Abstract
DNA methylation and copy number in the genomes of three immortalized prostate epithelial and five cancer cell lines (LNCaP, PC3, PC3M, PC3M-Pro4, and PC3M-LN4) were compared using a microarray-based technique. Genomic DNA is cut with a methylation-sensitive enzyme HpaII, followed by linker ligation, polymerase chain reaction (PCR) amplification, labeling, and hybridization to an array of promoter sequences. Only those parts of the genomic DNA that have unmethylated restriction sites within a few hundred base pairs generate PCR products detectable on an array. Of 2732 promoter sequences on a test array, 504 (18.5%) showed differential hybridization between immortalized prostate epithelial and cancer cell lines. Among candidate hypermethylated genes in cancer-derived lines, there were eight (CD44, CDKN1A, ESR1, PLAU, RARB, SFN, TNFRSF6, and TSPY) previously observed in prostate cancer and 13 previously known methylation targets in other cancers (ARHI, bcl-2, BRCA1, CDKN2C, GADD45A, MTAP, PGR, SLC26A4, SPARC, SYK, TJP2, UCHL1, and WIT-1). The majority of genes that appear to be both differentially methylated and differentially regulated between prostate epithelial and cancer cell lines are novel methylation targets, including PAK6, RAD50, TLX3, PIR51, MAP2K5, INSR, FBN1, and GG2-1, representing a rich new source of candidate genes used to study the role of DNA methylation in prostate tumors.
Keywords: promoter, microarray, DNA methylation, CpG islands, gene silencing
Introduction
Aberrant DNA methylation of CpG sites is among the earliest and most frequent alterations in cancer [1,2]. In many cases, DNA methylation at CpG, in or near the promoter or first exon of a gene, is associated with gene “silencing.” Multiple different methylases and proteins that either bind methylated DNA or unmethylated CpG are associated with: 1) transmitting the methylation status to other proteins in the chromatin; 2) maintaining the DNA methylation profile during replication; and 3) changing the methylation profile during differentiation of cells. Thus, as a result of methylation at multiple CpG sequences, chromatin structure in the promoter may be altered, preventing normal interaction with transcriptional machinery. If this occurs in genes critical to growth inhibition, the resulting silencing of transcription could promote tumor growth. Hypermethylation has been shown to be commonly associated with transcriptional inactivation for classic tumor suppressor genes, genes important for cell cycle regulation, and genes that mediate DNA mismatch repair [3].
At present, several molecular biology methods are routinely used to determine the methylation status of a CpG island. Among these, bisulfite nucleotide sequencing is a technique used for a detailed mapping of methylated cytosine residues within a gene promoter [4,5]. Restriction landmark genome scanning (RLGS) is a two-dimensional gel electrophoresis method that has been used to study genetic and epigenetic changes, including DNA methylation [6–8]. Microarrays allow many DNA sequences to be queried in parallel especially when the targets can be made into reduced complexity representations [9,10]. Using this method, the binding profile of proteins that interact specifically with methylated DNA sequences can be detected by chromatin immunoprecipitation (ChIP) [11–14]. Alternatively, DNA methylation can be detected directly by cleavage of the genome with a 5-methylcytosine-sensitive restriction enzyme. In one method, methylation at the methylation-sensitive restriction sites for BstUI and HpaII preserves certain methyl-insensitive MseI fragments that are otherwise cleaved if the site is unmethylated. Difference is amplified polymerase chain reaction (PCR) products indicate differences in BstUI and HpaII methylation [15–17].
