquiterpene profile, we expected the genes for synthesis of the class I compounds to be located on chromosome 6. To test this hypothesis, we analyzed a BC1S1 population, derived from a L. hirsutum × L. esculentum cross and segregating for chromosome 6, for cosegregation of a series of chromosome 6 restriction fragment length polymorphism markers with class I sesquiterpenes germacrene B, β-caryophyllene, and unknown III. However, not all sesquiterpenes could be reliably quantified. Because of its low abundance, germacrene D (class I) could not be monitored, and α-humulene (class I) could not be assayed quantitatively because of its coelution with β-bergamotene (class II; see Figures 1 and 2).
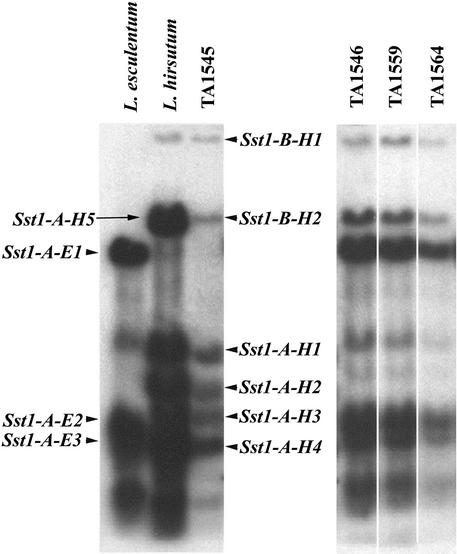
In addition, a 1-kb fragment of sesquiterpene synthase cDNA was amplified from L. esculentum for use as a restriction fragment length polymorphism marker (Figures 3 and 4; SST probe). Polymerase chain reaction (PCR) primers for the SST probe were based on the sequence of germacrene C synthase (GCS), a sesquiterpene synthase previously isolated from cherry tomato cv VFNT (Colby et al., 1998). Segregation analysis of the chromosome 6 markers and the SST candidate gene probe revealed two separate but linked (24 centimorgans [cM] apart) clusters of genes on chromosome 6, Sst1-A and Sst1-B (for sesquiterpene synthase1 A and B; Figures 3 and 4, and Table 2). The Sst1-A cluster showed the highest association with class I sesquiterpenes of all markers tested on chromosome 6, suggesting that Sst1-A genes are responsible for class I sesquiterpene synthases (Table 2). The Sst1-A cluster consists of at least four cosegregating L. hirsutum restriction fragments (with DraI) and three allelic fragments in L. esculentum. The Sst1-B cluster consists of two more weakly hybridizing bands, present only in L. hirsutum (Figure 3). No allelic restriction fragments for Sst1-B were found in L. esculentum under the stringency conditions used (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate, at 65°C).
Figure 3.
Identification of Two Sesquiterpene Synthase Clusters Sst1-A and Sst1-B.
Sst1-A and Sst1-B are identified by DNA gel blot analysis of parental lines and chromosome 6 NIL TA1545 and subNILs TA1546, TA1559, and TA1564. Sst1-A and Sst1-B suffixes indicate species (E and H) and DraI restriction fragment number. The occurrence of Sst1-A-H5 represents a nonisogenic allele in the parent L. hirsutum and has not been transferred to progeny (NILs and backcross populations). Sst1-A is responsible for accumulation of L. hirsutum sesquiterpenes, because only NIL TA1545 accumulates germacrene B and unknown III. NILs TA1546, TA1559, and TA1564 accumulate β-caryophyllene and α-humulene.
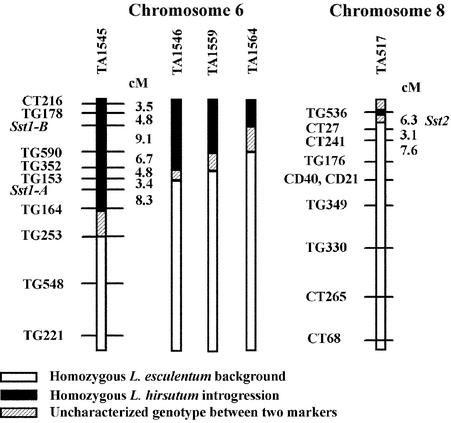
Figure 4.
Linkage Maps of TA1545 (Chromosome 6) and TA517 (Chromosome 8) Introgressions Based on Segregation in Backcross Populations.
Table 2 shows greatest significance for markers CT27 and Sst1-A cluster with the two classes of L. hirsutum sesquiterpenes. Genotypes of TA1545 subNILs TA1546, TA1559, and TA1564 do not contain Sst1-A and do not produce L. hirsutum class I sesquiterpenes.
Table 2.
Segregation Analysis of Sesquiterpene Biosynthesis in Backcross Populationsa
| Compound Line | Chromosome | Locus | P Value | R2 | Population | HHb | EHc | EEd |
|---|---|---|---|---|---|---|---|---|
| Germacrene B | 6 | Sst1-A | 0.00 | 0.27 | BC1S1 ‘98 | 7.87 | 4.95 | 0 |
| Unknown III | 6 | Sst1-A | 0.00 | 0.47 | BC1S1 ‘98 | 4.29 | 4.50 | 0 |
| β-Caryophyllene | 6 | Sst1-A | 0.00 | 0.21 | BC1S1 ‘98 | 0 | 0.71 | 4.52 |
| α-Santalene | 8 | CT27 | 0.00 | 0.29 | BC2S1 ‘99 | 6.05 | 2.80 | 0.01 |
| α-Bergamotene | 8 | CT27 | 0.00 | 0.28 | BC2S1 ‘99 | 2.52 | 1.19 | 0.01 |
Markers with highest significance for sesquiterpenes are listed. Genotypic means expressed as micrograms of sesquiterpene/100 mg leaf fresh weight. Sst1-A represents a cluster of four restriction fragments detected with the SST probe.
HH, homozygous L. hirsutum.
EH, heterozygotes.
EE, homozygous L. esculentum.
