Abstract
Transcriptional control is generally thought to operate as a binary switch, a behavior that might explain observations such as monoallelic gene expression, stochastic phenotypic changes and bimodal gene activation kinetics. By measuring the activity of the single-copy GAL1 promoter in single cells, we found that changes in the activities of either the transcriptional activator, Gal4 (by simple recruitment with synthetic ligands), or the transcriptional repressor, Mig1, generated graded (non-binary) changes in gene expression that were proportional to signal intensity. However, in the context of the endogenous glucose-responsive signaling pathway, these transcription factors formed part of a binary transcriptional response. Genetic studies demonstrated that this binary response resulted from regulation of a second repressor, Gal80, whereas regulation of Mig1 by a distinct signaling pathway generated graded changes in GAL1 promoter activity. Surprisingly, isogenetic cells can respond to glucose with either binary or graded changes in gene expression, depending on growth conditions. Our studies demonstrate that a given promoter can adapt either binary or graded behavior, and identify the Mig1 and Gal80 genes as necessary for binary versus graded behavior of the Gal1 promoter.
Keywords: CID/FK506/signaling/synthetic ligands/transcription
Introduction
An unresolved question at the interface of signaling and transcriptional regulation is whether regulators of transcription serve as binary switches between active and inactive promoter states or as graded regulators of transcription. We use the term ‘graded’ to describe a promoter response that achieves a continuous range of activity from fully on to fully off. In contrast we use ‘binary’ to refer to the situation when a promoter produces only two levels of activity: on and off.
When collections of cells are analyzed experimentally, either graded or binary patterns of transcriptional response would demonstrate similar increases in mRNA levels, but within single cells this distinction underlies fundamental aspects of gene regulation and cellular responses. Binary, all-or-none reactions by individual cells may ensure efficient cellular responses to fleeting signals, but may also limit the diversity of these responses. The binary pattern of response could also generate distinct populations within a group of cells: one consisting of responsive cells in which expression is turned on, and another comprised of unresponsive cells. Such responses could be critical to cell fate decisions and other all-or-none phenotypic responses to signals. Alternatively, graded alterations in gene expression may allow fine-tuned responses that permit proportionate responses to a stimulus. In the latter case, all cells in a population could respond with similar changes in expression and thereby maintain homogeneity.
The majority of existing evidence suggests that, within individual cells, a gene is either maximally expressed or is not expressed at all (Moreau et al., 1981; Weintraub, 1988; Fiering et al., 1990; Moon and Ley, 1990; Walters et al., 1995, 1996; Magis et al., 1996; Hollander et al., 1998). Under this binary model, transcriptional activators and their corresponding enhancers stimulate expression simply by increasing the likelihood that a promoter will be active, such that promoters containing enhancers express in a larger fraction of cells than promoters lacking enhancers. An important confirmation of this model is the observation that while the proportion of expression-positive cells differs in the presence and absence of the enhancer, the level of expression in individual expression-positive cells is similarly independent of the presence of the enhancer.
Transcriptional activators may also reduce ‘silencing’ of gene expression in a binary fashion (Magis et al., 1996; Walters et al., 1996). In cells that have been selected for active expression of an integrated reporter gene, removing the selective pressure for expression leads to the accumulation of cells that no longer express the reporter. Single-cell analysis of this process reveals two separate populations of cells: one expressing the reporter at maximal levels and one in which expression is silenced. The presence of enhancers or activators slows the silencing of reporter expression by maintaining the number of expression-positive cells without influencing the level of expression within these cells. These results have been interpreted as support for the idea that transcriptional activators produce binary changes in promoter activity.
Not all alterations in transcription factor activity generate binary changes in gene expression. In one important exception, mutations in components of the chromatin-remodeling transcription factor, Swi-Snf, reduce expression throughout the entire population of cells to a level that is intermediate to the fully activated and repressed states (Sudarsanam et al., 1999). Also, inducing the function of the MTF transcriptional activator increases expression of an MTF-responsive gene in a population of cells presumed to be universally expressing the reporter (Magis et al., 1996), suggesting that the activator produces graded changes in transcription under these conditions. Recently, the Tet repressor–VP16 fusion was shown to produce a graded response with increasing tetracycline concentrations (Rossi et al., 2000).
Presently, the mechanisms and molecular events underlying either graded or binary modes of transcription are unknown. We set out to study these mechanisms for the well studied Gal1 promoter by exploring the elements comprising the regulatory controls of this promoter, including the changing activities of transcriptional activators and repressors, and the influences of cell signaling. We show that graded and binary modes of transcriptional response are not intrinsic to the promoter, but are determined by specific regulatory mechanisms. Further more, we identify for the first time the genes essential to produce these two modes of promoter activity.