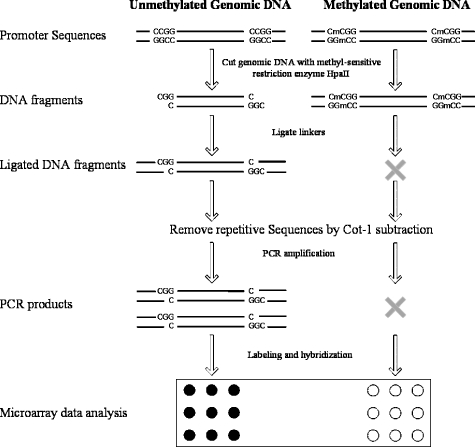
Another method is to cleave with a methyl-sensitive restriction enzyme, size fractionate, and hybridize fractions to a microarray. As methylation will change the size of cleavage products, they will be in a different fraction [18–20]. A protocol for detecting methylation differences between two genomes using this class of methods is outlined in Figure 1. The protocol relies on the occurrence of two methyl-sensitive cleavage sites in close proximity. If the restriction sites are both unmethylated, they can be cleaved and primers can be ligated. When the distance between the ligated primers is short enough, the fragment can be amplified efficiently by PCR. If, on other hand, the DNA is methylated at one of the cleavage sites, it will not be cut at that site and a longer fragment will be produced. In most cases, this longer fragment will be sufficiently long that the PCR of the fragment does not occur efficiently. During PCR, thousands of cleavage-ligation fragments from unmethylated parts of the genome amplify with varying efficiencies, and their representation in the final pool of amplified products depends on the efficiency of their amplification. However, the efficiency of amplification of any particular fragment should remain similar between experiments. Thus, in general, differences in the starting amount of a particular fragment will be preserved in the same ratio after PCR. The reduction in complexity while preserving ratios relies on the same principles as those previously published methods for comparative genomic hybridization (CGH) [9] and expression analysis [10]. These differences are measured on an array of genomic regions; in this case, we monitored methylation changes on an array 2732 promoter-first exon regions [21,22].
Figure 1.
Schematic of the protocol for detecting differences in HpaII fragment amplification between samples.
The method is applied to eight prostate epithelial cell lines—three immortalized epithelial lines and five lines derived from cancers. Differences in copy number can also be detected in this and all the other high-throughput methods referenced here. Here, copy number changes, which are also of interest, are distinguished from methylation changes by a variety methods, such as methylation-specific PCR (MSP), and by 5-aza-2′-deoxycytidine (DAC) treatment. Some of the methylation differences revealed have frequently been observed in prostate cancer, including CD44, CDKN1A, ESR1, PLAU, RARB, SFN, TNFRSF6, and TSPY, and others are previously known methylation targets in other cancers, including ARHI, bcl-2, BRCA1, CDKN2C, MTAP, PGR, SLC26A4, SPARC, SYK, TJP2, UCHL1, and WIT-1. However, most of the methylation candidates are potentially new targets that will need to be confirmed in tumors.
Materials and Methods
Cell Culture
Human prostate epithelial cell lines 267B1, RWPE-1, and Ml-csv40 were kindly provided by Dr. J. Rhim (U.S. Navy Hospital, Bethesda, MD). LNCaP was obtained from ATCC (Manassas, VA), and PC3, PC3M, PC3M-Pro4, and PC3M-LN4 were kindly provided by Dr. Isaiah J. Fidler (M. D. Anderson Hospital Cancer Center, Houston, TX). Cells were cultured in RPMI medium containing 10% fetal bovine serum and 4 mM l-glutamine. LNCaP was also cultured in the presence of mock (PBS) or DAC (1 µm, medium changed every 24 hours; Sigma-Aldrich Co., St. Louis, MO) and the cells were harvested at 24 hours after the third dose. Genomic DNA and total RNA were extracted from cell lines using DNeasy Tissue Kit and RNeasy Mini Kit (Qiagen, Inc., Valencia, CA), respectively.
Preparation of Promoter Microarray
A detailed description of the promoter array used here has been published [21,22]. Briefly, human promoter sequences (1000 bp upstream and 500 bp downstream from transcription initiation site) were retrieved batchwise from http://genome.ucsc.edu/. PCR primers were selected using an in-house version of Primer3 (http://www.broad.mit.edu/genome_software/other/primer3.html). Promoter fragments with an average length of 1.2 kb were amplified, purified, and spotted onto UltraGAPS-coated slides (Corning, Inc., Corning, NY) in the presence of 50% DMSO. The promoter microarray contains triplicate spots of 3083 promoter sequences (2732 when duplicates are considered), 787 nonpromoter controls, and 192 nonhuman controls. Many of the promoters on the array are from genes of particular relevance to cancer, and the array includes promoters from most of the genes that are known to be regulated by methylation in cancer. The array is freely available to collaborators.
