Abstract
The genus Borrelia includes the causative agents of Lyme disease and relapsing fever. An unusual feature of these bacteria is a genome that includes linear DNA molecules with covalently closed hairpin ends referred to as telomeres. We have investigated the mechanism by which the hairpin telomeres are processed during replication. A synthetic 140 bp sequence having the predicted structure of a replicated telomere was shown to function as a viable substrate for telomere resolution in vivo, and was sufficient to convert a circular replicon to a linear form. Our results suggest that the final step in the replication of linear Borrelia replicons is a site-specific DNA breakage and reunion event to regenerate covalently closed hairpin ends. The telomere substrate described here will be valuable both for in vivo manipulation of linear DNA in Borrelia and for in vitro studies to identify and characterize the telomere resolvase.
Keywords: Borrelia/DNA replication/hairpin DNA/linear DNA/telomere resolution
Introduction
The paradigm that eukaryotic organisms harbor linear chromosomes while bacterial replicons are circular has recently become more of a generality than a rule. A number of unrelated bacterial replicons are now known to be linear molecules (Hinnebusch and Tilly, 1993; Casjens, 1999; Volff and Altenbuchner, 2000). Among organisms containing such replicons are those in the genus Borrelia, which contains spirochetes causing Lyme disease and relapsing fever (for recent reviews see Schwan et al., 1999; Barthold, 2000; Nordstrand et al., 2000). Borrelia burgdorferi, one of several Lyme disease agents (Burgdorfer et al., 1982; Steere et al., 1983), was the first bacterium for which linear plasmids and a linear chromosome were reported (Barbour and Garon, 1987; Baril et al., 1989; Ferdows and Barbour, 1989). The genome of the prototype B.burgdorferi strain B31 is segmented and consists of a 911 kb linear chromosome as well as at least 12 linear and nine circular extrachromosomal elements (Fraser et al., 1997; Casjens et al., 2000). A unique characteristic of B.burgdorferi linear replicons is that they possess covalently closed hairpin ends with near perfect inverted repeats at the two ends, also referred to as telomeres (Hinnebusch and Barbour, 1991; Hinnebusch and Tilly, 1993; Casjens et al., 1997; Casjens, 1999). Linear DNA with hairpin telomeres has also been reported in other diverse organisms, such as poxviruses, African swine fever virus, Chlorella virus, yeast mitochondrial plasmids and the Escherichia coli phage N15 (see Hinnebusch and Tilly, 1993; Casjens, 1999).
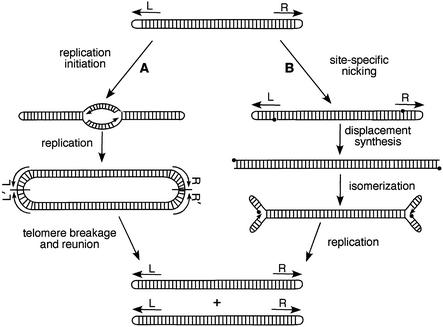
A pivotal area of inquiry in Borrelia biology is replication of the linear chromosome and plasmids. A number of models for replication of linear DNA with hairpin ends exist (see Casjens, 1999). They can be divided into those in which replication initiates internally and those in which initiation occurs at the telomeres. Figure 1 shows what we consider to be the two most likely models. In pathway A, initiation occurs internally, resulting in the generation of a head-to-head and tail-to-tail circular dimer, as suggested by the recent studies of Picardeau et al. (1999). This intermediate would then be processed by DNA breakage and reunion in the telomeric regions (telomere resolution) to regenerate covalently closed hairpin ends in the progeny molecules. In pathway B, initiation occurs by site-specific nicking in the telomeric repeats. The 3′ hydroxyl ends at the nick site are used as replication primers in displacement synthesis. Isomerization of the replicated telomeres creates new primers for DNA replication. Continuing replication then generates two progeny molecules, which are subsequently sealed.
Fig. 1. Two models for replication of linear DNA with hairpin ends. The arrows labeled L and R indicate the left and right inverted repeats, respectively, at the telomeres of linear Borrelia DNA. In pathway A, the line bisecting the telomere junctions in the replicated dimer carrying head-to-head (L′–L) and tail-to-tail (R–R′) junctions is an axis of 180° rotational symmetry. In pathway B, the closed circles denote 5′ phosphates at nick sites or at the DNA ends. A more complete set of models can be found in Casjens (1999).
Experiments presented below suggest that B.burgdorferi linear plasmid DNA replication makes use of a telomere resolution step (pathway A) involving a site-specific DNA breakage and reunion reaction.
