Abstract
BACKGROUND AND AIMS: Genomewide expression profiling has identified a number of genes expressed at higher levels in colorectal cancer (CRC) than in normal tissues. Our objectives in this study were: 1) to test whether genes were also distinct on the protein level; 2) to evaluate these biomarkers in a series of well-characterized CRCs; and 3) to apply hierarchical cluster analysis to the immunohistochemical data. METHODS: Tissue microarrays (TMAs) comprising 351 CRC specimens from 270 patients were constructed to evaluate the genes Adam10, CyclinD1, AnnexinII, NFKB, Casein-kinase-2-beta (CK2B), YB-1, P32, Rad51, c-fos, IGFBP4, and Connexin26 (Cx26). In total, 3,797 samples were analyzed. RESULTS: Unsupervised hierarchical clustering discovered subgroups of CRC that differed by tumor stage and survival. Kaplan-Meier analysis showed that reduced Cx26 expression was significantly associated with shorter patient survival and higher tumor grade (G1/G2 vs G3, P = .02), and Adam10 expression with a higher tumor stage (pT1/2 vs pT3/4, P = .04). CONCLUSIONS: Our study highlights the potential of TMAs for a higher-dimensional analysis by evaluating serial sections of the same tissue core (three-dimensional TMA analysis). In addition, it endorses the use of immunohistochemistry supplemented by hierarchical clustering for the identification of tumor subgroups with diagnostic and prognostic signatures.
Keywords: Connexin26 (Cx26), Adam10, colorectal cancer (CRC), hierarchical clustering, tissue microarray (TMA)
Introduction
Cancer of the colon and rectum is the second most prevalent cause of cancer deaths in men and the third most common in women [1]. Postoperative adjuvant chemotherapy improves outcome in stage III (Dukes stage C) colon cancer and is now widely accepted as standard therapy [2,3]. Many patients with stage II (Dukes stage B) disease are considered to be at high risk for recurrence and receive adjuvant therapy, although its benefit in such cases is uncertain. Markers that reliably predict survival are needed [2,4,5]. These biomarkers should support the clinical treatment of neoplastic processes (e.g., in selecting specific drug regimens).
Genomewide expression profiling has identified a number of genes expressed at higher levels in colorectal cancer (CRC) than in normal tissue, representing excellent candidates for diagnostic immunohistochemistry. Tissue microarrays (TMAs) can be used to test the prognostic significance of antibodies against proteins encoded by differentially expressed genes using large numbers of archival patient specimens. Our objectives in this study were: 1) to test whether genes found to be differentially expressed in CRCs by cDNA expression profiling were also distinct on the protein level; 2) to evaluate these potential immunohistochemical markers in a series of well-characterized CRCs including primary and metastatic tumors; and 3) to apply hierarchical cluster analysis to the semiquantitatively scored data and to determine whether the panel of markers allows a meaningful grouping of CRCs.
Materials and Methods
Tissue Array Construction
Two TMAs containing 351 samples from 270 patients were constructed. Tissue samples originated from surgical resections at the Department of Surgery of the Charité. On each TMA, six noncancerous normal mucosa specimens were included. The tumor collective and its clinicopathologic data are summarized in Table 1. One 0.6-mm core was taken from a representative area of the tumor and inserted into a recipient paraffin block to create the TMA [6]. We investigated serial slides cut consecutively and examined the same tumor region in three dimensions (Figure 1A). The investigation of one marker represents a two-dimensional (2D) analysis; additional sections can be viewed as a three-dimensional (3D) analysis of the tissue core in which each marker constitutes another dimension of evaluation. In total, 3797 specimens of colorectal tissue, including normal mucosa, were evaluated.
Table 1.
Cases Used for TMA.
| Number of specimens | 351 |
| Number of patients | 270 |
| Primary tumors | 155 |
| Metastases | 188 |
| Liver | 74 |
| Lymph nodes | 151 |
| Abdominal wall | 59 |
| Lung | 18 |
| Bone | 1 |
| Local recurrences | 11 |
| Normal colon mucosa | 6 |
Figure 1.