Results
Transcriptional regulation at the single-cell level was studied by dissecting the effects of transcriptional activators and repressors, either individually or in the context of intact signaling pathways, on the activity of a promoter on a cell-by-cell basis. To achieve this, we used both physiological and a small-molecule-based means of regulating the constituents that control expression from the Saccharomyces cerevisiae GAL1 promoter. In these experiments, expression was measured by fluorescence-activated cell sorting (FACS) analysis of cells carrying a single-copy, integrated reporter from which the full-length GAL1 promoter directed expression of green fluorescent protein (GAL1–GFP) (see Figure 1). Previous studies have demonstrated that GFP fluorescence closely correlates with expression levels from several yeast promoters (Figure 1A; Niedenthal et al., 1996; Sudarsanam et al., 1999).
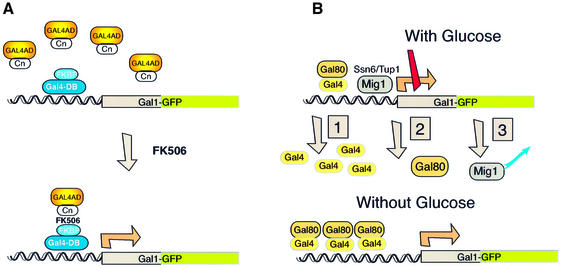
Fig. 1. Synthetic and natural modes of transcriptional activation of the integrated Gal1–GFP promoter. Regulation of a single-copy, integrated GAL1–GFP gene by FK506 and by glucose. (A) Activation with the synthetic ligand FK506. In cells expressing the Gal4BD–FKBP and Gal4AD–CN fusion proteins, FK506 generates an interaction between FKBP and calcineurin at the promoter, and thereby recruits the activation domain and stimulates transcription. The GAL1–GFP reporter, which consists of the full-length GAL1 promoter fused to the coding sequence of GFP, is shown schematically, with the binding sites for Gal4 and Mig1 indicated. (B) Activation by relief of glucose-induced repression. Glucose regulates GAL1 promoter activity by three mechanisms in wild-type cells (see text). The GAL1–GFP reporter, which consists of the full-length GAL1 promoter fused to the coding sequence of GFP, is shown schematically, with the binding sites for Gal4 and Mig1 indicated.
Using this approach, binary transcriptional responses would be expected to generate two peaks of GFP fluorescence: one of high intensity that corresponds to cells expressing GAL1–GFP, and one representing background fluorescence corresponding to cells in which GAL1–GFP is not expressed. In this case, increasing levels of expression would raise the height of the high-intensity peak as the number of expressing cells increased, but the positions (i.e. fluorescence intensities) of the two peaks would not change. Alternatively, graded transcriptional responses would generate a single fluorescence peak that increases in intensity and therefore changes position on a FACS plot as expression levels increase. In this case, the single peak would represent a homogeneous transcriptional response across an entire population of cells.
Direct control of Gal4 produced graded changes in GAL1–GFP expression
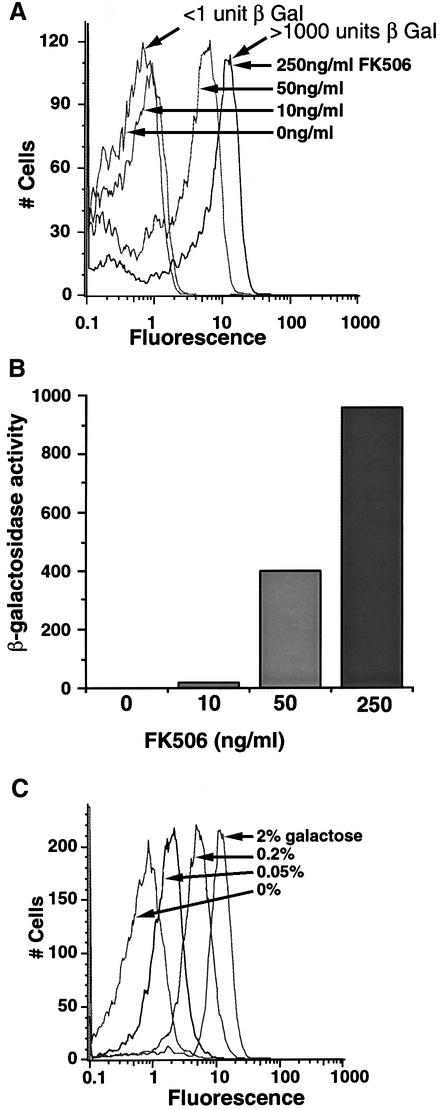
Initially, we addressed the effects of directly recruiting the transcriptional activator, Gal4, on GAL1–GFP expression. This approach was used to assay the behavior of an activator without the complication of endogenous cellular controls. To assay the behavior of an activator specifically, the Gal4 activation domain was directly recruited to the Gal4 DNA binding domain using FK506 (Figure 1A). Cells expressed the functional domains of Gal4 as two separate fusion proteins: a Gal4 DNA binding domain– FKBP12 (Gal4BD–FKBP) fusion and a Gal4 activation domain–calcineurin A (Gal4AD–CN) fusion. By generating an interaction between calcineurin and FKBP12, FK506 recruited the Gal4 activation domain to Gal4BD– FKBP bound at the GAL1 promoter and thereby activated transcription. FK506 was chosen because it generates little transcriptional response in yeast at nanomolar concentrations, as determined by studies using yeast whole-genome transcript arrays (Marton et al., 1998; K.Vogel and G.R.Crabtree, unpublished studies). Furthermore, the few genes that are affected are not part of the GAL1 regulatory network. Titrating FK506 levels produced dose-dependent stimulation of GAL1 expression (Clipstone et al., 1994; Ho et al., 1996; Biggar and Crabtree, 2000), and FACS analysis showed homogeneous (i.e. a single fluorescence peak) and continuous changes in GAL1–GFP expression across the entire population of cells (Figure 