To determine unambiguously whether the Sst1-A cluster was responsible for class I sesquiterpene biosynthesis, the two clusters Sst1-A and Sst1-B were genetically dissected using a set of chromosome 6 subNILs. As shown in Figures 3 and 4, subNILs TA1546, TA1559, and TA1564 contain only the L. hirsutum Sst1-B cluster, but their chemical profiles were identical with L. esculentum (data not shown). From these results, we concluded that only the Sst1-A cluster was responsible for biosynthesis of the chromosome 6–associated sesquiterpenes and that none of the sesquiterpenes assayed was associated with the Sst1-B cluster. The Sst1-A hybridization pattern suggested that a low-copy-number gene family was responsible for class I sesquiterpene synthesis in Lycopersicon.
Molecular Mapping of Genes Controlling Class II Sesquiterpene Biosynthesis
The same rationale and experimental approach with genomic markers were applied to analyze NIL TA517, which produces the class II sesquiterpenes α-santalene, α-bergamotene, and β-bergamotene. On a genomic DNA gel blot, the SST probe revealed no L. hirsutum alleles in TA517 (data not shown), indicating that the synthases responsible for class II sesquiterpenes had diverged to the extent that they no longer cross-hybridize with the class I synthase probe under moderately stringent washes (1 × SSC at 65°C). To test for the possibility that the chromosome 4 introgression of TA517 contains the class II synthases, we analyzed the BC1S1 plants for cosegregation of chromosome 4 markers with class II sesquiterpenes. Surprisingly, the chromosome 4 markers did not cosegregate with α-santalene and the bergamotenes as expected. Apparently, therefore, the class II sesquiterpene synthases are not located on the chromosome 4 introgressed segment. Because TA517 does accumulate L. hirsutum class II sesquiterpenes, it must contain a small, undetected contamination of the L. hirsutum genome elsewhere encoding the L. hirsutum synthase or synthases for α-santalene and the bergamotenes.
Therefore, another genetic approach was applied to identify the chromosomal location of the gene or genes responsible for class II sesquiterpene biosynthesis formed in TA517. A set of four segregating BC2S1 populations was selected in such a way that every chromosome segment of the L. hirsutum genome would segregate in at least one of the populations. Of the four BC2S1 populations, one (98T284) accumulated and segregated for the class II L. hirsutum sesquiterpenes α-santalene, α-bergamotene, and β-bergamotene. Previous genotyping, during construction of the NIL population, had demonstrated that population 98T284 would segregate for parts of chromosome 2, 5, and 8 not represented in the other BC2S1 populations (Monforte and Tanksley, 2000). Marker analysis of only these regions revealed several markers on chromosome 8 cosegregating with accumulation of the class II sesquiterpenes introgressed into TA517 (Table 2). Based on these results, NIL TA517 was probed with chromosome 8 markers and found to contain a small, previously undetected introgression near the centromere on chromosome 8, as revealed by probe TG536 (Figure 4). Because marker CT27 (6.3 cM from TG536) displayed the greatest association in the 98T284 population based on single-point quantitative trait loci analysis, we concluded that any class II sesquiterpene synthases referred to as the Sst2 locus are most likely located in the 6.3-cM interval between CT27 and TG536 on chromosome 8 (Figure 4).
The class II sesquiterpenes α-santalene, α-bergamotene, and β-bergamotene were always either present or absent together in individual plants from the segregating population, indicating that these sesquiterpenes accumulate as a result of either very closely linked genes or a single sesquiterpene synthase with multiple end products. Several plant sesquiterpene synthases are known to yield more than one end product. For example, grand fir (Abies grandis) has two multiple end product synthases, which produce 36 and 54 different end products, respectively (Bohlmann et al., 1998). Furthermore, Colby et al. (1998) isolated from cherry tomato cv VFNT and functionally expressed a germacrene C synthase (GCS), which produces germacrene C as well as smaller amounts of germacrene A, B, and D, in accordance with the sesquiterpene composition of this plant.
Isolation of Chromosome 6 Sesquiterpene Synthases and Heterologous Expression in Escherichia coli
In the hope of shedding light on how Sst1-A, the chromosome class I sesquiterpene synthase cluster on chromosome 6, controls class I sesquiterpene synthesis, we used the SST probe (previously used to detect the Sst1-A and Sst1-B clusters on chromosome 6) to isolate full-length cDNA transcripts from both L. esculentum and L. hirsutum cDNA libraries. Subsequent heterologous expression studies in E. coli and analyses of sesquiterpene end products could confirm the genetic association of class I sesquiterpenes with the Sst1 clusters on chromosome 6. Given the demonstration genetically that only the Sst1-A cluster is responsible for sesquiterpene biosynthesis, any isolated sesquiterpene synthase transcripts are most likely to be encoded by genes in this cluster.
The 10 largest cDNA clones from the L. hirsutum library were sequenced. They appeared to fall into two classes, representing transcripts from two different genes. Nine represented SSTLH1 (for sesquiterpene synthase Lycopersicon hirsutum1); only one represented SSTLH2 (Figure 5). Similarly, two different L. esculentum transcript types were isolated: six clones of SSTLE1 and four clones of SSTLE2. SSTLE1 and SSTLE2 are identical, except for the 5′ end of SSTLE2, which appears truncated at exactly the same position for all four of the SSTLE2 clones, possibly because of hairpin structures of the mRNAs during first-strand synthesis in cDNA library construction. Whether the two different L. esculentum transcripts represent different 5′-end splice variants of the same gene or are transcripts of two different genes is unclear.
Figure 5.
Amino Acid Alignment of L. esculentum Sesquiterpene Synthases SSTLE1-2 and SSTLH1-2 Hybridizing with Chromosome 6, and Germacrene C Synthase (Colby et al., 1998).
Amino acid residues discussed in the text are indicated with an asterisk. The black shading denotes the most common residue at any given position in the alignment.
Full-length coding sequences of SSTLH1, SSTLH2, and SSTLE1 were subcloned into pET11a expression vectors (Stratagene) and heterologously expressed in E. coli, from which protein extracts were assayed for sesquiterpene synthase activity (Figure 6). For SSTLE2, a methionine start codon was arbitrarily introduced at the 5′ end before the first coding amino acid, Leu (Figure 5), and was used for heterologous expression analysis in E. coli.