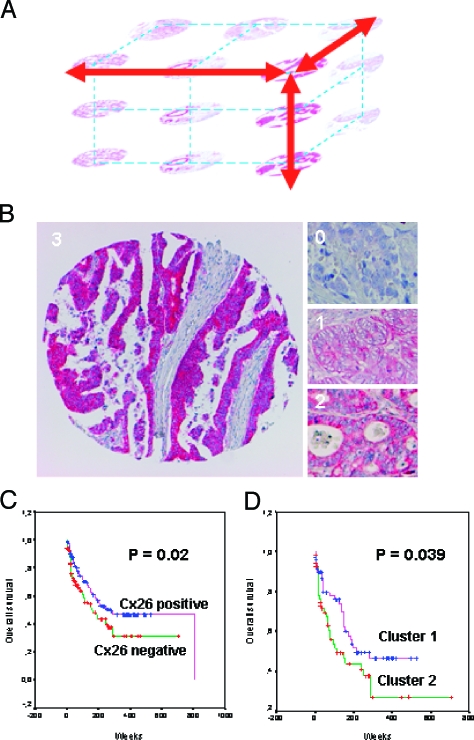
(A) Illustration of the 3D analysis of a tumor using TMAs. By sectioning one core, adjacent areas of a carcinoma potentially representing the same tumor cell clone are analyzed for multiple markers. (B) Examples of the immunohistochemical assessment of Cx26 staining in CRCs using TMAs. Negative staining of tumor cells (0), weakly positive (1), moderately positive (2), and strongly positive (3) (original magnification, x50 magnification). (C) Kaplan-Meier plot comparing disease-specific survival in patients with Cx26-positive colorectal tumors (n = 178) and patients with Cx26-negative tumors (n = 124) (P = .02). (D) Kaplan-Meier plot comparing disease-specific survival in cluster 1 “high tumor stage cluster” and cluster 2 “shorter survival cluster” (P = .039).
Immunohistochemistry
Commercial available antibodies against Adam10, Cyclin-D1, IGFBP4, NFKB, AnnexinII, Casein kinase 2 beta (CK2B), Rad51, YB-1, P32, c-fos, and Connexin26 (Cx26) were purchased. Antibody sources and staining conditions, including antigen retrieval methods, are summarized in Table 2. The genes were chosen in the following ways: first, by personal communication from collaborators who performed cDNA microarray analysis on primary and metastatic CRC; second, by review of the literature [7,8]; and, third, by the suitability of commercially available antibodies. Except for Cx26, only upregulated genes were investigated. Antigen retrieval was performed in a pressure cooker by boiling for 5 minutes then incubating for 25 minutes in a citrate buffer. Slides were stained manually using the Dako ChemMate TM Detection Kit Alkaline Phosphatase/Red Code No. K 5005 (Dako, Corporation Hamburg, Germany) following manufacturer's instructions. Dako Tris-buffered saline (TBS) was used as a washing buffer. For all antibodies, immunostaining of the cells was evaluated and scored semiquantitatively: (9) uninterpretable (missing spot, no tumor cells, or uninterpretable staining); (0) negative; (1+) weak; (2+) moderate; and (3+) strongly positive.
Table 2.
Antibodies for Immunohistochemistry.
| Antigen | Product Number | Supplier | Dilution | Pretreatment |
| Adam10 | Sc-16523 | Santa Cruz Biotechnology (Santa Cruz, CA) | 1:1000 | Microwave |
| Annexin II | Sc-9061 | Santa Cruz Biotechnology | 1:200 | Microwave |
| CK2B | Sc-12739 | Santa Cruz Biotechnology | 1:50 | Microwave |
| Cyclin D1 | 18-0220 | Zymed Laboratory, Inc. (South San Francisco, CA) | 1:50 | Microwave |
| NFKB | Sc-8008 | Santa Cruz Biotechnology | 1:400 | Microwave |
| IGFBP4 | AF-804 | R&D Systems, Inc. (Minneapolis, MN) | 1:50 | Microwave |
| c-fos | Ab-3 | Merck Biosciences, GmbH (Darmstadt, Germany) | 1:50 | Microwave |
| Rad 51 | NA71 | Merck Biosciences, GmbH | 1:200 | Microwave |
| YB-1 | Polyclonal, peptide-specific | Biogenes (Berlin, Germany) | 1:50 | Microwave |
| P32 | Polyclonal, peptide-specific | Biogenes | 1:200 | Microwave |
| Cx26 | CX-12H10 | Zymed Laboratory, Inc. | 1:100 | Microwave |
Hierarchical Clustering Analysis
Hierarchical clustering analysis of our TMA data was performed using the Cluster and TreeView software tool programs that were originally developed for analyzing cDNA microarray data (Gene Cluster 3.0 by Michel de Hoon). Cluster and TreeView software are freely available programs that can be accessed at http://rana.lbl.gov/EisenSoftware.htm. An Excel macro was designed for converting raw TMA staining data from a workbook with multiple worksheets in Excel into a tabular format compatible for use with Gene Cluster. Average linkage hierarchical clustering [9] was then performed on the reformatted data using the Cluster software, with filters set to require at least 80% interpretable immunostaining data for each specimen (n = 190) of 13 immunohistochemical evaluation methods (2470 datasets). Hierarchical clustering was carried out in two dimensions: tumors were grouped together based on the relatedness of their immunostaining profile, and antibodies were grouped based on which tumors they stain. The output was visualized using TreeView, which graphically displays the results of the cluster analysis as dendrograms and arrays, wherein the rows and columns correspond to the raw staining data, presented in the order determined by hierarchical clustering.