Methylation Microarray Analysis
HpaII (New England Biolabs, Beverly, MA) digestion was performed in 20 µl containing 0.5 µg of genomic DNA and 5 U of HpaII for 2 hours at 37°C. Ten-fold overdigestion, plus monitoring of the digestion of lambda DNA mixed with human genomic DNA in a parallel reaction, were used to minimize the possibility of partial digestion. The digested fragments are ligated with linker Meth_1a1/Meth_1a2/Meth_4a1/Meth_4a2 (20 pmol each) in the presence of 20 U/µl of T4 DNA ligase (New England Biolaboratories) and 1 mM ATP (Amersham Biosciences, Piscataway, NJ) at room temperature for 4 hours. The oligonucleotide sequences were as follows: Meth_1a1, 5′-AAGTATCGGAGGAGTCTTTGTTA-3′; Meth_1a2, 5′-phosphate-CGTAACAAAGACTCCTCCGATACTT-amine-3′; Meth_4a1, 5′-TCTCTTGAAGAGTAACTTGTTGG-3′; Meth_4a2, 5′-phosphate-CGCCAACAAGTTACTCTTCAAGAGA-amine-3′. Meth_1a1 and Meth_1a2 are mixed, heated to 95°C for 5 minutes, and cooled down to room temperature before ligation. Meth_4a1 and Meth_4a2 were treated similarly.
Repetitive DNA sequences were depleted from the ligated DNA using a previously described subtraction hybridization protocol [23] with some modification. Ten micrograms of human Cot1 DNA (Invitrogen Corporation, Carlsbad, CA) containing enriched repetitive sequences was biotin-labeled using the BiotinULS Labeling Kit (Fermentas Life Sciences, Hanover, MD) at 85°C for 30 minutes. Cot1 DNA was purified using ethanol precipitation and dissolved with 100 µl of binding buffer TEN100 (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl, pH 7.5). One hundred microliters (1 mg) of streptavidin magnetic particles was prepared according to the manufacturer's instructions (F. Hoffmann-La Roche Ltd., Alameda, CA), added to the biotin-labeled Cot1 DNA, and incubated at room temperature for 15 minutes. Tubes were applied to a magnetic particle separator (F. Hoffmann-La Roche Ltd.) and the supernatant was removed. Fresh binding buffer TEN100 was added, and incubation continued at room temperature for 15 minutes, followed by removal of the supernatant. The particles were washed twice with TEN100 (10 mM Tris-HCl, 1 mM EDTA, 1000 mM NaCl, pH 7.5). HpaII digested-ligated DNA was placed in the tube and adjusted to 100 µl with 6x SSC and 0.1% SDS. The mixture of ligated genomic DNA and biotinylated Cot1 DNA, attached to streptavidin beads, was denatured by boiling for 10 minutes and the mixture was hybridized at 65°C overnight in a rotating hybridization oven. After hybridization, 100 µl (1 mg) of streptavidin magnetic particles was prepared according to the manufacturer's instructions, added to the hybridization mixture with an extra 200 µl of TEN100, and incubated at room temperature for 30 minutes. Tubes were applied to a magnetic particle separator and the supernatant was aspirated. This supernatant was incubated with another 1 mg of streptavidin magnetic particles again at room temperature for 30 minutes. After the incubation, the supernatant was aspirated and purified using a MinElute PCR purification kit (Qiagen, Inc.).
Target DNA was amplified with 20 ng of DNA per 50 µl of reaction with 0.4 µM of each oligonucleotide, Meth_1p (5′-GTATCGGAGGAGTCTTTGTTACGG-3′) and Meth_4p (5′-CTTGAAGAGTAACTTGTTGGCGG-3′), with Ex Taq polymerase (Takara Bio, Inc., Madison, WI) at 60 seconds at 95°C; 20 cycles of 15 seconds at 95°C, 15 seconds at 63°C, and 30 seconds at 72°C; 7 min at 72°C; and then at 4°C. PCR products were purified using a MinElute PCR purification kit (Qiagen, Inc.).
Hybridization of Amplified HpaII Fragments to the Array and Data Analysis
PCR products were purified and 1 µg of DNA was labeled with Cy3 or Cy5 using Ready-To-Go DNA Labeling Beads (Amersham Biosciences). PCR products from 267B1 DNA were used as a reference and all other cell line PCR products were cohybridized with it. Labeling and hybridization were all duplicated with dye swapping, and experiments were repeated at least once from the very beginning. In addition, data were collected on three arrays per slide. The Cy3/Cy5-labeled targets were hybridized overnight to human promoter array slide at 42°C in the presence of 5x SSC, 0.1% SDS, 25% formamide, and Cot1 blocking solution. Slides were washed following the Corning protocol (Corning, Inc.) and scanned with a Perkin Elmer Scanarray Express Microarray Scanner (Perkin Elmer, Inc., Wellesley, MA). Microarray data were retrieved with a Quantarray Microarray Analysis Software (Perkin Elmer, Inc.). Methylation microarray data have been deposited in the Geo database (http://www.ncbi.nlm.nih.gov/geo/; GEO accession no. GSE951).