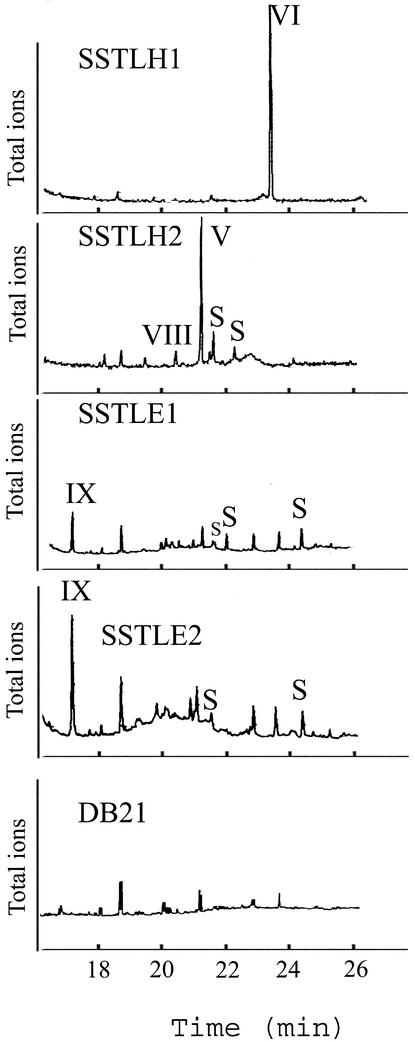
Figure 6.
Gas Chromatograms of Products of Heterologously Expressed Lycopersicon Sesquiterpene Synthases SSTLE1, SSTLE2, SSTLH1, SSTLH2 and Control DB21 (Cells).
Chromatograms were obtained in total ion mode. Reaction products correspond to roman numerals in Figure 2, whereas unidentified sesquiterpenes (with characteristic parental ion of m/z 204) are indicated with an S.
SSTLH1 catalyzed the conversion of the common C15 precursor farnesyl diphosphate to germacrene B in vitro, whereas SSTLH2 catalyzed primarily the conversion to germacrene D, with minor amounts of α-humulene and other unidentified sesquiterpenes (Figure 6, S) being produced. These results are consistent with the previously predicted scenario in which germacrene B and germacrene D are encoded by class I sesquiterpene synthases located in the Sst1-A cluster of sesquiterpene synthases on chromosome 6. In addition, the low abundance of germacrene D in planta in L. hirsutum is reflected in the isolation of nine SSTLH1 (germacrene B synthase) copies and only one SSTLH2 (germacrene D synthase).
The fact that germacrene D concentrations were undetectable in TA1545 could indicate that SSTLH2 is not present in TA1545, as one would expect if SSTLH1 and SSTLH2 were alleles of the same gene (despite their markedly different nucleotide sequence). However, specific PCR amplification and sequence verification of products showed that TA1545 contains both of the genes encoding SSTLH1 and SSTLH2. Because TA1545 is homozygous for this chromosome 6 region, SSTLH1 and SSTLH2 must be derived from nonallelic copies of a small gene family on chromosome 6 (Monforte and Tanksley, 2000).
Heterologously expressed SSTLE1 and SSTLE2 produced only minor amounts of δ-elemene (IX) in addition to even smaller amounts of other unidentified sesquiterpenes (Figure 6). However, these synthases were expected to produce β-caryophyllene and α-humulene, given the L. esculentum sesquiterpene content. An explanation for these results is that the in vitro assay conditions may be conducive to end product artifacts and that SSTLE1 and SSTLE2 are in fact responsible for β-caryophyllene and α-humulene biosynthesis in the plant. Alternately, the presence of other class I sesquiterpene synthases in L. esculentum, which either may be underrepresented in this L. esculentum cDNA library or is sufficiently diverged to preclude detection in this screen, cannot be excluded.
DISCUSSION
Genetic Control of Sesquiterpene Biosynthesis in L. hirsutum and L. esculentum
This study has provided important clues regarding the genetic organization and evolution of sesquiterpene biosynthesis in L. hirsutum and L. esculentum. The biosynthesis of two structurally distinct classes of sesquiterpenes in Lycopersicon species is controlled by loci on two different chromosomes. The genes for class I sesquiterpene synthases are found on chromosome 6 and consist of two low-copy clusters of sesquiterpene synthases, Sst1-A and Sst1-B, of which only Sst1-A is genetically associated with biosynthesis of class I sesquiterpenes. Class II sesquiterpenes are genetically associated with locus Sst2 near the centromere of chromosome 8, for which no putative sesquiterpene synthases have been isolated. Class I and class II sesquiterpene synthases have diverged to the extent that they no longer cross-hybridize on genomic DNA gel blots.
From the results of the hybridization analysis and expression studies, we conclude that the various sesquiterpenes in Lycopersicon originate from only a few expressed synthases, of which several are probably multiple end product synthases. In the L. hirsutum Sst1-A cluster, SSTLH1 and SSTLH2 are responsible for biosynthesis of germacrene B and germacrene D. However, neither of the L. hirsutum synthases produced unknown III in vitro. Therefore, does a third class I sesquiterpene synthase expressed in L. hirsutum account for this sesquiterpene, which may be either underrepresented in this cDNA library or sufficiently diverged to preclude detection in this screen? More probably, the in vitro conditions of the enzyme assay may cause a different end product specificity as compared with the in planta–expressed SSTLH1 or SSTLH2. In partial support of the latter is the high correlation (r > 0.93) between the accumulation of germacrene B and unknown III, which are always present or absent together in segregating BC1S1 plants, thus suggesting either a common multiple end product sesquiterpene synthase (i.e., SSTLH1) for these two compounds or tight linkage of distinct synthases.