Results
Function of a telomere resolution substrate in B.burgdorferi
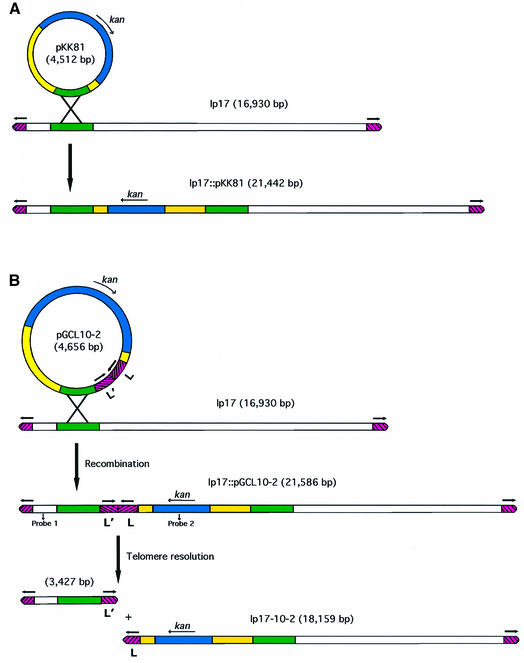
To probe the mechanism by which the linear replicons of B.burgdorferi are replicated, we constructed a putative telomere resolution substrate. We placed this substrate internally in lp17, and looked for telomere resolution as demonstrated by site-specific deletion formation at the substrate insertion site. The lp17 telomere was chosen because lp17 is the only replicon for which the DNA sequence around the hairpin telomeres has been reported (Hinnebusch and Barbour, 1991). A 140 bp replicated left-end telomere from lp17 [L′–L; see pathway A of Figure 1] was cloned into the E.coli plasmid pKK81 (see Figure 2A), which carries a kanamycin resistance gene under the control of the B.burgdorferi flgB promoter; this kan gene confers drug resistance in both E.coli and B.burgdorferi (Bono et al., 2000). The construct also carries a 1.9 kb fragment of lp17, which provides a target for homologous recombination, resulting in stable integration of the entire pKK81 plasmid into lp17 after transformation of B.burgdorferi with selection for kanamycin resistance (Figure 2A). Addition of the 140 bp replicated telomere (L′–L) to this plasmid (pGCL10-2) was similarly expected to give rise to a plasmid integrant in lp17 (Figure 2B). This integrant would carry the replicated telomere internally ∼3.4 kb from the left end of lp17. If pathway A from Figure 1 were correct, and if the putative substrate contained all the necessary information for functionality, the plasmid integrant would be processed into the two pieces shown in Figure 2B. In this scenario, the kan marker would segregate with the larger DNA fragment.
Fig. 2. Construction of linear plasmid integrants and predicted structure of telomere resolution products. (A) An E.coli plasmid (pKK81) carrying bp 1424–3315 from the left end of lp17 was used in a B.burgdorferi transformation to generate a plasmid integrant in lp17 via homologous recombin ation. (B) A linear plasmid integrant was constructed as in (A), but with an E.coli plasmid (pGCL10-2) that also carried the 140 bp replicated left-end telomere of lp17 corresponding to the L′–L junction (Figure 1). DNA breakage and reunion in the replicated telomere are expected to give rise to the two linear molecules shown; only the larger plasmid would contain the gene for kanamycin resistance (kan). Although the diagram has been drawn with homologous recombination preceding telomere resolution, a specific temporal order of events is not implied; telomere resolution of the transforming DNA followed by homologous recombination would yield the same set of final products. Blue denotes the kan gene, yellow represents the E.coli plasmid vector and green the lp17 sequences used for the recombination target. White indicates other lp17 sequences and red denotes telomeric regions. Probes 1 and 2 indicate the regions used as hybridization probes in Figure 3. The schematic is not drawn to scale.
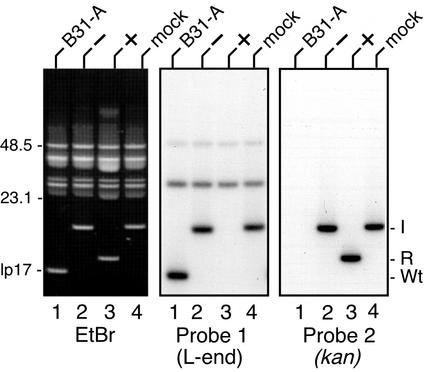
Borrelia burgdorferi strain B31-A was transformed with ∼6 µg of the E.coli plasmids shown in Figure 2. Over 103 transformants were recovered in a single experiment using the telomere-containing plasmid pGCL10-2. Total plasmid DNA from several transformants was analyzed using field inversion gels. In the conditions used, the migration of circular DNA molecules was retarded, and they were separated from the linear plasmids in the size range used for this study (Figure 3). Integration of pKK81 (minus telomere) into lp17 conferred the expected change in lp17 migration (lane 2), compared with the wild-type plasmid (lane 1). Transformation with the plasmid carrying the 140 bp replicated telomere (plus telomere, lane 3), however, did not give rise to a full-length lp17 integrant; instead, a plasmid of the size expected from a telomere resolution event was observed. A control plasmid carrying a 140 bp inverted repeat with the same base composition as the B.burgdorferi telomere, but with an unrelated sequence, was also constructed and used for transformation (mock telomere, lane 4). The resulting integrant was full sized, indicating that the presumptive telomere resolution we observed in B.burgdorferi was dependent upon a specific DNA sequence in the telomeric region.
Fig. 3. Analysis of plasmid DNA from B.burgdorferi transformants. Left panel: an ethidium bromide-stained field inversion gel of plasmid DNA from untransformed B31-A (lane 1) or from B31-A transformed with pKK81 (lane 2; minus the lp17 telomere), pGCL10-2 (lane 3; plus the 140 bp replicated telomere) and pGCL13–1 (lane 4; with a 140 bp replicated mock telomere). The migration positions of 48.5 and 23.1 kb markers and wild-type lp17 are shown. Center panel: a Southern blot of the gel in the left panel, hybridized with Probe 1, corresponding to bp 414–1231 from the left end of lp17 (Figure 2). Right panel: a Southern blot of the gel in the left panel, hybridized with Probe 2 from the kan gene. The migration positions of intact lp17 integrants (I), resolved integrants (R) and wild-type lp17 (Wt) are shown on the right side of the figure.
To analyze the lp17 constructs further, the gel in the left panel of Figure 3 was blotted and hybridized with an lp17 left-end probe (Probe 1, Figure 2) and a kan gene probe (Probe 2). Probe 1 hybridized with all the constructs, except the one carrying the resolved telomere (lane 3, center of Figure 3), indicating that, as expected (Figure 2B), this region was missing from the processed lp17. Several higher molecular weight bands also hybridized to Probe 1. These correspond to lp28-3 and lp56, which share sequence homology to lp17 near the left end (Casjens et al., 2000). As expected, the resolved telomere substrate did, however, hybridize with the kan probe (lane 3, right panel), indicating that it arose from a plasmid integration event.