Statistical Analysis
Fisher's exact test was used to determine the strength of association between the investigated parameters. P ≤ .05 was considered significant. All calculations were performed on a PC using the statistical software package SPSS (Munich, Germany). Clinicopathologic parameters including follow-up were available for all specimens with a mean follow-up period of 108 weeks. The differences of the Kaplan-Meier survival curves were tested for statistical significance with the log rank test, and the 95% confidence intervals were calculated. For each tumor specimen, the date of operation, date of last follow-up, and vital status at last follow-up (i.e., living or deceased) were recorded. Disease-specific survival was calculated.
Multivariate analyses were performed with a proportional hazard model (i.e., Cox regression), and stepwise backward/forward procedures provided by SPSS software were used to reduce the number of variables in the Cox models. For assessing and comparing the Cox models, a Wald test with significance level of .05 was used for both inclusion and exclusion of variables.
Results
Immunohistochemistry
In total, immunohistochemical data from 3797 colorectal tissue spots of CRC and normal colon mucosa were acquired using 11 different antibodies. The normal colon mucosa samples showed no relevant staining for all investigated genes except for Cx26, for which we observed a weak cytoplasmic staining in two of six cases. Sometimes the mucin of the goblet cells was stained, but we did not consider this as specific staining. The results of the entire tumor collective and all antibodies are summarized in Table 3. The expression was scored semiquantitatively by a fourtier scale (0—negative, 1—weak, 2—moderate, 3—strongly positive; Figure 1B) for the clustering analysis. This was reduced to a two-tier system (0/1—negative, 2/3—positive) for the independently performed statistical analysis of single genes and their correlation with clinicopathologic parameters including survival (Figure 1C). For the markers YB-1 and CK2B, nuclear and cytoplasmic stainings were evaluated independently.
Table 3.
Investigated Specimens with Immunohistochemical Results.
| Evaluation Method | All Evaluated Specimens (n = 3791) | |||
| Low (n = 1924) | High (n = 1867) | |||
| n | % | n | % | |
| Cx26 | 124 | 41 | 178 | 59 |
| Adam10 | 90 | 28 | 236 | 72 |
| Annexin II | 140 | 44 | 177 | 56 |
| CK2B cytoplasm | 100 | 33 | 206 | 67 |
| CK2B nuclear | 245 | 80 | 61 | 20 |
| Cyclin D1 | 207 | 60 | 138 | 40 |
| NFKB | 135 | 45 | 167 | 55 |
| IGFBP4 | 173 | 79 | 46 | 21 |
| Rad 51 | 67 | 27 | 180 | 73 |
| YB-1 cytoplasm | 255 | 86 | 43 | 14 |
| YB-1 nuclear | 158 | 53 | 139 | 42 |
| P32 cyt | 106 | 32 | 221 | 68 |
| c-fos | 124 | 62 | 75 | 38 |
Hierarchical Clustering Analysis of TMA Immunostains
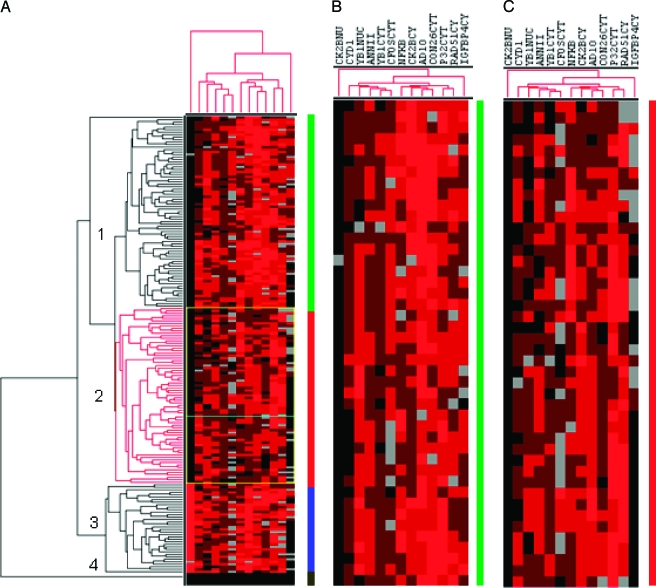
An unsupervised hierarchical clustering algorithm allowed us to cluster the specimens on the basis of their similarities measured over the 13 immunohistochemical evaluations. Requiring 80% interpretable immunostaining results for each specimen, in total, 2470 data points were included in the analysis. The expression of the antibodies was clustered on the basis of their similarities measured over the group of 190 tumors (Figure 2). In dendrograms shown in Figure 2, the length and subdivision of the branches display the relatedness of the colorectal tumors (left) and gene expressions (top). Unsupervised hierarchical clustering was able to distinguish colorectal specimens in four groups (Figure 2A). As expected, the normal tissue showed a separate cluster (bottom). The other tumors were divided into three clusters on the basis of this set of antibodies. For each of the antibodies indicated at the top of the figure, strong positive staining is indicated by a red square, moderate positive staining in dark brown, weak staining by light brown, absence of staining as black, and no available data as grey.