2A). Since GFP fluorescence is not quantitative, we determined the magnitude of the difference between the response achieved with no FK506 and that seen with 250 ng/ml (240 nM) FK506 by assaying the relative β-galactosidase levels expressed from a GAL1–LacZ reporter also carried by these cells (Figure 2B). Since the two assays could be run on the same population of cells, this allowed us to determine accurately the background level of expression with no FK506. Comparing Figure 2A with B indicates that the background level is undetectable and that the fluorescence seen in Figure 2A in the absence of drug is simply the normal cellular fluorescence. A similar conclusion was reached by FACS analysis of cells not containing a GFP gene (data not shown). Thus, recruitment of the transcriptional activator with FK506 generated graded changes in gene expression over a several thousand-fold range, such that in each cell, GAL1–GFP was expressed at levels proportional to drug dose.
Fig. 2. Single-cell GAL1–GFP expression in response to varying the activity of the transcriptional activator Gal4. (A) Single-cell analysis of the response to directly increasing activity of the Gal4 activator with FK506. Yeast (YSB30) carrying integrated GAL1–GFP and GAL1–LacZ reporter genes and expressing Gal4BD–FKBP and Gal4AD–CN were treated with 0, 10, 50 and 250 ng/ml FK506. Cells were then analyzed for GFP expression by FACS and for (B) β-galactosidase expression in the entire population of cells. (C) Single-cell expression in response to different galactose concentrations and endogenous regulation. Yeast (YSB31) carrying the integrated GAL1–GFP reporter were grown in glucose media, washed, and then exposed to the indicated concentrations of galactose in 2% raffinose media. Cells were harvested at mid-log phase to avoid nutrient depletion and then analyzed for GFP expression by FACS.
To examine the type of control found under physiological regulation, we examined transcriptional control exerted by the endogenous GAL1 regulatory mechanisms, including wild-type GAL4. Through the action of distinct signals, wild-type yeast repress GAL1 expression in glucose media and induce GAL1 expression in galactose (Lohr et al., 1995) (Figure 2C). In the absence of glucose, galactose stimulates GAL1 expression by increasing the function of Gal4 as a transcriptional activator (Figure 1B). Thus, GAL1–GFP expression was measured in cells exposed to varying concentrations of galactose. Similarly to the experiment presented above, in which FK506 directly controlled Gal4 activity (see Figure 2A), physiological regulation of Gal4 activity with galactose produced graded changes in GAL1 promoter activity over at least a 500-fold range, with the entire populations of cells homogeneously expressing GFP at higher levels in response to increasing galactose concentrations (Figure 2C).
The repressor, Mig1, uniformly reduced GAL1–GFP expression in all cells
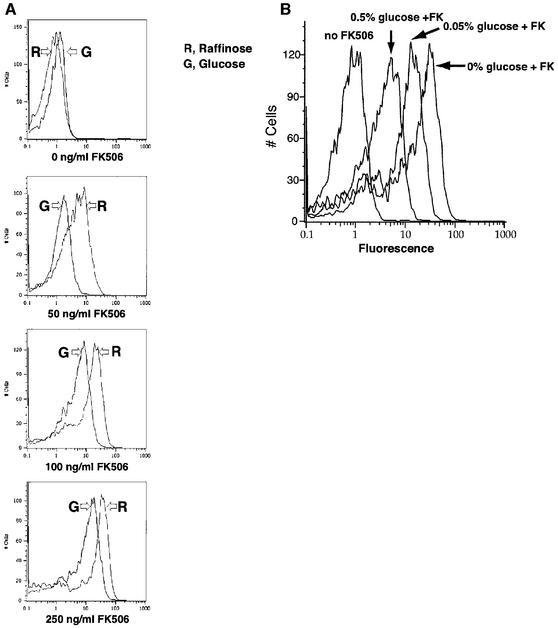
As mentioned, glucose represses GAL1 transcription. To explore the effects of transcriptional repression on gene expression at the single-cell level, yeast carrying the Gal4BD–FKBP and Gal4AD–CN fusions were treated with FK506 to induce GAL1–GFP expression and then exposed to glucose. In these cells, glucose directly inhibits GAL1–GFP expression by inducing the function of Mig1, a transcriptional repressor that binds to the GAL1 promoter (Figure 1B; Nehlin et al., 1991). Constitutive expression of the Gal4BD–FKBP and Gal4AD–CN fusion proteins, combined with a deletion of GAL80, prevented indirect effects of glucose on GAL1 expression (Lamphier and Ptashne, 1992; Johnston et al., 1994; Biggar and Crabtree, 2000). Comparing GFP levels in cells induced with FK506 and grown in either 2% raffinose (0% glucose) or 2% glucose showed that glucose shifted the peak of expression downward without silencing the promoter (Figure 3A). This result was seen at all FK506 concentrations tested. Furthermore, in cells treated with a given amount of FK506 (150 ng/ml), increasing glucose concentrations shifted the peak of GFP expression downward (Figure 3B). Thus, similar to the transcriptional activator, the actions of the transcriptional repressor, Mig1, produced graded changes in gene expression.