Our analysis of plasmid DNA from eight B.burgdorferi transformants with pGCL10-2 (telomere substrate) resulted in recovery of four insertions with resolved telomeres in lp17, with the other four in lp56 (data not shown). The left end of the latter plasmid shares sequence homology with the left end of lp17 for about the first 2.5 kb, explaining how lp56 could also serve as a target for homologous recombination with the transforming plasmid. Although lp28-3 also shares sequence homology at the left end with lp17, this homology only extends ∼190 bp into the recombination target; no insertions into lp28-3 were recovered. DNA from a library of >1000 B.burgdorferi transformants with pGCL10-2 was also analyzed and found to contain about a 50:50 ratio of resolved insertions into lp17 and lp56. Other events, such as autonomous replication of the transforming plasmid or integration into other B.burgdorferi plasmids, were not observed (data not shown).
Structural integrity of the resolved telomere
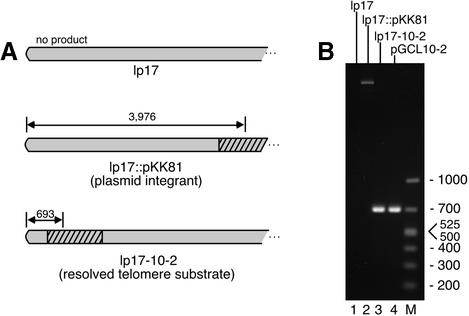
Borrelia burgdorferi transformants were analyzed further to probe the structure of plasmids with a resolved synthetic telomere. PCR reactions were performed using total B.burgdorferi plasmid DNA as template with a 37 bp left primer corresponding to the telomere and a right primer from the kan gene (Figure 4). The expected and observed results were no product for wild-type lp17, a 4 kb product for the plasmid integrant without the telomere (lane 2) and a 693 bp product from the resolved telomere substrate (lane 3). A reaction using the E.coli plasmid construct carrying the replicated telomere as a template provided a marker for the expected product (lane 4), and displayed identical migration to the product from the resolved substrate. The PCR products from several resolved lp17 and lp56 constructs were also sequenced, confirming the expected structure. The DNA sequencing revealed a T insertion at position 44 from the left-end terminal nucleotide in all cases. The insertion was also present in the parent E.coli plasmid, but not in the original oligonucleotide used to construct the telomere.
Fig. 4. Telomere structure of lp17 constructs in B.burgdorferi transformants. (A) Predicted PCR products using left-end primer OGCB5 and kan primer pOK.2 are shown above each telomere. The hatched region indicates the kan gene. (B) Ethidium bromide-stained 1.4% agarose gel containing PCR reactions using the indicated DNAs. The sizes (in bp) of molecular weight markers in lane M are indicated.
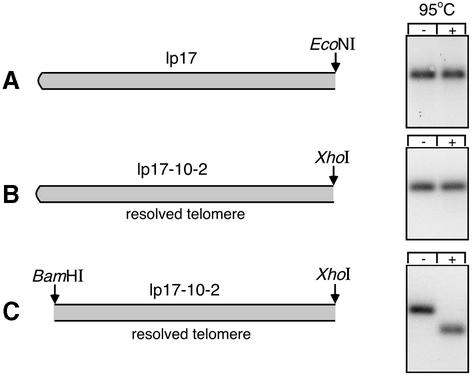
To confirm the presence of a hairpin telomere in the resolved substrate, we looked for snap-back of the denatured DNA. Total plasmid DNA from B.burgdorferi transformants was digested with restriction endonucleases and analyzed by agarose gel electrophoresis, either with or without prior heat treatment of the samples at 95°C. The gel was blotted and hybridized as shown in Figure 5. In the case of the positive control (wild-type lp17, which is known to carry a hairpin telomere), snap-back of the DNA occurred after heat treatment, resulting in no difference in migration compared with the untreated sample (Figure 5A). Similarly, the resolved telomere displayed identical behavior before and after heat treatment (Figure 5B). In contrast, a negative control in which the hairpin was severed by BamHI cleavage showed the expected increase in mobility characteristic of single-stranded DNA (Figure 5C). These results indicate that the resolved telomere substrate is maintained with a hairpin telomere. This conclusion was corroborated by the requirement for prior mung bean nuclease treatment of the resolved telomere for effective use as a PCR template. Failure to snip the hairpin with the single-strand-specific nuclease resulted in snap-back of the template and little or no PCR product (data not shown).
Fig. 5. Test for hairpin telomere at the left end in the resolved substrate. (A) The schematic shows the telomeric structure of wild-type lp17 and the position of an EcoNI site located 2639 bp from the left end. The right panel shows a Southern blot of a 1% agarose gel containing EcoNI-digested B.burgdorferi plasmid DNA hybridized with Probe 1. Duplicate samples were loaded without (–) or with (+) heat treatment at 95°C for 6 min, followed by incubation at 0°C, as indicated. (B) The schematic shows the position of an XhoI site 2538 bp from the left end of the resolved telomere substrate. The blot on the right contains XhoI-digested plasmid DNA from a B.burgdorferi transformant carrying the resolved telomere substrate. The blot was hybridized with Probe 2 (kan). (C) The schematic shows the resolved telomere substrate cut with both XhoI and BamHI to remove the hairpin end on a 70 bp fragment. The blot on the right was also hybridized with Probe 2.