Figure 2.
(A) Hierarchical cluster analysis of CRC TMA immunostaining results. For each of the antibodies indicated at the top of the figure, strong positive staining is indicated by a red square, moderate staining by a light brown, weak positive staining by dark brown, absence of staining as black, and no available data as grey. The dendogram at the top shows the clustering of antibodies based on the relatedness of tumors stained by each antibody. The dendogram on the left side shows the clustering of the tumors based on the degree of similarity of their immunohistochemical staining results. (B) Enlarged portion from the so-called “high tumor stage cluster” with prominent Adam10 staining (see vertical line). (C) Enlarged portion of the “shorter survival stage cluster” with prominently reduced Cx26 staining (see vertical line). Number and different colors: cluster group 1 (green), cluster group 2 (red), cluster group 3 (blue), cluster group 4 normal tissue (black).
Tumor group 1 was dominated by an increased Adam10 and IGFBP4 expression, whereas no nuclear CK2B staining was detectable (Figure 2B). Within this subgroup, 85% of cases showed either pT3 or pT4 tumor stage. Tumor group 2 showed lower levels of Cx26, Adam10, and IGFBP4 expressions than group 1 tumors, whereas nuclear CK2B was similarly negative (Figure 2C). In contrast, group 3 tumors were essentially distinguishable by the positive nuclear expression of the CK2B antigen.
Immunohistochemistry and Clinicopathologic Parameters
Survival analysis Exploratory analysis was conducted to correlate the outcome of patients monitored during the 809-week period with the immunohistochemistry results. The analysis was restricted to disease-specific survival and was performed on all specimens. Overexpression (scores 2+ and 3+) of Cx26 was found in 124 (41%) specimens. A total of 178 (59%) exhibited no relevant Cx26 staining (scores 0 and 1+). The corresponding survival curves according to Cx26 expression are shown in Figure 1C. The median survival time of the patients was 194,68 weeks. Statistical analysis showed that patients with high Cx26 expression tumors had significantly longer survival rates than patients with low Cx26 expression (P = .02). All other investigated antigens showed no prognostic relevance (P > .05). Comparing the cluster group 1 with the cluster group 2 in all investigated samples, we could demonstrate a significant difference (P = .039) with shorter survival in cluster 2 (Figure 1D).
TNM parameters Cx26 overexpression (scores 2+ and 3+) was significantly linked to G1/G2 tumors when compared to G3 carcinomas (P = .026). Adam10 overexpression was linked to a higher tumor stage (pT1/2 vs pT2/3, P = .04). No other significant correlation with antibody expression could be demonstrated. The significant results of Cx26 and Adam10 are summarized in Table 4. When performing multivariate analysis comparing Cx26 with the parameters tumor stages (pT), grading (G), and nodal stage (pN), Cx26 was not an independent parameter. However, excluding the grading and the tumor stage, Cx26 was an independent factor in multivariate analysis and showed a higher significance (P = .022) than nodal status (pN0 vs pN1/2).
Table 4.
Immunohistochemical Results with Cx26 and Adam10 in CRC.
| Cx26 (n = 302) | Adam10 (n = 326) | |||
| Low (n = 124) | High (n = 178) | Low (n = 90) | High (n = 236) | |
| Primary tumor | 43 | 71 | 33 | 85 |
| Metastasis | 81 | 107 | 57 | 151 |
| Liver | 22 | 26 | 17 | 36 |
| Lymph nodes | 35 | 43 | 19 | 71 |
| Abdominal wall | 12 | 24 | 10 | 29 |
| Lung | 7 | 10 | 9 | 7 |
| Bone | 0 | 1 | 0 | 1 |
| Stage (P = .04) | ||||
| pT1/2 | 19 | 101 | 8 | 82 |
| pT3/4 | 32 | 145 | 42 | 190 |
| Tumor differentiation (P = .026) | ||||
| G1/G2 | 77 | 32 | 58 | 23 |
| G3 | 141 | 31 | 167 | 60 |
| Nodal status | ||||
| pN0 | 16 | 104 | 14 | 5 |
| pN1/2 | 33 | 137 | 39 | 186 |
| Metastasis | ||||
| M0 | 42 | 81 | 29 | 60 |
| M1 | 73 | 104 | 89 | 146 |
We could find differences between the primary tumor and the metastases from the same patient, but the percentage was very low (e.g., regarding Cx26, we found from 55 paired samples the same expression pattern of Cx26 in the primary tumor and metastases in 41 patients, a negative expression in metastases and a high expression in the primary tumor in eight patients, and a positive expression in the primary tumor and a negative expression in metastases in six patients). The differences were not significant in the Wilcoxon test (P = 0.79), especially suited for paired samples.