Statistical microarray data analysis was performed using R-language and the Limma package from www.bioconductor.org [24]. Microarray data were normalized with print-tip loess or composite within-array normalization, followed by between-array normalization, before statistical analysis [25]. A moderated t-test was used for statistical analysis [26]. Genesis software was used to cluster genes as well as to visualize and represent data [27].
Print-tip loess normalization was applied based on the assumption that the spot intensities generated from two samples were similarly distributed. However, in the demethylation experiments using DAC, all the changes should be in one direction due to demethylation. Composite normalization used 88 promoters that showed significantly increased hybridization signals in LNCaP when compared with at least one of the three normal prostate cell lines: These already “unmethylated” promoters should not change during demethylation treatment.
The microarray hybridization data were sorted based on their chromosome location and analyzed as CGHs. The CGH microarray data were subjected to statistical analysis as described by Clark et al. [28] with some modification. We first constructed a quadratic loess curve, which can be viewed as a locally weighted polynomial regression curve through each data set. We then identified those regions in which contiguous segments of the loess curve were consistently greater than (or less than) 1.8 SD away from the mean of the all the data points. Having located these regions of interest, we used the Mann-Whitney U test to determine whether each selected region differed significantly (P < .001) from the set of data points from regions that had not been selected for examination by this test. Alternative analysis methods are also available (e.g., Ref. [29]).
MSP
The methylation status of 14 randomly picked genes, which were observed as differential methylated between PC3M and 267B1, was also determined by MSP. In brief, 2 µg of genomic DNA was treated with sodium bisulfite for 16 hours using an EZ DNA Methylation Kit (Zymo Research, Orange, CA). After purification, 1 µl of the aliquot was used as a template for each PCR reaction. The MSP primers were designed with the MethPrimer program [30] and primer information is presented in Table 1. Semiquantitative PCR was performed in an ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). Each 15 µl of reaction contained 100 ng of DNA, 1x HotStartTaq PCR Buffer (with 1.5 mM MgCl2; Qiagen, Inc.), 1:25,000 dilution of SybrGreen I (Molecular Probes, Eugene, OR), 0.35 µM 6-ROX (Molecular Probes), 0.2 mM dNTPs, 4 mM MgCl2, 0.025 U/µl HotStartTaq DNA polymerase (Qiagen, Inc.), and 0.8 µM of each primer. Semiquantitative PCR was performed at 95°C for 15 minutes, 50 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 30 seconds; followed by a dissociation stage (95°C for 15 seconds, 60°C for 15 seconds, and 95°C for 15 seconds).
Table 1.
Primers for Methylation-Specific Semiquantitiative PCR.