Conversion of a circular plasmid to a linear form
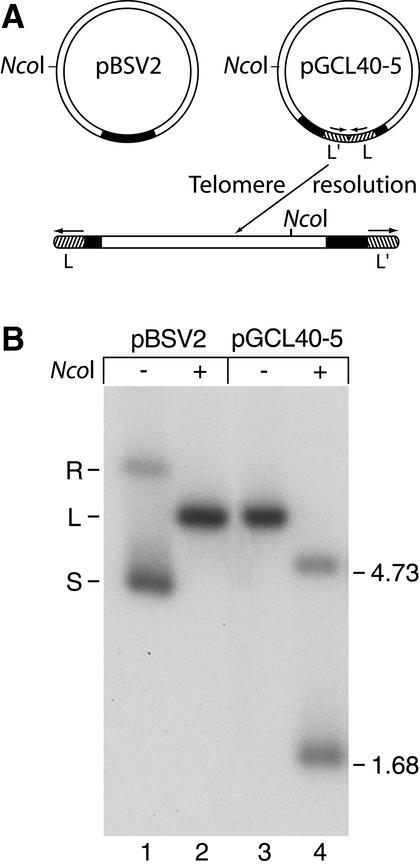
To study the telomere resolution reaction further, we next asked whether we could convert a circular B.burgdorferi plasmid into a linear form (Figure 6A). The 140 bp replicated telomere used in the lp17 experiments above was cloned directly into the BamHI site of the polylinker of pBSV2, a circular 6.4 kb E.coli–B.burgdorferi shuttle vector derived from cp9 (Stewart et al., 2001). The circular plasmid construct (pGCL40-5) was generated in E.coli and used to directly transform B.burgdorferi B31-A. DNA was isolated from 10 KmR transformants and all contained pGCL40-5 in an apparently linear form. Southern blot analysis (Figure 6B) was performed on total plasmid DNA from transformants with either the shuttle vector or the shuttle vector carrying the replicated telomere. While uncut pBSV2 was found as supercoiled and relaxed circular forms (lane 1), uncut pGCL40-5 (lane 3) was found only as a band migrating in the linear position, indicating efficient telomere resolution. Cleavage of the DNA with the single-hit enzyme NcoI converted pBSV2 into a linear molecule (lane 2). In contrast, NcoI converted pGCL40-5 (lane 4) into two discrete bands of 1.7 and 4.8 kb, as expected for a linear plasmid resulting from telomere resolution. Additional analyses by PCR and DNA sequencing confirmed that the circular B.burgdorferi shuttle vector had indeed been converted into a linear plasmid with a Borrelia telomere at each end (data not shown). Finally, it is noteworthy that the linear plasmid forms of the shuttle vector had a copy number that was reduced by a factor of five compared with the circular form (discussed below).
Fig. 6. Conversion of a circular plasmid to a linear form. (A) Schematic showing the shuttle vector without (pBSV2) or with (pGCL40-5) the 140 bp replicated telomere (L′–L) inserted into the polylinker (solid line). The circular plasmid pGCL40-5 is expected to be converted to a linear form if telomere resolution occurs. (B) Southern blot of a 0.7% agarose gel containing total B.burgdorferi plasmid DNA from strain B31-A transformed with the circular shuttle vector (pBSV2) or the shuttle vector carrying the replicated telomere (pGCL40-5). DNA samples were run before and after digestion with NcoI. Five times more DNA was used for the pGCL40-5 transformant because of the 5-fold reduction in copy number. The blot was hybridized with a probe made from the backbone (pOZK) of the shuttle vector (Stewart et al., 2001). The migration positions of the supercoiled, linear and relaxed forms of pBSV2 are denoted by S, L and R, respectively. The migration positions of 1.68 and 4.73 kb markers (not shown) are noted on the right side of the figure.
Discussion
A telomere resolution step in the replication of linear DNA in Borrelia
In the work presented here, we have generated a functional telomere resolution substrate composed of a 140 bp replicated telomere from the left end of the linear B.burgdorferi plasmid lp17. When placed internally in lp17 or lp56, this site was specifically recognized as a target for a reaction that generated a hairpin telomere. From these results, coupled with other published data, we suggest that the overall mechanism of DNA replication for lp17, lp56, and probably other linear replicons in Borrelia species, is as shown in pathway A of Figure 1. Initiation of bidirectional DNA replication occurs internally in a linear molecule and proceeds to the ends, where a DNA breakage and reunion reaction processes the replicated telomeres to recreate linear DNA molecules with hairpin ends. This general scheme for the replication of linear molecules in Borrelia is supported by the physical mapping of the bidirectional origin of replication in the B.burgdorferi chromosome at the approximate midpoint of the linear molecule (Picardeau et al., 1999). Furthermore, analysis of the DNA sequence of the linear plasmids for AT and CG skew (Picardeau et al., 2000) also suggests an internal location for the replication origins of these plasmids.
Models such as pathway B of Figure 1, and other models involving the initiation of replication at or near the hairpin telomeres (Casjens, 1999), seem unlikely in the light of recently published data (Picardeau et al., 1999, 2000) coupled with the findings presented here. Nonetheless, pathway B can not be completely ruled out by our experiments. The replicated telomere substrate used here is not the natural substrate for replication via a pathway involving initiation at or near the telomeres. Yet, it is at least theoretically possible for this substrate to be resolved into a hairpin telomere via a pathway B process. However, this assumes that sequence-specific recognition and nicking could occur in the replicated telomere that lacks a hairpin end. While the likelihood of such a reaction occurring can not be assessed, we nonetheless present two additional arguments for why we feel that pathway B is not likely. (i) Use of pathway B would be expected to result in autonomous replication of a linear form of a telomere-containing plasmid upon transformation of B.burgdorferi. Such autonomous replication was not observed for pGCL10-2 either in the library of transformed B31-A or following transformation of a B.burgdorferi strain where integrative recombination could not occur (lacking the target plasmids lp17 and lp56; data not shown). (ii) Initiation of replication at or near the telomeres would be expected to give rise to replication of the left-end fragment of lp17 resulting from the telomere resolution process (see bottom left of Figure 2B). We attempted to recover the left end of lp17 by inverting the lp17 recombination target in pGCL10-2 (green region, Figure 2B). This resulted in a plasmid construct in which the kan marker would segregate with the left end, rather than the internal fragment of lp17, after telomere resolution. No KmR transformants were recovered after transformation of B.burgdorferi with this plasmid, again suggesting that pathway B was not operative.