The comparison between proximal (cecum, colon ascendens, and colon transversum) and distal carcinomas (rectum, sigma, and colon descendens) showed a significant correlation with positive NFKB carcinomas in distal colorectal tumors (P = .032).
Typical expression patterns of individual genes are available as supplementary data on the publisher's web site and our Berlin-TMA web portal (http://pathoweb.charite.de/tmaportal).
Discussion
This study is the first comprehensive and the largest analysis of different immunohistochemical biomarkers associated with clinicopathologic parameters in CRCs using the synergy of TMA and hierarchical clustering. We were able to investigate 3797 specimens. As a result, new biomarkers in the progression of CRC were detected.
In the past, whole mount sections were used to validate new biomarkers. They have the advantage of providing an overview on a larger tumor area, thus enabling particularly the evaluation of the presence and extent of heterogeneity in protein expression within a carcinoma. The major disadvantage, however, is the large amount of tissues, reagents, and labor that is needed for the investigation of only one marker. Similarly, the identification and evaluation of a specific tumor area within a whole mount section are cumbersome. The problem of expression heterogeneity can be compensated by the analysis of a large tumor collective and the use of multiple cores representing the same tumor within a TMA [10]. Furthermore, TMAs provide a simple mean to analyze adjacent areas by sectioning one core with a high probability to evaluate tumor cells of the same clone. In addition to the two dimensions of one TMA spot, similar tumor cells can be investigated for multiple markers in a 3D analysis (Figure 1A). Hierarchical clustering and other statistical means provide an even higher-dimensional analysis of a tumor collective.
Hierarchical clustering has been so far mainly applied to expression profiling data, which constitutes a quantitative numerical measure on the amount of mRNA within a tumor. Immunohistochemical data are, on one hand, simpler because the expression level can only be scored semiquantitatively; on the other hand, it is far more sophisticated because it visualizes the complexity of protein expression between different cell types (e.g., tumor versus stromal cells) and cell compartments (e.g., cytoplasmic, nuclear, and membranous). Only a few studies combined TMA technology and hierarchical clustering [11–13] so far using a three-tier (absent, weakly, and strongly positive) as well as a four-tier system similar to ours. There are yet no clear criteria and final arguments to favor one or the other system [14]. We were mainly influenced by conventional practice (i.e., scoring of HER/NEU2 expression in four scales) and the view that one additional grade may enhance the semiquantitative representation of the protein level. Further studies will demonstrate whether a three-tier or a four-tier system is advantageous. We evaluated only tumor cell expression and scored cytoplasmic and nuclear staining independently for two markers.
The synergy of hierarchical clustering and TMA immunohistochemistry (i.e., the combination of a high-throughput technology with an elegant statistical method) carries the potential to improve our understanding of immunoprofiles and gene signatures. Unsupervised hierarchical clustering allowed us to subgroup 190 CRC specimens on the basis of their similarities in 13 gene expressions (Figure 2). Hierarchical clustering was able to distinguish colorectal specimens in four groups. As expected, the normal tissue showed a separate cluster (bottom) for the upregulated genes. The other tumors were divided into three clusters on the basis of this set of genes. Notably, in the upper cluster, 85% of the tumors are pT3/4 carcinomas, which thus represents a “high tumor stage cluster.” This cluster is supported by a high Adam10 staining, which is significantly correlated with a higher tumor stage. In the second cluster, the so-called “shorter survival cluster,” 79% of the patients survived for shorter than 200 weeks and a relatively high percentage of the patients (39%) carried high-grade tumors (G3), which is represented by the prominently reduced Cx26 staining. The third cluster is distinguished by the high nuclear CK2B expression. Until now, the role of CK2B in carcinogenesis remains unclear. The CK2B overexpression in the cytosol is clustered together with Adam10, Cx26, and NFKB, which might be of biologic relevance. However, the overall number of tumors within this cluster is relatively low (n = 36) and the results should thus be interpreted cautiously.