Fig. 3. Single-cell GAL1–GFP expression in response to varying the activity of the transcriptional repressor Mig1. (A) Glucose represses at all levels of activation by FK506. Cells carrying an integrated GAL1–GFP reporter (YSB30) and expressing Gal4BD–FKBP and Gal4AD–CN were grown in either 2% glucose or 2% raffinose media and treated with the indicated concentrations of FK506. Cells were then analyzed for GFP expression by FACS. (B) Glucose repression produces graded transcriptional repression of Gal1 transcription. Cells carrying an integrated GAL1–GFP reporter (YSB30) and expressing Gal4BD–FKBP and Gal4AD–CN were treated with 150 ng/ml FK506 (to induce Gal1–GFP maximally) in media containing the indicated concentrations of glucose and 2% raffinose. Cells were harvested at mid-log phase and then analyzed for GFP expression by FACS.
Physiological regulation of GAL1 generated binary changes in GAL1–GFP expression
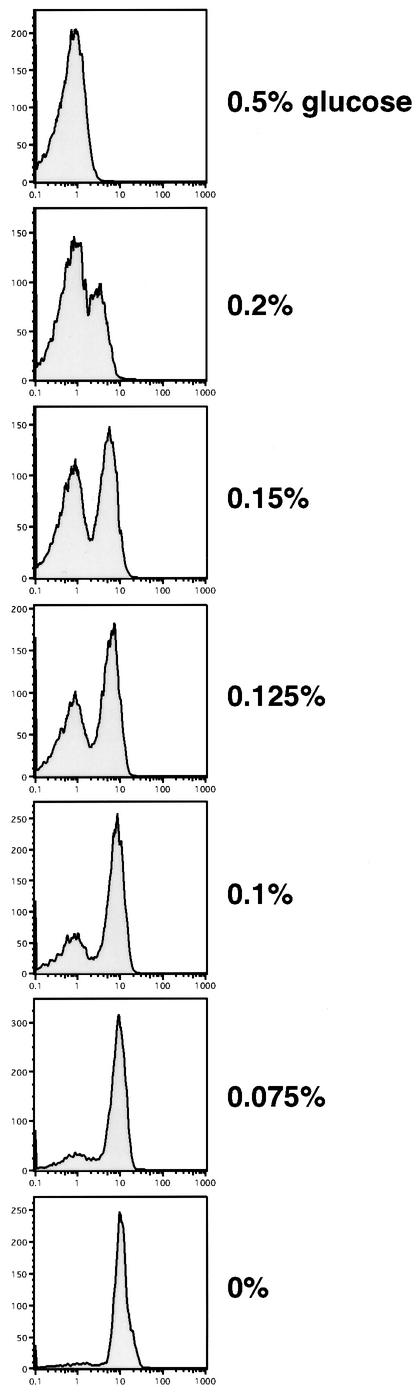
The results of these studies of transcription factors differ from the binary transcriptional responses seen in other systems in response to extracellular signals (Fiering et al., 1990; Walters et al., 1996). We therefore set out to address how, in individual cells, GAL1–GFP expression responded to signaling cascades that culminate by altering the activities of both the transcriptional activator, Gal4, and the transcriptional repressor, Mig1. For these experiments, yeast carrying the single-copy, integrated GAL1–GFP reporter and the endogenous GAL1 regulatory network were studied. In wild-type cells, glucose both suppresses the function of the activator, Gal4, and stimulates the function of the repressor, Mig1 (Figure 1; Lamphier and Ptashne, 1992; Johnston et al., 1994). To study the transcriptional response to glucose, cells were grown in 2% galactose media containing varying amounts of glucose. FACS analysis showed that, in contrast to the graded changes seen above, glucose generated a binary response in GFP expression (Figure 4). As the glucose concentration decreased, an accumulating fraction of cells expressed GFP, and these populations of GFP-positive cells expressed GAL1–GFP at levels similar to those in fully activated cells (0% glucose). Conversely, a second subpopulation of cells, which failed to express GFP, declined in number as glucose levels fell. Again, as in previous figures, the quantitative differences in expression from 0.5% glucose to no glucose were at least 500-fold, as measured by β-galactosidase levels. Thus, unlike the graded influences observed when studied in isolation, GAL4 and MIG1 combined in the glucose-mediated signaling pathways to form part of a binary control over GAL1–GFP expression.