It should be noted that arguments (i) and (ii) above are dependent upon the assumption that the protein(s) involved in telomere processing can act in trans. This is likely to be the case for two reasons. First, the high degree of sequence similarity observed in Borrelia telomeres (Casjens et al., 1997; Casjens, 1999) suggests that a single enzymatic activity is involved in processing the telomeres carried on all the different linear replicons. Secondly, our ability to convert the non-essential, circular, cp9-derived shuttle vector into a linear form through the addition of only the 140 bp telomere resolution substrate.
In summary, to this point, the work presented here coupled with recent published findings (Picardeau et al., 1999, 2000) suggest that the final step in replication of the linear replicons of B.burgdorferi and probably other Borrelia species is a telomere resolution reaction, as depicted in pathway A of Figure 1.
The mechanism of telomere resolution
The molecular details by which telomere resolution occurs remain to be established. Our data indicate, however, that the target site for the reaction is specific: an inverted repeat of the same size and base composition as the lp17 left telomere, but with an unrelated sequence, was completely inactive in telomere resolution (Figure 3). However, both the authentic and the mock-replicated telomere spontaneously formed an extruded cruciform when carried on a supercoiled plasmid (data not shown). From this we conclude that the telomere resolvase is not a generalized Holliday junction-resolving enzyme (see West, 1997; White et al., 1997; Lilley and White, 2000) that simply recognizes an extruded cruciform structure in the circular dimeric replication intermediate. Instead, an element of specific DNA sequence recognition must be involved in the process. This contention is also supported by the high degree of sequence conservation in the first two dozen or so nucleotides of the telomeres from all sequenced chromosomes and linear plasmids from various Borrelia species (Casjens et al., 1997; Casjens, 1999). The actual sequence recognized by the telomere resolvase remains to be established. Studies to define a minimally sized yet functional telomere are currently in progress. In addition to the 140 bp replicated telomere reported here, we have constructed and tested a replicated telomere of 70 bp, corresponding to a resolved telomere of 35 bp. This substrate was resolved as efficiently as the 140 bp replicated telomere (data not shown).
Sequence-specific telomere resolution may occur through a conservative site-specific recombination type pathway (see Hallet and Sherratt, 1997; Mizuuchi, 1997 for recent reviews). Site-specific recombinases perform a two-step transesterification that involves cleavage of the scissile phosphate by the hydroxyl group from an active site serine or tyrosine. To conserve the energy from the broken phosphodiester bond, the amino acid acting as the nucleophile becomes covalently linked at the cleavage site as a reaction intermediate. Resealing of the DNA and release of the covalently linked enzyme occur by reversal of the cleavage step subsequent to strand exchange. Alternatively, telomere resolution might occur through a two-step process that does not conserve phosphodiester bond energy. In this case, site-specific endonucleolytic cleavage would be followed by DNA ligase-mediated sealing in the presence of a high-energy cofactor. Finally, it is worthy of mention that non-replicative DNA transposition utilizes a hairpin intermediate (Kennedy et al., 1998; Bhasin et al., 1999; Turlan and Chandler, 2000). The first two steps of this reaction are cleavage of the 3′ end of the transposon, followed by nucleophilic attack of the 3′ hydroxyl end on the opposite DNA strand to generate a hairpin intermediate. In this process, the bond energy from the second phosphodiester bond that is broken is conserved in the bond that produces the hairpin. At first glance, this hairpin-generating mechanism makes a plausible model for the Borrelia telomere resolution reaction; however, the energetic requirements of the reaction would allow for the formation of only one of the two hairpins during resolution of a replicated telomere. Formation of the first hairpin would also result in the production of a linear DNA molecule without a hairpin, with free 5′ and 3′ ends instead. Formation of a second hairpin by a transposition mechanism is now precluded, since there is no longer a way to capture the necessary bond energy for hairpin formation.
The process of telomere resolution has also been reported as a step in the replication of poxvirus DNA with hairpin ends (DeLange et al., 1986; Merchlinsky and Moss, 1986); however, the biochemical mechanism of this process in poxviruses remains to be established. A virus-encoded topoisomerase can perform the reaction in vitro, but at a low efficiency (Sekiguchi et al., 1996; Palaniyar et al., 1999). A Holliday junction-resolving enzyme has also been suggested as a possible player in telomere resolution in poxviruses (Garcia et al., 2000). The E.coli phage N15 also uses a telomere resolution step, and a phage-encoded breakage and reunion activity has recently been purified (Deneke et al., 2000). The protein that performs this function appears to belong to the family of site-specific recombinases with an active site tyrosine. Close scrutiny of the B.burgdorferi sequence has suggested a possible member of the tyrosine recombinase family (BBB03 carried on cp26) that might perform the telomere resolution reaction (Rybchin and Svarchevsky, 1999). The work described here, in which we have defined a functional telomere resolution substrate, now opens the door for in vitro studies to identify and characterize the telomere resolvase from B.burgdorferi.
Linear and circular replicons in Borrelia
By introducing a telomere resolution substrate onto a circular B.burgdorferi plasmid derived from cp9, we were able to convert the circular replicon to a linear form. These results extend our lp17 experiments by demonstrating that a linear replicon with 70 bp telomeres at both ends is functional. In addition, our demonstration of telomere resolution with the circular shuttle vector rules out the possibility that resolution events observed with lp17 might have resulted from homologous recombination between directly repeated copies of the left-end telomere on the plasmid integrant (Figure 2B).