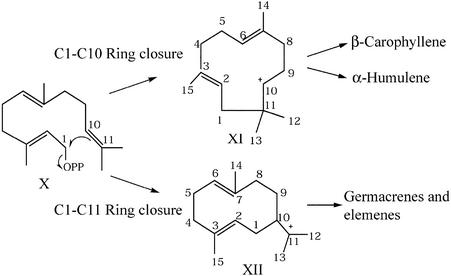
Turning to the L. esculentum alleles at Sst1-A, heterologously expressed SSTLE1 and SSTLE2 produced only minor amounts of δ-elemene (IX) in addition to even smaller amounts of other unidentified sesquiterpenes (Figure 6). However, these synthases were expected to produce β-caryophyllene and α-humulene, given the L. esculentum sesquiterpene content. As for unknown III, perhaps the in vitro assay conditions are conducive to end product artifacts. In support of this idea, the biosynthesis of the structurally related class I germacrenes, elemenes, and caryophyllenes (β-caryophyllene and α-humulene) occurs through a similar reaction mechanism. After the initial generation of a carbocation on C1 through release of the diphosphate moiety from the farnesyl diphosphate (X) substrate, the carbocation attacks the π electrons on C10 to C11, resulting in a C1 to C10 ring closure in the biosynthesis of germacrene and elemene and a C1 to C11 ring closure for β-caryophyllene and α-humulene (Figure 7; Starks et al., 1997; Colby et al., 1998). Imprecisions in the ring closure are most likely responsible for the phenomenon of multiple end product synthases and could lead to artifactual end products in vitro. Considering these mechanisms and the genetic data, it is possible that these sesquiterpene synthases are responsible for β-caryophyllene and α-humulene production in planta (Van der Hoeven, 1999). Transgenic strategies are the next step in resolving questions involving the isolated synthases with respect to multiple end product specificity and in vitro end product artifacts.
Figure 7.
C1 to C10 and C1 to C11 Ring Closure in the Biosynthesis of Germacrenes, Elemenes, and Caryophyllenes.
Release of the diphosphate (OPP) from the linear precursor farnesyl diphosphate (X), followed by attack of the C1 carbocation on the C10 to C11 π electrons, results in two possible carbocation intermediates (XI and XII). After any further carbocation-induced reactions, carbocation intermediates are quenched through release of proton-yielding sesquiterpene hydrocarbons.
Both SSTLE1 and SSTLE2 are highly similar to GCS from VFNT Cherry (Table 3 and Figure 5; Colby et al., 1998). SSTLE1 differs at only four nucleotide positions, which results in four amino acid substitutions in GCS (Figure 5): Ser497, Ser528, Thr529, and Pro538. When expressed in E. coli, GCS produces the class I compounds germacrene C and smaller amounts of germacrene B and D (Colby et al., 1998). Thus, one or more of the four substituted residues in SSTLE1 and SSTLE2 are involved in determining the stereochemistry of the enzyme's active site and consequently affect end product specificity. Sequence comparison of GCS and SSTLE1 with the three-dimensional structure of tobacco epi-aristolochene synthase shows that Ser528 and Thr529 are located directly next to Tyr527 and form a hydrophobic pocket with Trp272, which allows substrate binding and shields the carbocation substrate in the active site from quenching by the aqueous solvent (Figure 5; Starks et al., 1997). Apparently, therefore, these two residues in particular may affect the orientation of Tyr527 in the active site and thereby affect the folding of the linear substrate, resulting in the formation of different sesquiterpene end products. Similar inferences on the contribution of individual residues on end product specificity can be made for SSTLH1 and SSTLH2, but given the high number of amino acid differences between these two, predictions would be highly speculative (Table 3 and Figure 5; Van der Hoeven, 1999).
Table 3.
Percentage of Amino Acid (above the Diagonal as Indicated by Asterisks) and Nucleotide (below the Diagonal) Similarity between cDNAs of Several Lycopersicon Sesquiterpene Synthases
| GCS | SSTLE1 | SSTLE2 | SSTLH1 | SSTLH2 | |
|---|---|---|---|---|---|
| GCS | * | 99.3 | 98.7 | 89.0 | 87.2 |
| SSTLE1 | 99.7 | * | 99.5 | 89.7 | 87.9 |
| SSTLE2 | 99.3 | 93.9 | * | 89.2 | 87.4 |
| SSTLH1 | 93.6 | 93.6 | 93.6 | * | 98.9 |
| SSTLH2 | 92.7 | 93.1 | 92.7 | 93.3 | * |
Evolution of Sesquiterpene Biosynthesis in L. hirsutum and L. esculentum
The L. hirsutum clones display ∼93% nucleotide similarity with the L. esculentum clones as well as between themselves (Table 3). This suggests that the divergence of the two L. hirsutum synthases began approximately at the time of L. esculentum and L. hirsutum speciation. In contrast, the relatively minor sequence differences between SSTLE1, SSTLE2, and GCS (VFNT Cherry) seem to indicate a much more recent divergence. In terms of structural divergence of end products of L. esculentum synthases (GCS, SSTLE1, SSTLE2), the evolution of new class I sesquiterpenoid compounds apparently can be achieved by substituting for a very few amino acids, perhaps only one. However, the much greater sequence divergence observed between L. hirsutum and L. esculentum class I synthases does not result in biosynthesis of compounds that are structurally strikingly different from the class I sesquiterpenes (germacrenoids and caryophyllenes). For example, both the L. esculentum VFNT Cherry synthase GCS and the L. hirsutum synthases SSTLH1 and SSTLH2 produce germacrenoid compounds. In conclusion, from the results presented here, there is no clear correlation between the extent of sequence divergence and structural divergence within the family of class I synthases on chromosome 6.
A puzzling finding is that the substituted residues between GCS and the SSTLE clones (Figure 5)—Ser497, Ser528, Thr529, and Pro538 in GCS—are all nonrandomly distributed at the C terminus, with two residing directly next to a residue constituent of the active site; this raises questions about the spatial distribution of mutations underlying the evolution sesquiterpene synthases. These L. esculentum synthases provide an excellent starting point for studying the individual effects of residue substitutions on active site stereochemistry and end product specificity.