Fig. 4. Single-cell GAL1–GFP expression in response to varying glucose levels in cells carrying the entire GAL1 regulatory network. Cells carrying the integrated GAL1–GFP reporter (YSB31) were grown in 2% glucose media, washed, and transferred to media containing the indicated concentrations of glucose and 2% galactose for 14 h. Cells were harvested at mid-log phase and then analyzed for GFP expression by FACS.
GAL80 is both necessary and sufficient for a binary transcriptional response
To understand further the mechanism underlying the binary character to the transcriptional response, we tested the effects of glucose under a variety of conditions. Glucose represses GAL1 expression by three established mechanisms in wild-type cells (Figure 1B; Lamphier and Ptashne, 1992; Johnston et al., 1994). As discussed, glucose activates Mig1 to directly repress transcription from the GAL1 promoter. Activation of Mig1 by glucose also represses expression of the activator, Gal4. Lastly, glucose activates the expression and function of Gal80, which binds and inactivates Gal4. In cells lacking MIG1, GAL80 assumes the major role in mediating glucose repression of GAL1 (data not shown; Nehlin et al., 1991). Thus, to test specifically the effects of GAL80 on expression at the single-cell level, mig1Δ cells were grown in media containing varying concentrations of glucose and analyzed for GAL1–GFP expression. Similarly to wild-type cells, mig1Δ mutants showed a binary transcriptional response to glucose (Figure 5A), which when quantitated had a range of several hundred-fold. Thus, GAL80 sufficed to establish an all-or-none pattern of expression from the GAL1 promoter. To test whether GAL80 was also required for this pattern of response, GAL1–GFP expression was analyzed in gal80Δ mutants grown in media with varying concentrations of glucose. In this case, FACS analysis showed that glucose induced graded changes in GAL1–GFP expression, with a single peak of GFP expression that increased in fluorescence intensity as glucose levels declined (Figure 5B). Thus, GAL80 was both necessary and sufficient for a binary transcriptional response to glucose.
Fig. 5. Single-cell GAL1–GFP expression in response to glucose in cells lacking MIG1 or GAL80. (A) Binary responses to glucose in mig1Δ mutants. mig1Δ cells carrying the integrated GAL1–GFP reporter (YSB32) were grown in 2% glucose media, washed, and transferred to media containing the indicated concentrations of glucose and 2% galactose for 14 h. Cells were harvested at mid-log phase and then analyzed for GFP expression by FACS. (B) Graded responses to glucose in gal80Δ mutants. gal80Δ cells carrying the integrated GAL1–GFP reporter (YSB35) were grown in 2% glucose media, washed, and transferred to media containing the indicated concentrations of glucose and 2% galactose for 14 h. Cells were then analyzed for GFP expression by FACS.
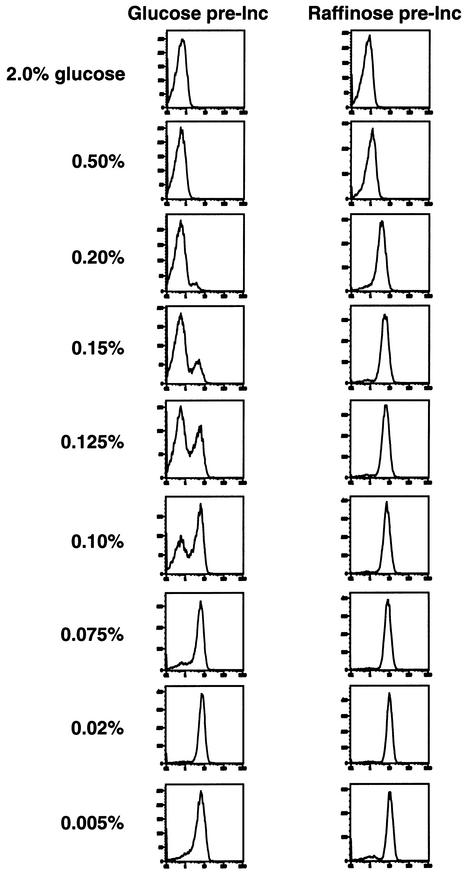
Growth conditions can dictate graded versus binary behavior of the Gal1 promoter
In the experiments described thus far, cell numbers were grown under repressing conditions in standard media containing 2% glucose prior to exposure to experimental conditions. In a parallel study, cells were grown in non-glucose (2% raffinose) media. In this setting, all three arms of glucose repression were inactive (see Figure 1), but the absence of galactose prevented GAL1–GFP expression because Gal80 binds and inactivates Gal4 in the absence of galactose. These cells were then exposed to galactose and varying concentrations of glucose under conditions similar to those in Figure 3, and GFP expression was measured by FACS. Unlike the binary response seen when cells were grown in glucose media (Figures 3 and 6, left panels), cells grown in raffinose media responded to glucose with graded changes in gene expression (Figure 6). Thus, when pre-grown under different conditions, isogenic cells displayed distinct patterns of GAL–GFP expression in response to the same glucose signal.