Interestingly, the transition of pBSV2 from a circular to a linear form was accompanied by a reduction in copy number by a factor of five (see Figure 6). Moreover, attempts to convert a circular cp26 to a linear plasmid were only successful when the linear product existed in the presence of a cp26 recombinant dimer (data not shown). The reason for these peculiarities is not known, but they hint at underlying differences in the replication or partitioning process for linear and circular Borrelia replicons. Conversion of a linear Borrelia hermsii plasmid to a circle (Ferdows et al., 1996) and of a linear B.burgdorferi plasmid to a linear head-to-head dimer (Marconi et al., 1996) have been reported, but a change from circular to linear has not been observed previously.
Engineering of linear replicons in Borrelia
In the work reported here, we have shown that insertion of a functional telomere resolution site internally in lp17 results in a deletion of the genetic material from that site to the original end of the plasmid. This finding opens the door to a new method of manipulation of the linear replicons in Borrelia. It should be possible to design deletions of any linear molecule by inserting a telomere resolution site at a desired position in a given replicon, such that the selectable marker segregates with the portion of the linear molecule that is to survive (Figure 2). A limitation of this approach is that deletion of essential genes, including replication sites and functions, would result in lethality; however, this restriction applies to deletion formation in general. In particular, the generation of deletions of increasing size from the left and right ends of linear replicons should be invaluable for mapping replication origins and essential functions, as well as delineating any other genes with a discernible phenotype. This methodology may be particularly useful in infectious Borrelia strains, which have very low transformation efficiencies (Tilly et al., 2000). We have successfully created deletions at the left end of lp17 and lp56 in an infectious low-passage culture of the type strain B31 (data not shown). In an organism in which genetic manipulation has been difficult, this new deletion approach adds a powerful new method to the arsenal of genetic tools available.
The ability to construct deletions in an infectious strain resulted from a 7- to 15-fold stimulatory effect on transformation (data not shown) that the replicated telomere had when recombination was targeted to a region near the left end of lp17 (see Figure 2). This stimulation was not observed with the mock telomere or with pKK81, and is therefore dependent upon a sequence within the wild-type telomere. Whether the stimulation has a distance dependence from the telomere is not yet known. While the mechanism for the increased recombination frequency is not understood, it is likely to be related to the DNA breakage and reunion reaction involved in telomere resolution. DNA breakage near the end of a linear replicon could present an opportunity for strand invasion events with a resulting increase in recombination frequency. The alternative possibility, that extrusion of the inverted repeat in a replicated telomere as a cruciform could act as a target site for the recruitment of recombination proteins, seems less likely, since the mock telomere does not stimulate recombination. The observed increase in recombination may help to explain some of the proposed exchange of genetic information between the linear chromosome and linear plasmids of B.burgdorferi (Casjens et al., 1997; Casjens, 1999). DNA breakage events are known to stimulate not only homologous recombination, but illegitimate exchange as well (Paques and Haber, 1999; Flores-Rozas and Kolodner, 2000).
In summary, our results suggest that B.burgdorferi linear plasmid replication uses a telomere resolution step involving a site-specific DNA breakage and reunion reaction to regenerate hairpin ends. We have defined a functional telomere resolution substrate that will be useful both for in vivo manipulation of linear DNA in Borrelia and for in vitro studies to identify and characterize the telomere resolvase.
Materials and methods
Plasmid constructs
For construction of pKK81 (see Figure 2), the region from 1424 to 3296 of the complete lp17 sequence (including the telomeres) was amplified using primers L1 (1424–1443; 5′ attactggggcactatttgg) and L2 (3315–3296; 5′ ccataagataaaccaacact), and cloned into the vector pCR2.1-TOPO (Invitrogen). The L1–L2 region was recovered by cleavage with BamHI and XhoI, and cloned into BamHI–XhoI-digested pJLB12g, which carries the kanamycin resistance gene from Tn903 under the control of the B.burgdorferi flgB promoter (Bono et al., 2000). For the construction of pGCL10-2, the 140 bp replicated telomere from the left end of lp17 was cloned into the BamHI site of pKK81. The inverted repeat was constructed from two synthetic 72mers (Genosys) that had been gel purified: OGCB2 (5′GATCcaagttaaagttagcaatttaaagggtaaagttttgagtca aaatactctatactaataaaaaattat) and OGCB3 (5′ATATataattttttattagtatagagt attttgactcaaaactttaccctttaaattgctaactttaacttg). The BamHI sticky end in OGCB2 and the sticky end in OGCB3 that extends two nucleotides beyond the axis of symmetry in the replicated telomere are in upper case letters.
These two oligos were phosphorylated with polynucleotide kinase, annealed and ligated, digested with BamHI, spermine precipitated and ligated into pKK81. The ligated DNA was used to transform E.coli strain DB1256, which is defective in the sbc operon, allowing propagation of inverted repeats (DeLange et al., 1986). For pGCL13-1, the equivalent of pGCL10-2 but carrying a mock telomere, the same construction approach was used with the following oligos: OGCB8 (5′GATCcttcaatttcaatcg ttaaatttcccatttcaaaactcagttttatgagatatgattattttttaata) and OGCB9 (5′TAT Atattaaaaaataatcatatctcataaaactgagttttgaaatgggaaatttaacgattgaaattgaag). Similarly, pGCL15-6, carrying a 70 bp replicated telomere, was constructed as described above using the oligos OGCB4 (5′GAT Cgagtcaaaatactctatactaataaaaaattat) and OGCB5 (5′ATATataattttttatta gtatagagtattttgactc). For this construct, the ligated oligo was cleaved with Sau3AI before ligation to the BamHI site in the vector.