Classes I and II sesquiterpene synthases must be highly diverged from each other, given the lack of SST probe hybridization with any putative class II sesquiterpene synthases on chromosome 8. We propose a scenario in which divergence of two different lineages of class I and class II synthases began well before the speciation of L. hirsutum and L. esculentum and resulted in two different lineages that encode the biosynthesis of structurally quite different sesquiterpenes (classes I and II; see Figure 2). The considerable genetic divergence between class I and class II sesquiterpene synthases is reflected in the structural divergence of their end products. Cloning the L. hirsutum sesquiterpene synthase (or synthases) for α-santalene, α-bergamotene, and β-bergamotene is essential for determining the molecular basis for the absence of cognate L. esculentum sesquiterpenes associated with chromosome 8 and for studying the evolutionary relationship between the two different lineages of class I and class II synthases. Identification of the actual class II sesquiterpene synthases also would help in assessing whether the absence of class II sesquiterpenes in L. esculentum can be viewed as a direct consequence of domestication on sesquiterpene composition.
The combined approach of genetics and biochemistry in this study has proved particularly insightful in explaining the naturally occurring variation in sesquiterpene biosynthesis in Lycopersicon. Further studies may eventually allow for a deeper understanding of this variation in all Solanaceae and provide the tools for transgenic and nontransgenic approaches toward numerous breeding objectives for crop improvement.
METHODS
Plant Material
Three plants of each of 38 near-isogenic lines (NILs; Monforte and Tanksley, 2000), Lycopersicon esculentum E6203 and L. hirsutum LA1777, were grown under greenhouse conditions during winter 1997–1998. (The genotypes of the NILs are available from the authors.) Greenhouses were lit for 14 hr/day with 400-W high-pressure sodium lights. Temperature was controlled to maintain 28°C during the day and 18°C during the night. A BC1S1 population (98T324; 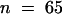 ) and parental control plants were grown under field conditions in summer 1998 in Ithaca, New York. In summer 1999, four BC2S1 populations (99T279, 99T280, 99T281, and 99T284) were grown under field conditions. Only population 99T284 (
) and parental control plants were grown under field conditions in summer 1998 in Ithaca, New York. In summer 1999, four BC2S1 populations (99T279, 99T280, 99T281, and 99T284) were grown under field conditions. Only population 99T284 (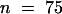 ) segregated for α-santalene, α-bergamotene, and β-bergamotene, and it was used for chemotypic and genotypic analysis. All populations were grown in a completely randomized design. For chemotypic analysis, samples were taken from whole leaves to ensure that sesquiterpenes in both trichomes and the leaf itself were quantified.
) segregated for α-santalene, α-bergamotene, and β-bergamotene, and it was used for chemotypic and genotypic analysis. All populations were grown in a completely randomized design. For chemotypic analysis, samples were taken from whole leaves to ensure that sesquiterpenes in both trichomes and the leaf itself were quantified.
Bulk Extraction for Chemical Compound Identification
Leaf material for chemical identification from L. hirsutum LA1777 and L. esculentum was harvested in mid-August 1998 after a 3-month growth period in the field. Leaf tissue was pooled from several plants of each line (800 g from each line) for bulk extraction. Tissue was coarsely ground in liquid N2 and transferred to a 4-liter Erlenmeyer flask, to which 2 liters of CH2Cl2 was added immediately. The tissue was extracted for 3 hr at room temperature with continuous shaking. The extract was filtered through Whatman (no. 4) filter paper, followed by evaporation of the solvent under vacuum. A small amount of the viscous residue was redissolved in CH2Cl2 and analyzed by gas chromatography–mass spectrometry (GC-MS); the remainder was further purified for NMR analysis.
Compound Purification
The bulk of the residue was dissolved in a minimal amount of hexane/ethyl ether (97:3 [v/v]) and used for flash chromatography (22-mm-diameter column, silica AgNO3 15%, hexane/ethyl ether 97:3; Still et al., 1978). Ten-milliliter fractions were collected, from which 5-μL aliquots were taken for separation on thin-layer chromatography (TLC) plates (Kieselgel 60 F; Merck) eluted with hexane/ethyl ether (97:3). A small amount of phosphomolybdic acid (TLC spray reagent; Applied Science Laboratories) was sprayed onto the TLC plate to visualize the sesquiterpenes, which were heated until the blue color was fully developed. The fractions indicated by TLC to contain sesquiterpenes were checked for purity by GC. The fractions contained the following compounds of interest in the quantities stated at the corresponding purities: α-bergamotene (5.5 mg, 39%; contaminated with 33% α-santalene), α-santalene (5.5 mg, 99%), β-bergamotene (1.8 mg, >92%), and germacrene B (8 mg, >92%). Quantities and purities were determined based on GC analysis of a series of dilutions of β-caryophyllene (Fluka, Neu-Alm, Switzerland) as a reference.
GC and GC-MS
For analysis of samples by GC and GC-MS, we used a DB-FFAP capillary column (30 m × 0.24 mm diameter; JandW Scientific, Folsom, CA) on a Hewlett-Packard 5890 GC equipped with either a flame ionization detector or a Hewlett-Packard 5970 Mass Selective Detector in the total ion chromatogram mode. The oven was programmed to start at 40°C and then increase at 10°C/min to 250°C. The injector port was set at 250°C, and the detector was kept at 300°C. Searches of the Wiley Registry of Mass Spectral Data were performed with BenchTop/PBM Software (Palisade Mass Spectrometry, Newfield, NY) for the peaks of interest. Spectra were confirmed by reference to data in the literature. β-Caryophyllene (Fluka) was used as a reference compound to quantify the sesquiterpene content of the samples by flame ionization detector peak area integration. Ethylcaproate (0.25 mM final concentration) served as an internal standard to correct for solvent evaporation.
The following mass spectra (GC-MS; 70 eV), were obtained:
α-Santalene, m/z 204 (22), 189 (21), 161 (17), 133 (13), 121 (37), 107 (38), 105 (29), 94 (100), 93 (92), 79 (40), 77 (35), 69 (33), 55 (22), 53 (17). Identification was confirmed by comparing the retention time with that for α-santalene from a sample of sandalwood oil (Santalum album; Adams et al., 1975).