| RefSeq ID | Gene Symbol | Expression Ratio (Log2) | Primer Information* |
| Putative hypermethylated promoters in PC3M relative to 267B1 | |||
| NM_000082 | CKN1 | -0.75 | M-FW: GTTAATTTTCGAGAAAGGAATTAGC |
| RW: AAAATATCTTCAACGCCTCGAC | |||
| U-FW: ATGTTAATTTTTGAGAAAGGAATTAGTG | |||
| RW: AAAAAAAATATCTTCAACACCTCAAC | |||
| NM_001008 | RPS4Y† | -6.36 | M-FW: GTTATTTAGGTTGGAGTGTAGTGGC |
| RW: GAATCACGAAATCAAAAAATCG | |||
| U-FW: GTTATTTAGGTTGGAGTGTAGTGGTG | |||
| RW: CAAATCACAAAATCAAAAAATCAAA | |||
| NM_003118 | SPARC†‡ | -7.16 | M-FW: GATATTTTCGTTTACGTCGTTAGTTC |
| RW: AAAAAATAAAAAAATACTCCCCCG | |||
| U-FW: GATATTTTTGTTTATGTTGTTAGTTTGT | |||
| RW: AAAAATAAAAAAATACTCCCCCAAA | |||
| NM_003206 | TCF21 | NA | M-FW: AATATGTTTATCGGTTTTTTTAGCG |
| RW: TTAAAACTCTCCTCGATACTCTCGT | |||
| U-FW: TTTAAATATGTTTATTGGTTTTTTTAGTGA | |||
| RW: CAATTAAAACTCTCCTCAATACTCTCATT | |||
| NM_003999 | OSMR | -2.52 | M-FW: ATTTTGGTTAATACGGTGAAATTTC |
| RW: CCAAACTAAAATACAATAACGCGAT | |||
| U-FW: TTTTGGTTAATATGGTGAAATTTTGT | |||
| RW: TCACCCAAACTAAAATACAATAACACA | |||
| NM_004701 | CCNB2 | -1.78 | M-FW: GTTAAAATTTAGAGGCGTTTTACGT |
| RW: ACGTTTAATTATCACAACAACCGAT | |||
| U-FW: TTTTGTTAAAATTTAGAGGTGTTTTATGT | |||
| RW: CACATTTAATTATCACAACAACCAAT | |||
| NM_005509 | DMXL1 | -1.37 | M-FW: ATTTCGTTTAGGGATTTGGAAATAC |
| RW: AAACTACAAATCCCAATATACACCG | |||
| U-FW: TTTTGTTTAGGGATTTGGAAATATG | |||
| RW: AAACTACAAATCCCAATATACACCACT | |||
| NM_005732 | RAD50 | -2.69 | M-FW: ATTTTTTTGATTTTGAGATTCGC |
| RW: GATCCGAAACATATTTACAAACGTT | |||
| U-FW: ATTTTTTTGATTTTGAGATTTGTGG | |||
| RW: TCAATCCAAAACATATTTACAAACATT | |||
| NM_005983 | SKP2 | -1.72 | M-FW: TATTTCGTGGGTCGATTAGTTTC |
| RW: ACTAAAAATTATAATTTCCGTCCCG | |||
| U-FW: TATTTTGTGGGTTGATTAGTTTTGT | |||
| RW: ACTAAAAATTATAATTTCCATCCCACT | |||
| NM_006479 | PIR51 | -1.97 | M-FW: GTATAAATTCGGTTTTGGTGGATC |
| RW: CAAATTCTTATTAACTTCAACGACGA | |||
| U-FW: GTATAAATTTGGTTTTGGTGGATTG | |||
| RW: TTCTCAAATTCTTATTAACTTCAACAACA | |||
| NM_014350 | GG2-1 | -1.94 | M-FW: GTTTGGAGTATTAGTGTTCGTTCG |
| RW: CGAAACCTTTTAAAAAAAATAAAACG | |||
| U-FW: GTTTGGAGTATTAGTGTTTGTTTGG | |||
| RW: CAAAACCTTTTAAAAAAAATAAAACAAC | |||
| NM_021025 | TLX3 | NA | M-FW: GTTGTGGTTCGGGTTTTAATATTC |
| RW: CTACCGCAACCATTAACTACGAT | |||
| U-FW: GTTGTGGTTTGGGTTTTAATATTTG | |||
| RW: TCCTACCACAACCATTAACTACAAT | |||
| NM_024501 | HOXD1 | -1.61 | M-FW: TTTTAGTGAAAGTAAGCGTCGTATC |
| RW: CTATCCCTCGCAATTTATAACGA | |||
| U-FW: TTTTTAGTGAAAGTAAGTGTTGTATTGG | |||
| RW: TCTTCTATCCCTCACAATTTATAACAAC | |||
| Putative hypomethylated promoters in PC3M relative to 267B1 | |||
| NM_006142 | SFN‡ | 5.60 | M-FW: TAAGTTGGTAGAGTAGGTCGAACGT |
| RW: CTAAAAACAAATTTCGCTCTTCG | |||
| U-FW: GGTTAAGTTGGTAGAGTAGGTTGAATG | |||
| RW: CTACTAAAAACAAATTTCACTCTTCACA | |||
M: primer designed to amplify methylated DNA; U: primer designed to amplify unmethylated DNA.
Gene that does not have a CpG island within the amplified promoter region.
Gene already known as a methylation target in cancer.