The B.burgdorferi shuttle vector construct carrying the 140 bp lp17 left-end replicated telomere (pGCL40-5) was constructed by ligation of the synthetic replicated telomere into the BamHI site of pBSV2 (Stewart et al., 2001).
Oligonucleotide primers and PCR
Probe 1, for hybridization of Southern blots, was generated by random priming of the PCR product generated with primers OGCB6 from position 1231 to 1207 of lp17 (5′ ataataaatcatatatttggactgg) and OGCB7 from 414 to 438 of lp17 (5′ acacgttcaaccattattactaacc). Probe 2, for the kanamycin resistance gene, was made with kan5′ + NdeI (5′ catatgagccatattcaacgggaaacg) and kan3′ (5′ tcatatcaggattatcaatacc). For PCR analysis of the lp17 and lp54 constructs in B.burgdorferi DNA, the primers OGCB5 (see above) from the left-end telomere and pOK.2 from the kan gene (5′ atgacgagcgtaatggctgg) were used. These last two primers were also used for DNA sequencing of resolved telomeres, as were pOK.7 and pOK.8 (Bono et al., 2000).
For PCR, all templates were first treated with mung bean nuclease to snip the hairpin telomeres (Hinnebusch et al., 1990). Approximately 200 ng of DNA were incubated in 20 µl of 30 mM NaOAc pH 5.0 containing 50 mM NaCl, 1 mM ZnCl2, 10 mM MgCl2 and 5% (v/v) glycerol. Mung bean nuclease (3 U; Stratagene) was added and the reactions were incubated at 30°C for 30 min. One microliter of a mixture of 1 M Tris–HCl pH 8.0 and 50 mM EDTA was added. The treated template was added directly to PCR reactions at a final concentration of 4 ng per 40 µl reaction. PCR reactions were run for 25 cycles of 94°C for 30 s, 50°C for 30 s and 68°C for 1 min.
Borrelia burgdorferi strains, growth and transformation
All Borrelia transformations were carried out with strain B31-A (Bono et al., 2000), a high-passage, non-infectious clone of the prototype B.burgdorferi sensu stricto isolate B31 (Burgdorfer et al., 1982). Methods for cultivation, transformation and plating of B.burgdorferi B31-A have been described previously (Bono et al., 2000; Stewart et al., 2001).
Preparation and analysis of DNA
Initial analysis of DNA from B.burgdorferi transformations was performed on DNA isolated from 5 ml cultures in early stationary phase. Total DNA was purified using Wizard genomic preps (Promega). For further analysis, total plasmid DNA was purified from 100–125 ml stationary Borrelia cultures using Qiagen plasmid midi prep kits with a Qiagen-tip 100 column. All agarose gels were run in 40 mM Tris base containing 20 mM acetic acid and 1 mM EDTA. For field inversion gel separation of B.burgdorferi plasmids, a 150 ml, 0.65% agarose gel (15 cm wide × 20 cm long) was used. Approximately 200 ng of DNA were loaded per well. Gels were run at 80 V for 40 min, at which time Program 0 was initiated on an MJ Research PPI-200 programmable power inverter at 80 V for 21 h with buffer recirculation.
Acknowledgments
Acknowledgements
The authors would like to thank Abdallah Elias for helpful suggestions, discussions and laboratory materials during the course of this work. Comments on the manuscript by David Haniford, Joe Hinnebusch, Kerri Kobryn and Colin Coros are greatly appreciated. The authors would also like to thank Anita Mora, Gary Hettrick and Ian Craig for the preparation of figures. Most of this work was completed while G.C. was on sabbatical at the Rocky Mountain Laboratories. G.C. gratefully acknowledges sabbatical leave support by a Fellowship from the John Simon Guggenheim Memorial Foundation and salary support through a Distinguished Scientist Award from the Canadian Institutes of Health Research.
References
- Barbour A.G. and Garon,C.F. (1987) Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science, 237, 409–411. [DOI] [PubMed] [Google Scholar]
- Baril C., Richaud,C., Baranton,G. and Saint Girons,I.S. (1989) Linear chromosome of Borrelia burgdorferi. Res. Microbiol., 140, 507–516. [DOI] [PubMed] [Google Scholar]
- Barthold S.W. (2000) Lyme borreliosis. In Nataro,J.P., Blaser,M.J. and Cunningham-Rundles,S. (eds), Persistent Bacterial Infections. ASM Press, Washington, DC, pp. 281–304.