α-Bergamotene, m/z 204 (4), 189 (4), 161 (10), 119 (92), 107 (31), 105 (24), 93 (100), 91 (41), 79 (30), 77 (30), 69 (45), 55(26).
β-Bergamotene, m/z 204 (11), 189 (3), 161 (33), 133 (41), 120 (30), 119 (20), 105 (22), 93 (70), 91 (37), 79 (41), 69 (100), 55 (30).
α-Humulene, m/z 204 (12), 189 (6), 161(5), 147(29), 121(36), 119(14), 107 (17), 105 (15), 93 (100), 91(32), 79 (39), 67 (27), 55(15), 53 (18).
β-Caryophyllene, m/z 204 (15), 189 (25), 175 (16), 161 (44), 148 (29), 147 (35), 133 (98), 120 (50), 119 (46), 107 (44), 105 (65), 93 (96), 91 (100), 79 (88), 77 (58), 69 (81), 67 (50), 55 (46), 53 (48). β-Caryophyllene (peak 1) was identified by coinjection with a pure β-caryophyllene standard (Fluka). α-Humulene and β-caryophyllene are reported to constitute the major sesquiterpene hydrocarbons present in the trichome secretions of L. esculentum varieties (Andersson et al., 1980; Urbasch, 1985; Colby et al., 1998).
δ-Elemene, m/z 204 (9), 189 (6), 161 (29), 136 (58), 121 (100), 105 (27), 93 (82), 91 (36), 78 (20), 77 (30), 55 (20).
Germacrene B: m/z 204 (27), 189 (26), 161 (48), 147 (24), 133 (35), 121 (100), 107 (52), 105 (65), 93 (84), 91 (52), 81 (42), 79 (35), 77 (32), 67 (52), 55 (32), 53 (32).
Unknown III, m/z 204 (12), 189 (26), 161 (20), 133 (50), 120 (57), 105 (74), 93 (52), 91 (96), 79 (100), 69 (63), 67 (80), 55 (65), 53 (57).
Germacrene D, m/z 204 (21), 161 (100), 133 (20), 119 (40), 105 (46), 93 (27), 91 (43), 81 (27), 79 (23), 77 (23), 55 (18).
Nuclear Magnetic Resonance
The hexane/ethyl ether solvent of the fractions was evaporated under vacuum, and the fractions were resolubilized in CDCl3 or C6D6. A 400-MHz Varian spectrometer was used for 1H nuclear magnetic resonance (NMR) and 13C NMR. The following chemical shifts were obtained and compared with previously published spectra:
β-Bergamotene, 1H NMR (400 MHz, CDCl3): δ 1.11 (m, 2H), 1.23 (s, 3H), 1.42 (d, 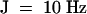 , 1H), 1.59 (s, 3H), 1.68 (s, 3H), 1.82 (m, 2H), 2.02 (m, 1H), 2.29 (m, 2H), 2.52 (t,
, 1H), 1.59 (s, 3H), 1.68 (s, 3H), 1.82 (m, 2H), 2.02 (m, 1H), 2.29 (m, 2H), 2.52 (t, 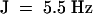 , 2H), 4.59 (s, 1H), 4.66 (s, 1H), 5.07 (t, 1H). This spectrum matches the spectrum published by Colby et al. (1998).
, 2H), 4.59 (s, 1H), 4.66 (s, 1H), 5.07 (t, 1H). This spectrum matches the spectrum published by Colby et al. (1998).
Endo-α-bergamotene 1H NMR (400 MHz, CDCl3): δ 1.59 (s, 3H), 1.68 (s, 3H), 1.71 (d, 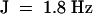 , 3H), 1.88 (m, 2H), 1.98 (dt,
, 3H), 1.88 (m, 2H), 1.98 (dt, 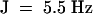 , 2H), 2.13 (m, 1H), 2.19 (t,
, 2H), 2.13 (m, 1H), 2.19 (t, 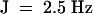 , 2H), 2.34 (m, 2H), 5.08 (t, J
, 2H), 2.34 (m, 2H), 5.08 (t, J 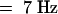 , 1H), 5.21 (q,
, 1H), 5.21 (q, 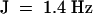 , 1H).
, 1H).
α-Santalene, 1H NMR (400 MHz, CDCl3): δ 0.84 (s, 5H), 1.00 (s, 3H), 1.60 (s, 1H), 1.62 (s, 3H), 1.69 (s, 3H), 1.90 (m, 2H), 5.13 (t, 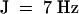 , 1H). The identification of α-santalene by 1H NMR was ambiguous because the chemical shift of the vinylic proton differed from the 300-MHz spectrum published by Coates et al. (1988). But considering two 80-MHz spectra of α-santalene, indicating shifts at ∼5.11, and the exact match–spectral match of this compound (both mass spectrum and coelution exactly matching those of α-santalene from sandalwood oil), the chemical shift reported by Coates et al. (1988) is probably incorrect for this particular proton (Monti and Larsen, 1978; Christenson and Willis, 1980).
, 1H). The identification of α-santalene by 1H NMR was ambiguous because the chemical shift of the vinylic proton differed from the 300-MHz spectrum published by Coates et al. (1988). But considering two 80-MHz spectra of α-santalene, indicating shifts at ∼5.11, and the exact match–spectral match of this compound (both mass spectrum and coelution exactly matching those of α-santalene from sandalwood oil), the chemical shift reported by Coates et al. (1988) is probably incorrect for this particular proton (Monti and Larsen, 1978; Christenson and Willis, 1980).
Germacrene B, 13C NMR (400 MHz, C6D6): δ 14.26, 16.34, 20.47, 20.80, 22.70 (very weak), 26.14, 32.87, 34.4 (very weak), 39.25, 126.82, 127.88, 128.12, 128.50, 131.55. Germacrene B was isolated in sufficient amounts for 13C NMR. The 13C spectrum matched literature values for germacrene B (Breeden et al., 1996); however, one carbon (δ 128.42) was obscured by one of the three benzene solvent peaks (δ 127.76, δ 128.00, and δ 128.24). The 1H NMR spectrum for this compound is also identical to that of germacrene B (Clark et al., 1987).