To the best of our knowledge, this is the first study showing that reduced Cx26 expression is significantly associated with a shorter patient survival and with a higher grading of CRC. Interestingly, our results are in accordance with observations in human bladder, breast, and lung carcinomas in which Cx26 expression was found to be reduced [15,16]. The Cx26 gene was previously isolated as a candidate tumor-suppressor gene for breast cancer [15,17]. Connexins are a family of transmembrane proteins that enable gap junctional intercellular communication (GJIC), which mediates the transfer of ions, metabolites, and small regulatory molecules between cells [18]. Gap junctions, specialized clusters of intercellular channels, allow adjacent cells to directly share ions and hydrophilic molecules of up to ∼1 kDa in size [19]. This process, known as GJIC, is thought to control homeostasis and coordination of cellular activities in both excitable and nonexcitable tissues [20]. In all, 20 human connexin genes have been identified so far, usually classified by their molecular weight, ranging between 25 and 62 kDa in size [21].
One of the longstanding interests in the field has been the role of GJIC in carcinogenesis. There is substantial evidence that GJIC and/or connexins may act as tumor suppressors [16]. Reduced or aberrant GJIC or connexin expression has been found in many tumors and tumor cell lines [22,23]. Restoration of GJIC in tumor cell lines by connexin transfection can reduce growth and tumorigenicity [24–27]. Cx32-null mice have an increased incidence of hepatocarcinogenesis [28], whereas fibroblasts derived from Cx43-null mice display increased growth rate, loss of cell adhesion, altered morphology, and other properties associated with a transformed phenotype [29]. Recently, identical mutations in Cx26 have been shown to underlie the keratitis-ichthyosis-deafness (KID) [30] and hystrix-like ichthyosis-deafness (HID) syndromes [31,32], and these patients have an increased incidence of squamous cell carcinoma (SCC). In vitro transfection of connexin in HeLa and breast cancer cells significantly reduces cell growth both in vitro an in vivo [24, 32].
Overexpression of another candidate gene, Adam10, showed significant correlation with a higher tumor stage. The “a disintegrin and metalloprotease” (ADAM) family contributes to the regulation of cell-cell and cell-matrix interactions that are critical determinants of malignancy. The recently discovered ADAM family of proteins is unique in that the members have the potential to regulate both extracellular matrix remodeling and cell migration [33,34]. Some ADAM proteins interact with integrins and thus may also play a role in metastasis of cancer cells. In fact, expression of some ADAM proteins is increased in malignant cell populations [35,36], including cells obtained from pancreatic and hepatocellular carcinomas [37,38] and from breast cancers [39]. In one of our former studies, we associated the overexpression of the chromosomal region at 15q22-23 with a shorter patient survival and tumor progression [40,41]. Interestingly, the Adam10 gene is located in this region.
In conclusion, we show in this study that reduced Cx26 expression in CRC is significantly associated with shorter patient survival and a higher tumor grade, and that Adam10 is correlated with a higher tumor stage. Furthermore, we were able to show that hierarchical clustering of TMAs is a useful, promising, and very powerful tool for further investigations, and will lead us to a diagnostic and prognostic signature of different carcinomas.
Acknowledgements
The provision of tumor samples with follow-up by the Department of Surgery at Campus Buch (P. M. Schlag) is gratefully acknowledged. We are much indebted to Martina Eickmann for secretarial assistance and critical reading.
Footnotes
This work was supported by the research fund of the Charité University Hospital (grant no. 281-2003).