- Bhasin A., Goryshin,I.Y. and Reznikoff,W.S. (1999) Hairpin formation in Tn5 transposition. J. Biol. Chem., 274, 37021–37029. [DOI] [PubMed] [Google Scholar]
- Bono J.L., Elias,A.F., Kupko,J.J.,III, Stevenson,B., Tilly,K. and Rosa,P. (2000) Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol., 182, 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W., Barbour,A.G., Hayes,S.F., Benach,J.L., Grunwaldt,E. and Davis,J.P. (1982) Lyme disease—a tick-borne spirochetosis? Science, 216, 1317–1319. [DOI] [PubMed] [Google Scholar]
- Casjens S. (1999) Evolution of the linear DNA replicons of the Borrelia spirochetes. Curr. Opin. Microbiol., 2, 529–534. [DOI] [PubMed] [Google Scholar]
- Casjens S., Murphy,M., DeLange,M., Sampson,L., van Vugt,R. and Huang,W.M. (1997) Telomeres of the linear chromosomes of Lyme disease spirochaetes: nucleotide sequence and possible exchange with linear plasmid telomeres. Mol. Microbiol., 26, 581–596. [DOI] [PubMed] [Google Scholar]
- Casjens S. et al. (2000) A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol., 35, 490–516. [DOI] [PubMed] [Google Scholar]
- DeLange A.M., Reddy,M., Scraba,D., Upton,C. and McFadden,G. (1986) Replication and resolution of cloned poxvirus telomeres in vivo generates linear minichromosomes with intact viral hairpin termini. J. Virol., 59, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneke J., Ziegelin,G., Lurz,R. and Lanka,E. (2000) The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl Acad. Sci. USA, 97, 7721–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdows M.S. and Barbour,A.G. (1989) Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc. Natl Acad. Sci. USA, 86, 5969–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdows M.S., Serwer,P., Griess,G.A., Norris,S.J. and Barbour,A.G. (1996) Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J. Bacteriol., 178, 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H. and Kolodner,R.D. (2000) Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci., 25, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.M. et al. (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature, 390, 580–586. [DOI] [PubMed] [Google Scholar]
- Garcia A.D., Aravind,L., Koonin,E.V. and Moss,B. (2000) Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl Acad. Sci. USA, 97, 8926–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallet B. and Sherratt,D.J. (1997) Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol. Rev., 21, 157–178. [DOI] [PubMed] [Google Scholar]
- Hinnebusch J. and Barbour,A.G. (1991) Linear plasmids of Borrelia burgdorferi have a telomeric structure and sequence similar to those of a eukaryotic virus. J. Bacteriol., 173, 7233–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch J. and Tilly,K. (1993) Linear plasmids and chromosomes in bacteria. Mol. Microbiol., 10, 917–922. [DOI] [PubMed] [Google Scholar]
- Hinnebusch J., Bergstrom,S. and Barbour,A.G. (1990) Cloning and sequence analysis of linear plasmid telomeres of the bacterium Borrelia burgdorferi. Mol. Microbiol., 4, 811–820. [DOI] [PubMed] [Google Scholar]
- Kennedy A.K., Guhathakurta,A., Kleckner,N. and Haniford,D.B. (1998) Tn10 transposition via a DNA hairpin intermediate. Cell, 95, 125–134. [DOI] [PubMed] [Google Scholar]
- Lilley D.M. and White,M.F. (2000) Resolving the relationships of resolving enzymes. Proc. Natl Acad. Sci. USA, 97, 9351–9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi R.T., Casjens,S., Munderloh,U.G. and Samuels,D.S. (1996) Analysis of linear plasmid dimers in Borrelia burgdorferi sensu lato isolates: implications concerning the potential mechanism of linear plasmid replication. J. Bacteriol., 178, 3357–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M. and Moss,B. (1986) Resolution of linear minichromosomes with hairpin ends from circular plasmids containing vaccinia virus concatemer junctions. Cell, 45, 879–884. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. (1997) Polynucleotidyl transfer reactions in site-specific DNA recombination. Genes Cells, 2, 1–12. [DOI] [PubMed] [Google Scholar]
- Nordstrand A., Barbour,A.G. and Bergstrom,S. (2000) Borrelia pathogenesis research in the post-genomic and post-vaccine era. Curr. Opin. Microbiol., 3, 86–92. [DOI] [PubMed] [Google Scholar]
- Palaniyar N., Gerasimopoulos,E. and Evans,D.H. (1999) Shope fibroma virus DNA topoisomerase catalyses Holliday junction resolution and hairpin formation in vitro. J. Mol. Biol., 287, 9–20. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardeau M., Lobry,J.R. and Hinnebusch,B.J. (1999) Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol. Microbiol., 32, 437–445. [DOI] [PubMed] [Google Scholar]
- Picardeau M., Lobry,J.R. and Hinnebusch,B.J. (2000) Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res., 10, 1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybchin V.N. and Svarchevsky,A.N. (1999) The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol., 33, 895–903. [DOI] [PubMed] [Google Scholar]
- Schwan T.G., Burgdorfer,W. and Rosa,P.A. (1999) Borrelia. In Murray,P.R., Baron,E.J., Pfaller,M.A., Tenover,F.C. and Yolken,R.H. (eds), Manual of Clinical Microbiology. ASM Press, Washington, DC, pp. 746–758.
- Sekiguchi J., Seeman,N.C. and Shuman,S. (1996) Resolution of Holliday junctions by eukaryotic DNA topoisomerase I. Proc. Natl Acad. Sci. USA, 93, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere A.C., Grodzicki,R.L., Kornblatt,A.N., Craft,J.E., Barbour,A.G., Burgdorfer,W., Schmid,G.P., Johnson,E. and Malawista,S.E. (1983) The spirochetal etiology of Lyme disease. N. Engl. J. Med., 308, 733–740. [DOI] [PubMed] [Google Scholar]
- Stewart P.E., Thalken,R., Bono,J.L. and Rosa,P. (2001) Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol., 39, 714–721. [DOI] [PubMed] [Google Scholar]
- Tilly K., Elias,A.F., Bono,J.L., Stewart,P. and Rosa,P. (2000) DNA exchange and insertional inactivation in spirochetes. J. Mol. Microbiol. Biotechnol., 2, 433–442. [PubMed] [Google Scholar]
- Turlan C. and Chandler,M. (2000) Playing second fiddle: second-strand processing and liberation of transposable elements from donor DNA. Trends Microbiol., 8, 268–274. [DOI] [PubMed] [Google Scholar]
- Volff J.N. and Altenbuchner,J. (2000) A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol. Lett., 186, 143–150. [DOI] [PubMed] [Google Scholar]
- West S.C. (1997) Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet., 31, 213–244. [DOI] [PubMed] [Google Scholar]
- White M.F., Giraud-Panis,M.J., Pohler,J.R. and Lilley,D.M. (1997) Recognition and manipulation of branched DNA structure by junction-resolving enzymes. J. Mol. Biol., 269, 647–664. [DOI] [PubMed] [Google Scholar]