Chemotypic Screening of NILs and Segregating Populations
From each plant, 200 mg of leaf sample (approximately two leaflets) was used for sample preparation. Material was harvested from fully expanded young leaves of 6- to 8-week-old plants, homogenized in 800 μL of CH2Cl2, and partitioned against 5 M NaCl. One or 2 μL of sample was used for GC analysis.
Probe Preparation, Library Construction and Screening, and Heterologous Expression of Sesquiterpene Synthases
L. esculentum E6203 and L. hirsutum LA1777 plants were grown under greenhouse conditions. Tissue was collected from small expanding leaves, of which ∼1 g was used for RNA extraction. Poly(A)+ RNA was isolated by using the Poly-Attract mRNA isolation kit IV (Promega). cDNA libraries were constructed by using the lambda ZAPExpress system (Stratagene) according to the manufacturer's recommendations. A small aliquot of double-stranded cDNA obtained during library construction was used to amplify a 1.4-kb fragment of a sesquiterpene synthase. Primers (forward, 5′-AACGAT-TGGGAGTGGCTTAT-3′; reverse, 5′-GTTTTCCTTTGGCGTTTGT-3′) were designed based on those for germacrene C synthase (GCS) from cherry tomato cv VFNT (Colby et al., 1998). This probe subsequently was used to screen both libraries (106 plaques each) and used as a molecular marker in mapping studies. Thirty positively hybridizing clones were selected from the L. esculentum library and 40 clones from the L. hirsutum library. Sequence analysis of these clones showed that for both libraries, two different types of sesquiterpene synthase cDNAs were obtained. The two L. esculentum cDNAs, SSTLE1 and SSTLE2 (GenBank accession numbers AF279453 and AF279454, respectively), and the two L. hirsutum cDNAs, SSTLH1 and SSTLH2 (GenBank accession numbers AF279455 and AF279456, respectively), were subcloned in the expression vector pET11 (Stratagene) by using the sticky-end polymerase chain reaction (PCR) approach and transformed into DB21 cells (Zeng, 1998). Further details of the cloning procedure can be obtained from the authors.
Protein expression assays were essentially as described in Back et al. (1994) and Back and Chappell (1995). Liquid cultures of the clones in 250 mL of Luria broth were grown to an OD600 of 1.0. Then, isopropyl-β-d-thiogalactopyranoside was added to a concentration of 3 mM, and the cultures were incubated for 4 hr at 25°C without shaking. The cells were collected by centrifugation and resuspended in 1 mL of lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 100 mM NaCl, and 1 mM DTT). After the lysozyme (Sigma) was added to a concentration of 1 mg/mL, the samples were incubated on ice for 20 min. The lysozyme-treated suspensions then were subjected to two freeze–thaw cycles, freezing the samples with dry ice and thawing them in a 25°C water bath. The viscous suspension was centrifuged at 30,000g in a Sorvall SS34 rotor for 20 min at 4°C. The cleared supernatant was extracted twice with pentane and added to 9 mL of assay buffer (200 mM Tris-HCl, pH 7.5, and 40 mM MgCl2) in a Pyrex 10-mL centrifuge tube. Forty micrograms of farnesyl diphosphate (Sigma) was added, and samples were incubated overnight at room temperature, overlaid with ∼1 mL of pentane, and tightly sealed to prevent evaporation. The next day, the pentane layer was removed and reduced to ∼15 μL under a gentle stream of N2. One or 2 μL of this sample was used for GC-MS analysis.
Molecular Marker Analysis
DNA from NILs and segregating populations was extracted; digested with EcoRI, HindIII, DraI, and EcoRV; subjected to DNA gel blot hybridization; and probed with genomic markers. Markers were selected according to the high-density molecular map of Tanksley et al. (1992). Marker data were analyzed using the Mapmaker 3.0 software package (Lander and Botstein, 1989) for construction of a linkage map. Marker-trait associations were investigated by regression analysis with the Qgene software package (Nelson, 1994).
Acknowledgments
We thank Nancy Eanetta for help with harvesting and chemical analysis of plants in the summer of 1999. We also thank Dr. Rodney Croteau for very helpful advice about the expression of sesquiterpene synthases in E. coli.
References
- Adams, D.R., Bhatnagar, S.P., and Cookson, R.C. (1975). Sesquiterpenes of Santalum album and Santalum spicatum. Phytochemistry 14, 1459–1460. [Google Scholar]
- Andersson, B.A., Holman, R.T., Lundgren, L., and Stenhagen, G. (1980). Capillary gas chromatograms of leaf volatiles: A possible aid to breeders for pest and disease resistance. J. Agric. Food Chem. 28, 985–989. [Google Scholar]
- Back, K., and Chappell, J. (1995). Cloning and bacterial expression of a sesquiterpene cyclase from Hyoscyamus muticus and its molecular comparison to related terpene cyclases. J. Biol. Chem. 270, 7375–7381. [DOI] [PubMed] [Google Scholar]
- Back, K., Yin, S., and Chappell, J. (1994). Expression of a plant sesquiterpene cyclase gene in Escherichia coli. Arch. Biochem. Biophys. 315, 527–532. [DOI] [PubMed] [Google Scholar]
- Bernacchi, D., Beck Bunn, T., Eshed, Y., Lopez, Y., Petiard, V., Uhlig, J., Zamir, D., and Tanksley, S. (1998). Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor. Appl. Genet. 97, 381–397. [Google Scholar]
- Bohlmann, J., Meyer-Gauen, G., and Croteau, R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden, D.C., Young, T.E., Coates, R.M., and Juvik, J.A. (1996). Identification and bioassay of kairomones for Helicoverpa zea. J. Chem. Ecol. 22, 513–539. [DOI] [PubMed] [Google Scholar]