Fig. 6. Single-cell GAL1–GFP expression in response to glucose in cells expanded in non-glucose media. Cells carrying the integrated GAL1–GFP reporter (YSB31) were grown overnight in 2% raffinose media, washed, and transferred to media containing the indicated concentrations of glucose and 2% galactose for 14 h. Cells were harvested at mid-log phase and then analyzed for GFP expression by FACS.
Low-density cultures were used to measure transcriptional responses
We considered the possibility that our results would be affected by varying cell densities and degrees of glucose depletion from the media. For this reason, cells were grown logarithmically to low densities (<0.075) at the time of harvest. As a result, continued growth in culture beyond the time points presented here (14 h) produced only minor changes in GFP expression profiles (data not shown). In addition, the observation that mig1 mutants displayed binary behavior, while gal80 mutants displayed graded behavior under similar conditions, indicated that binary and graded behavior were not due to culture conditions.
Discussion
Our data demonstrate that cells are equipped with the flexibility to generate either cell-by-cell binary changes in gene expression or population-wide graded changes. We find that specific regulation of a transcriptional activator or a repressor produced graded effects on gene expression. On the other hand, depending on growth conditions, these same transcription factors formed part of either binary or graded transcriptional responses when they functioned as parts of endogenous signaling pathways. Although the integrated glucose-mediated controls over GAL1 promoter activity are significantly more complicated than specific regulation of individual transcription factors, our studies suggest that the regulation of GAL80 within the glucose-response pathway imparts binary character to transcriptional responses.
At least two molecular mechanisms could explain the combination of binary and graded characteristics of promoter activity. Levels of initiation at a promoter could be pulsatile and set to function at only fully on or fully off, and graded regulation would be achieved by controlling the pulse length. Alternatively, and perhaps more likely, rates of initiation could simply vary continuously over a wide range, and binary responses depend on all-or-none changes in the functions of trans-regulatory factors. Present technical approaches do not permit these differences to be distinguished. We noted that under conditions that produced binary changes in expression, the peak of expression in GFP-positive cells shifted slightly (∼3-fold) with varying glucose concentrations (Figures 4, 5A and 6). Thus, a binary response may not exclude underlying graded changes in promoter activity.
Our data indicate that different signaling mechanisms impart distinct modes of transcriptional response. Glucose-mediated repression of the GAL1 promoter depends almost entirely on two transcriptional repressors, MIG1 and GAL80 (data not shown; Nehlin et al., 1991), and distinct signaling pathways control the functions of these two repressors in response to glucose (reviewed by Carlson, 1999). A balance between a kinase and a phosphatase controls Mig1 function (Jiang and Carlson, 1996). Regulation of Gal80 by glucose is less well defined, but appears to involve changes in the levels and activities of Gal80, Gal3 and Gal1 (Zenke et al., 1996). Interest ingly, cells lacking GAL80 responded to glucose in a graded fashion, while cells lacking MIG1 responded in a binary way. In separate experiments, titrating the function of Gal80 with galactose produced graded changes in expression, demonstrating that binary responses were not due to properties inherent to Gal80 itself. Instead, the results suggest that the two glucose-mediated signaling pathways controlling Mig1 and Gal80 ultimately produce either gradual changes in gene expression or all-or-none changes, respectively.
Binary changes in gene expression may result from the catalytic nature of many signaling cascades. Hence, the signal amplification that results may lead to activation of an entire cellular pool of transcription factors. Thus, only cells that initiate the signaling cascade will alter expression, and they will do so with a maximal response, thereby generating a binary pattern of transcriptional regulation. An all-or-none response to graded signaling could also arise from the positive and negative feedback mechanisms that exist in many signaling pathways. Similarly, transcriptional activators stimulate promoters in a synergistic fashion, with minimal or no expression seen when one activator binds a promoter, but with an unexpectedly high level of expression seen when several activators bind (Carey et al., 1990). Even with gradual or graded regulation of transcription factor activity, this synergy among activators may lead to apparent binary responses in gene expression, since transcription will reflect the probability of the filling of independent transcription factor binding sites.