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Galanis E, Alberts SR, O'Connel MJ. New adjuvant therapy for colon cancer: justified hope or commercial hype. Surg Oncol Clin North Am. 2000;9:813–823. [PubMed] [Google Scholar]
- 3.Macdonald JS. Adjuvant therapy of colon cancer. CA Cancer J Clin. 1999;49:202–219. doi: 10.3322/canjclin.49.4.202. [DOI] [PubMed] [Google Scholar]
- 4.O'Connel MJ, Schaid DJ, Ganju V, Cunningham J, Kovach JS, Thibodeau SN. Current status of adjuvant chemotherapy for colorectal cancer: can molecular markers play a role in predicting prognosis? Cancer. 1992;(Suppl 70):1732–1739. doi: 10.1002/1097-0142(19920915)70:4+<1732::aid-cncr2820701614>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Chung DC. Molecular prognostic markers and colorectal cancer: the search goes on. Gastroenterology. 1998;114:1330–1332. doi: 10.1016/s0016-5085(98)70441-x. [DOI] [PubMed] [Google Scholar]
- 6.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 7.Hegde P, Qi R, Gaspard R, Abernathy K, Dharap S, Earle-Hughes J, Gay C, Nwokekeh NU, Chen T, Saeed AI, et al. Identification of tumor markers in models of human colorectal cancer using a 19,200-element complementary DNA microarray. Cancer Res. 2001 Nov 1;61(21):7792–7797. [PubMed] [Google Scholar]
- 8.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001 Apr 1;61(7):3124–3130. [PubMed] [Google Scholar]
- 9.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Kochli OR, Mross F, Dieterich H, Moch H, Mihatsch M, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159:2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen TO, Hsu FD, O'Connell JX, Gilks CB, Sorensen PH, Linn S, West RB, Liu CL, Botstein D, Brown PO, et al. Tissue microarray validation of epidermal growth factor receptor and SALL2 in synovial sarcoma with comparison to tumors of similar histology. Am J Pathol. 2003;163:1449–1456. doi: 10.1016/S0002-9440(10)63502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Rijn M, Gilks CB. Applications of microarrays to histopathology. Histopathology. 2004;44:97–108. doi: 10.1111/j.1365-2559.2004.01766.x. [DOI] [PubMed] [Google Scholar]
- 13.Makretsov NA, Huntsman DG, Nielsen TO, Yorida E, Peacock M, Cheang MC, Dunn SE, Hayes M, van de Rijn M, Bajdik C, et al. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin Cancer Res. 2004;10:6143–6151. doi: 10.1158/1078-0432.CCR-04-0429. [DOI] [PubMed] [Google Scholar]
- 14.Knösel T, Yu Y, Stein U, Schwabe H, Schluns K, Schlag PM, Dietel M, Petersen I. Overexpression of cyclooxygenase-2 correlates with chromosomal gain at the cyclooxygenase-2 locus and decreased patient survival in advanced colorectal carcinomas. Dis Colon Rectum. 2004;47:70–77. doi: 10.1007/s10350-003-0008-7. [DOI] [PubMed] [Google Scholar]
- 15.Grossman HB, Liebert M, Lee IW, Lee SW. Decreased connexin expression and intercellular communication in human bladder cancer cells. Cancer Res. 1994;54:3062–3065. [PubMed] [Google Scholar]
- 16.Chen Y, Hühn D, Knösel T, Pacyna-Gengelbach M, Deutschmann N, Petersen I. Downregulation of connexin 26 in human lung cancer is related to promoter methylation. Int J Cancer. 2005;113:14–21. doi: 10.1002/ijc.20498. [DOI] [PubMed] [Google Scholar]
- 17.Lee SW, Tomasetto C, Sager R. Positive selection of candidate tumor-suppressor genes by subtractive hybridization. Proc Natl Acad Sci USA. 1991;88:2825–2829. doi: 10.1073/pnas.88.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knuechel R, Siebert-Wellnhofer A, Traub O, Dermietzel R. Connexin expression and intercellular communication in two- and three-dimensional in vitro cultures of human bladder carcinoma. Am J Pathol. 1996;49:1321–1332. [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar NM, Gilula NB. The gap junction communication channel. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- 20.Simon AM, Goodenough DA. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 1998;8:477–483. doi: 10.1016/s0962-8924(98)01372-5. [DOI] [PubMed] [Google Scholar]
- 21.Eiberger J, Degen J, Romualdi A, Deutsch U, Willecke K, Sohl G. Connexin genes in the mouse and human genome. Cell Commun Adhes. 2001;8:163–165. doi: 10.3109/15419060109080717. [DOI] [PubMed] [Google Scholar]
- 22.Saitoh M, Oyamada M, Oyamada Y, Kaku T, Mori M. Changes in the expression of gap junction proteins (connexins) in hamster tongue epithelium during wound healing and carcinogenesis. Carcinogenesis. 1997;18:1319–1328. doi: 10.1093/carcin/18.7.1319. [DOI] [PubMed] [Google Scholar]