- Carter, C.D., Gianfagna, T.J., and Sacalis, J.N. (1989. a). Sesquiterpenes in glandular trichomes of a wild tomato species and toxicity to the Colorado Potato Beetle. J. Agric. Food Chem. 37, 1425–1428. [Google Scholar]
- Carter, C.D., Sacalis, J.N., and Gianfagna, T.J. (1989. b). Zingiberene and resistance to Colorado potato beetle in Lycopersicon-hirsutum-f.-hirsutum. J. Agric. Food Chem. 37, 206–210. [Google Scholar]
- Christenson, P.A., and Willis, B.J. (1980). East Indian sandalwood oil: Stereoselective synthesis of (+)-epi-beta-santalene and (+)-epi-beta-santalol. J. Org. Chem. 45, 3068–3072. [Google Scholar]
- Clark, B.C.J., Chamblee, T.S., and Iacobucci, G.A. (1987). HPLC isolation of the sesquiterpene hydrocarbon germacrene B from lime peel oil and its characterization as an important flavor impact constituent. J. Agric. Food Chem. 35, 514–518. [Google Scholar]
- Coates, R.M., Denissen, J.F., Juvik, J.A., and Babka, B.A. (1988). Identification of alpha-santalenoic and endo-beta-bergamotenoic acids as moth oviposition stimulants from wild tomato leaves. J. Org. Chem. 53, 2186–2192. [Google Scholar]
- Colby, S.M., Crock, J., Dowdle-Rizzo, B., Lemaux, P.G., and Croteau, R. (1998). Germacrene C synthase from Lycopersicon esculentum cv. VFNT cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc. Natl. Acad. Sci. USA 95, 2216–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, P.R., and Raven, P.H. (1964). Butterflies and plants: A study in co-evolution. Evolution 18, 586–608. [Google Scholar]
- Eigenbrode, S.D., Trumble, J.T., Millar, J.G., and White, K.K. (1994). Topical toxicity of tomato sesquiterpenes to the beet armyworm and the role of these compounds in resistance derived from an accession of Lycopersicon hirsutum f. typicum. J. Agric. Food Chem. 42, 807–810. [Google Scholar]
- Gould, F. (1988). Genetics of pairwise and multispecies plant–herbivore coevolution. In Chemical Mediation of Coevolution, K.C. Spencer, ed (San Diego, CA: Academic Press), pp. 13–55.
- Harborne, J.B. (1993). Introduction to Ecological Biochemistry. (London: Academic Press).
- Juvik, J.A., Babka, B.A., and Timmerman, E.A. (1988). Influence of trichome exudates from species of Lycopersicon on oviposition behaviour of Heliothis zea Boddie. J. Chem. Ecol. 14, 1261–1278. [DOI] [PubMed] [Google Scholar]
- Lander, E., and Botstein, D. (1989). MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- Lesburg, A.C., Guangzhi, Z., Cane, D.E., and Christianson, D.W. (1997). Crystal structure of pentalenene synthase: Mechanistic insights on terpenoid cyclization reactions in biology. Science 277, 1820–1827. [DOI] [PubMed] [Google Scholar]
- Lundgren, L., Norelius, G., and Stenhagen, G. (1985). Leaf volatiles from some wild tomato species. Nord. J. Bot. 5, 315–320. [Google Scholar]
- Monforte, A.J., and Tanksley, S.D. (2000). Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in an L. esculentum background: A tool for gene mapping and gene discovery. Genomics, in press. [PubMed]
- Moniz, M., and Drouin, G. (1996). Phylogeny and substitution rates of angiosperm actin genes. Mol. Biol. Evol. 13, 1198–1212. [DOI] [PubMed] [Google Scholar]
- Monti, S.A., and Larsen, S.D. (1978). Total synthesis of racemic alpha-santalene and of racemic teresantalic acid. J. Org. Chem. 43, 2282–2284. [Google Scholar]
- Nelson, C.J. (1994). Molecular Mapping in Bread Wheat. Ph.D. Dissertation (Ithaca, NY: Cornell University).
- Rahimi, F.R., and Carter, C.D. (1993). Inheritance of zingiberene in Lycopersicon. Theor. Appl. Genet. 87, 593–597. [DOI] [PubMed] [Google Scholar]
- Sinha, N.K., and Juvik, J.A. (1987). Resistance among accessions of Lycopersicon hirsutum to Heliothis zea the tomato fruitworm. Hortscience 22, 1105. [Google Scholar]
- Starks, C.M., Back, K., Chappell, J., and Noel, J.P. (1997). Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277, 1815–1820. [DOI] [PubMed] [Google Scholar]
- Still, W.C., Kahn, M., and Mitra, A. (1978). Rapid chromatographic technique for preparative separations with moderate resolution. J. Org. Chem. 43, 2923–2925. [Google Scholar]
- Synder, J.C., Guo, Z., Thacker, R., Goodman, J.P., and Pyrek, J.S. (1993). 2,3-Dihydrofarnesoic acid, a unique terpene from trichomes of Lycopersicon hirsutum, repels spider mites. J. Chem. Ecol. 19, 2981–2997. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D., Ganal, M.W., Prince, J.P., De Vicente, M.C., Bonierbale, M.W., Broun, P., Fulton, T.M., Giovannoni, J.J., and Grandillo, S. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbasch, I. (1985). Vergleichende Analyse der atherischen ole in blattdrusenhaaren verschiedener Kultur- und Wild-tomatenpflanzen (Lycopersicon spp.). Planta Med. 12, 58–60. [Google Scholar]
- Van der Hoeven, R.S. (1999). Functional expression of four members of a tomato sesquiterpene cyclase family isolated from L. esculentum E6203 and L. hirsutum LA1777: Mechanistic studies of farnesyldiphosphate cyclization reactions. In Biochemical and Genetic Studies of Sesquiterpene and Sugar Polyester Biosynthetic Pathways in Solanaceous Species. Ph.D. Dissertation (Ithaca, NY: Cornell University), pp. 105–147.
- Weston, P.A., Johnson, D.A., Burton, H.T., and Snyder, J.C. (1989). Trichome secretion composition trichome densities and spider mite resistance of ten accessions of Lycopersicon hirsutum. J. Am. Soc. Hort. Sci. 114, 492–498. [Google Scholar]
- Zeng, G. (1998). Sticky-end PCR: New method for subcloning. BioTechniques 25, 206–208. [DOI] [PubMed] [Google Scholar]