Chromatin remodeling may be an additional cause of binary transcriptional responses. Promoters bound into chromatin become unresponsive to the transcriptional machinery (Weintraub, 1985; Lorch et al., 1987; Matsui, 1987; Workman and Roeder, 1987), and the change between the repressed and remodeled configurations of chromatin may serve as a switch between transcriptionally incompetent and competent states. This switch could impart a binary character to transcriptional responses involving alterations in chromatin structure, such as responses at the GAL1 promoter (Axelrod et al., 1993; Cavalli and Thoma, 1993; Lohr and Lopez, 1995). However, mutations in components of the chromatin remodeling transcription factor Swi-Snf reduced expression in entire populations of cells in a graded fashion (Sudarsanam et al., 1999). Thus, the role of chromatin in determining binary responses in gene expression may be limited.
Recently, Blau and colleagues have shown that competition between an activator and a repressor can produce a binary pattern of transcription (Rossi et al., 2000). They found that viral infection of cells with a TetR–VP16 fusion and a TetR can generate binary patterns of expression, while the activator alone produces graded changes in expression. This is thought to reflect the probability of occupancy of the single site for a repressor or activator. In our case, the repressor (Mig1) and the activator (Gal4) bind at separate sites on the Gal1 promoter. Hence, competition between activators and repressors in a population of cells is unlikely to account for the two forms of regulation that we describe and the shift between the graded and binary behavior found with growth conditions.
GAL80 is necessary and sufficient to impart a binary response to glucose, but the aspect(s) of GAL80 regulation responsible for binary changes in transcription remains unknown. Possibilities include all-or-none propagation of cellular signals that control Gal80 function, binary changes in expression of GAL80 itself, and/or binary interactions between Gal80 and Gal4. Interestingly, unlike cells expanded in glucose media, cells grown in raffinose media showed graded responses to glucose signaling. Gal80 was still active in cells grown in raffinose because GAL1–GFP expression required galactose (data not shown). Furthermore, the differences between growing cells in glucose or raffinose did not result only from inactivation of Mig1 in raffinose, since mig1Δ cells showed binary responses to glucose. Thus, pre-growth in the absence of glucose appeared to alter the context in which glucose-mediated signals control Gal80 function, resulting in a switch between binary and graded modes of regulation. This difference in response patterns should provide a means of defining the component(s) of Gal80 regulation responsible for binary transcriptional responses.
In summary, analysis of the regulation of a single-copy GAL1 promoter in individual yeast cells has identified some of the molecular components of binary and graded transcriptional control. Most surprisingly, isogenetic cells possess the flexibility to generate either binary or graded changes in gene expression in response to the same glucose signal.
Materials and methods
Yeast strains and growth conditions
Cells were grown in standard media at 30°C (Guthrie and Fink, 1991). YSB30 (Matα leu2-3,112 ura3-52 trp1-901 his3-Δ200 ade2-101 gal4Δ gal80Δ fpr1Δ::ADE2 TOR1-1 LY2::GAL→HIS3 GAL→LacZ URA3:: GAL1→GFP) was derived by transforming YSB7 (Biggar and Crabtree, 2000) with pGAL-GFP cut with NcoI with the URA3 gene. YSB31 (Matα ura3-52 lys2-801 ade2-101 trp1-63 his3-Δ200 leu2-Δ1 URA3:: GAL1→GFP) was derived by transforming YPH499 with pGAL-GFP cut with NcoI with the URA3 gene. YSB32 (YSB31 mig1Δ::TRP1) was derived by transforming YSB31 with pMIG1-ΔTRP1 cut with EcoRI. YSB35 (Mata ura3-52 lys2-801 ade2-101 trp1 his3-200 leu2-1 metΔ can1Δ gal80Δ538 GAL1-GFP::URA3 Gal-LacZ::LEU2) was derived from a cross between YSB36 [YPH500 (Sikorski and Hieter, 1989) carrying pGAL-GFP] and YM3733 (Lutfiyya and Johnston, 1996).
Plasmids
To create the GAL–GFP reporter, enhanced GFP was cloned as a XhoI–EcoRI fragment into pRS306 (Sikorski and Hieter, 1989) along with an EcoRI–NotI fragment containing the ADC1 transcriptional terminator to form pYSB113. The full-length GAL1/10 promoter was cloned as a KpnI fragment into pYSB113 to generate pGAL-GFP. Orientation of the GAL1/10 promoter fragment placed GFP under the GAL1 promoter. pMIG1-ΔTRP1 replaced +170 to +1430 of the MIG1 open reading frame with TRP1.
FACS analysis
All yeast strains were grown for at least 14 h under inducing conditions prior to FACS. Cells were sonicated briefly prior to analysis, and at least 10 000 cells were analyzed per sample. β-galactosidase levels were measured using methyl-umbilliferol-galactoside (MUG) (Biggar and Crabtree, 1999).