- 23.Laird DW, Fistouris P, Batist G, Alpert L, Huynh HT, Carystinos GD, Alaoui-Jamali MA. Deficiency of connexin 43 gap junctions is an independent marker for breast tumors. Cancer Res. 1999;59:4104–4110. [PubMed] [Google Scholar]
- 24.Hirschi KK, Xu CE, Tsukamoto T, Sager R. Gap junction genes Cx26 and Cx43 individually suppress the cancer phenotype of human mammary carcinoma cells and restore differentiation potential. Cell Growth Differ. 1996;7:861–870. [PubMed] [Google Scholar]
- 25.Huang RP, Fan Y, Hossain MZ, Peng A, Zeng ZL, Boynton AL. Reversion of the neoplastic phenotype of human glioblastoma cells by connexin 43 (Cx43) Cancer Res. 1998;58:5089–5096. [PubMed] [Google Scholar]
- 26.Zhang ZQ, Zhang W, Wang NQ, Bani-Yaghoub M, Lin ZX, Naus CC. Suppression of tumorigenicity of human lung carcinoma cells after transfection with connexin 43. Carcinogenesis. 1998;19:1889–1894. doi: 10.1093/carcin/19.11.1889. [DOI] [PubMed] [Google Scholar]
- 27.Saunders MM, Seraj MJ, Li Z, Zhou Z, Winter CR, Welch DR, Donahue HJ. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001;61:1765–1767. [PubMed] [Google Scholar]
- 28.Temme A, Buchmann A, Gabriel HD, Nelles E, Schwarz M, Willecke K. High incidence of spontaneous and chemically induced liver tumors in mice deficient for connexin 32. Curr Biol. 1997;7:713–716. doi: 10.1016/s0960-9822(06)00302-2. [DOI] [PubMed] [Google Scholar]
- 29.Martyn KD, Kurata WE, Warn-Cramer BJ, Burt JM, TenBroek E, Lau AF. Immortalized connexin 43 knockout cell lines display a subset of biological properties associated with the transformed phenotype. Cell Growth Differ. 1997;8:1015–1027. [PubMed] [Google Scholar]
- 30.Richard G, Rouan F, Willoughby CE, Brown N, Chung P, Ryynanen M, Jabs EW, Bale SJ, DiGiovanna JJ, Uitto J, et al. Missense mutations in GJB2 encoding connexin-26 cause the ectodermal dysplasia keratitis-ichthyosis-deafness syndrome. Am J Hum Genet. 2002;70:1341–1348. doi: 10.1086/339986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Geel M, van Steensel MA, Kuster W, Hennies HC, Happle R, Steijlen PM, Konig A. HID and KID syndromes are associated with the same connexin 26 mutation. Br J Dermatol. 2002;146:938–942. doi: 10.1046/j.1365-2133.2002.04893.x. [DOI] [PubMed] [Google Scholar]
- 32.Mesnil M, Krutovskikh V, Piccoli C, Elfgang C, Traub O, Willecke K, Yamasaki H. Negative growth control of HeLa cells by connexin genes: connexin species specificity. Cancer Res. 1995;55:629–939. [PubMed] [Google Scholar]
- 33.Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131:275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Black RA, White JM. ADAMs: focus on the protease domain. Curr Opin Cell Biol. 1998;10:654–659. doi: 10.1016/s0955-0674(98)80042-2. [DOI] [PubMed] [Google Scholar]
- 35.Wu E, Croucher PI, McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of haematological malignancies. Biochem Biophys Res Commun. 1997;235:437–442. doi: 10.1006/bbrc.1997.6714. [DOI] [PubMed] [Google Scholar]
- 36.Iba K, Albrechtsen R, Gilpin BJ, Loechel F, Wewer UM. Cysteine-rich domain of human ADAM 12 (meltrin alpha) supports tumor cell adhesion. Am J Pathol. 1999;154:1489–1501. doi: 10.1016/s0002-9440(10)65403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grutzmann R, Foerder M, Alldinger I, Staub E, Brummendorf T, Ropcke S, Li X, Kristiansen G, Jesnowski R, Sipos B, et al. Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch. 2003;443:508–517. doi: 10.1007/s00428-003-0884-1. [DOI] [PubMed] [Google Scholar]
- 38.Tannapfel A, Anhalt K, Hausermann P, Sommerer F, Benicke M, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Identification of novel proteins associated with hepatocellular carcinomas using protein microarrays. J Pathol. 2003;201:238–249. doi: 10.1002/path.1420. [DOI] [PubMed] [Google Scholar]
- 39.O'Shea C, McKie N, Buggy Y, Duggan C, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Expression of ADAM-9 mRNA and protein in human breast cancer. Int J Cancer. 2003;105:754–761. doi: 10.1002/ijc.11161. [DOI] [PubMed] [Google Scholar]
- 40.Knösel T, Schlüns K, Stein U, Schwabe H, Schlag PM, Dietel M, Petersen I. Genetic imbalances with impact on survival in colorectal cancer patients. Histopathology. 2003;43:323–331. doi: 10.1046/j.1365-2559.2003.01720.x. [DOI] [PubMed] [Google Scholar]
- 41.Knösel T, Schlüns K, Stein U, Schwabe H, Schlag PM, Dietel M, Petersen I. Chromosomal alterations during lymphatic and liver metastasis formation of colorectal cancer. Neoplasia. 2004;6:23–28. doi: 10.1016/s1476-5586(04)80050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]