References
- Axelrod J.D., Reagan,M.S. and Majors,J. (1993) GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev., 7, 857–869. [DOI] [PubMed] [Google Scholar]
- Biggar S.R. and Crabtree,G.R. (1999) Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J., 18, 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar S.R. and Crabtree,G.R. (2000) Chemically regulated transcription factors reveal the persistence of repressor-resistant transcription after disrupting activator function. J. Biol. Chem., 275, 25381–25390. [DOI] [PubMed] [Google Scholar]
- Carey M., Lin,Y.-S., Green,M.R. and Ptashne,M. (1990) A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature, 345, 361–364. [DOI] [PubMed] [Google Scholar]
- Carlson M. (1999) Glucose repression in yeast. Curr. Opin. Microbiol., 2, 202–207. [DOI] [PubMed] [Google Scholar]
- Cavalli G. and Thoma,F. (1993) Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J., 12, 4603–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipstone N.A., Fiorentino,D.F. and Crabtree,G.R. (1994) Molecular analysis of the interaction of calcineurin with drug–immunophilin complexes. J. Biol. Chem., 269, 26431–26437. [PubMed] [Google Scholar]
- Fiering S., Northrop,J.P., Nolan,G.P., Mattila,P.S., Crabtree,G.R. and Herzenberg,L.A. (1990) Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev., 4, 1823–1834. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, New York, NY.
- Ho S.N., Biggar,S.R., Spencer,D.M., Schreiber,S.L. and Crabtree,G.R. (1996) Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature, 382, 822–826. [DOI] [PubMed] [Google Scholar]
- Hollander G.A. et al. (1998) Monoallelic expression of the interleukin-2 locus. Science, 279, 2118–2121. [DOI] [PubMed] [Google Scholar]
- Jiang R. and Carlson,M. (1996) Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev., 10, 3105–3115. [DOI] [PubMed] [Google Scholar]
- Johnston M., Flick,J.S. and Pexton,T. (1994) Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 3834–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphier M.S. and Ptashne,M. (1992) Multiple mechanisms mediate glucose repression of the yeast GAL1 gene. Proc. Natl Acad. Sci. USA, 89, 5922–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. and Lopez,J. (1995) GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J. Biol. Chem., 270, 27671–27678. [DOI] [PubMed] [Google Scholar]
- Lohr D., Venkov,P. and Zlatanova,J. (1995) Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J., 9, 777–787. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe,J.W. and Kornberg,R.D. (1987) Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell, 49, 203–210. [DOI] [PubMed] [Google Scholar]
- Lutfiyya L.L. and Johnston,M. (1996) Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol., 16, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magis W., Fiering,S., Groudine,M. and Martin,D.I. (1996) An upstream activator of transcription coordinately increases the level and epigenetic stability of gene expression. Proc. Natl Acad. Sci. USA, 93, 13914–13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton M.J. et al. (1998) Drug target validation and identification of secondary drug target effects using DNA microarrays. Nature Med., 4, 1293–1301. [DOI] [PubMed] [Google Scholar]
- Matsui T. (1987) Transcription of adenovirus 2 major late and peptide IX genes under conditions of in vitro nucleosome assembly. Mol. Cell. Biol., 7, 1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A.M. and Ley,T.J. (1990) Conservation of the primary structure, organization and function of the human and mouse β-globin locus-activating regions. Proc. Natl Acad. Sci. USA, 87, 7693–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P., Hen,R., Wasylyk,B., Everett,R., Gaub,M.P. and Chambon,P. (1981) The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res., 9, 6047–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J.O., Carlberg,M. and Ronne,H. (1991) Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J., 10, 3373–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal R.K., Riles,L., Johnston,M. and Hegemann,J.H. (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast, 12, 773–786. [DOI] [PubMed] [Google Scholar]
- Rossi F.M., Kringstein,A.M., Spicher,A., Guicherit,O.M. and Blau,H.M. (2000) Transcriptional control: rheostat converted to on/off switch. Mol. Cell, 6, 723–728. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P., Cao,Y., Wu,L., Laurent,B.C. and Winston,F. (1999) The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J., 18, 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M.C., Fiering,S., Eidemiller,J., Magis,W., Groudine,M. and Martin,D.I. (1995) Enhancers increase the probability but not the level of gene expression. Proc. Natl Acad. Sci. USA, 92, 7125–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M.C., Magis,W., Fiering,S., Eidemiller,J., Scalzo,D., Groudine,M. and Martin,D.I. (1996) Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev., 10, 185–195. [DOI] [PubMed] [Google Scholar]
- Weintraub H. (1985) Assembly and propagation of repressed and depressed chromosomal states. Cell, 42, 705–711. [DOI] [PubMed] [Google Scholar]
- Weintraub H. (1988) Formation of stable transcription complexes as assayed by analysis of individual templates. Proc. Natl Acad. Sci. USA, 85, 5819–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J.L. and Roeder,R.G. (1987) Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell, 51, 613–622. [DOI] [PubMed] [Google Scholar]
- Zenke F.T., Engles,R., Vollenbroich,V., Meyer,J., Hollenberg,C.P. and Breunig,K.D. (1996) Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p [published erratum appears in Science, 1996, 273, 417]. Science, 272, 1662–1665. [DOI] [PubMed] [Google Scholar]