Gene Expression Profiling
Affymetrix U133A chips (Affymetrix, Inc., Santa Clara, CA) were used to profile gene expression levels for the 267B1 and PC3M cell lines. Ten micrograms of total RNA of 267B1 and PC3M was prepared using a Qiagen RNeasy Mini kit. Labeling and scanning procedure followed Affymetrix's standard procedure (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). A gene was considered to be differentially expressed between two different samples when expression was “called present” in at least one sample under default parameters and there was a twofold or greater change in net fluorescence between samples.
Results and Discussion
Immortalized Normal Prostate Epithelial Cells and Cancer Cell Lines
Total genomic DNA from eight prostate epithelial cell lines were compared using the protocol in Figure 1: DNA was digested with HpaII, then primers were ligated and PCR-amplified. The PCR products were hybridized to an array of promoter sequences. Mlcsv40 and 267B1 are neonatal prostate lineages immortalized with the SV40 early region genes (unpublished and Ref. [31]), RWPE-1 is an adult human prostatic epithelial cell line immortalized with human papillomavirus 18 [32]. PC3M is a liver metastatic derivative of one of the most heavily studied prostate cancer cell lines PC3 [33]. PC3M-LN4 and PC3M-Pro4 are metastatic and less metastatic derivatives, respectively, of PC3M [34]. LNCaP is a human prostate cancer cell line established from a lymph node metastasis [35]. All these lines were expected to resemble each other in overall methylation profile because all cell lines are derived from prostate epithelium. However, we expected to uncover some DNA methylation differences, as the cell lines are clearly different in phenotype, with some being derived from either neonate or adult normal epithelium and some from adult epithelial tumors. Furthermore, a recent study has indicated that prostate cancer cell lines may possess the same “hypermethylation fingerprint” as primary and metastatic prostate cancers [36].
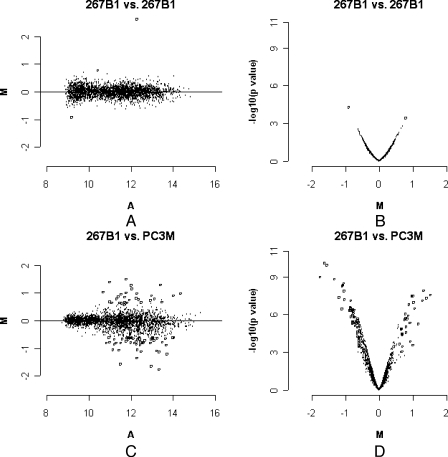
To ensure the reproducibility of the protocol, identical samples of genomic DNA from the same cell line were processed separately on separate days. A comparison for 267B1 is presented as an example in Figure 2, A and B. Figure 2A is an M-A plot, which were constructed with M = log2R - log2G and A = (log2R + log2G) / 2, where R is the intensity of the scanner output signal for the experimental sample fluorophore (channel 2, Ch2) and G is the scanner output signal for the reference sample fluorophore (channel 1, Ch1) on the background-subtracted, normalized, and scaled channel intensities. Statistically significant, differentially hybridized DNA was identified by a t-test. The distribution of the corresponding P values as a function of M was summarized in a volcano plot (M-p plot; Figure 2B). Essentially 99.9% (2729/2732) of the ratios between the two channels was less than 1.5-fold and had P values less than .001. This experiment was used to set a threshold for other experiments in which a ratio of at least 1.5-fold and a P value less than .001 were used. This conservative threshold ensures that the hybridization differences between cell lines that are discussed here are likely to be real, although these stringent criteria also mean that many real differences in methylation may not be noted, as they are below this threshold.
Figure 2.
Estimation of data reproducibility and significance of differences. M: log base 2 ratio of each spot after print-tip loess normalization and scale betweenarray normalization. (A) Average of two channels' log intensities of each spot; a measurement of the overall brightness of the spot. All data involved at least six arrays. The P value is for the moderated t-test. The most significant changes are represented by open circles. (A) M-A plot, hybridization of amplified HpaII fragments from 267B1 vs 267B1. (B) M-p plot, hybridization of amplified HpaII fragments from 267B1 vs 267B1. (C) M-A plot, hybridization of amplified HpaII fragments from PC3M vs 267B1. (D) M-p plot, hybridization of amplified HpaII fragments from PC3M vs 267B1.
Differential Array Hybridization between Cell Lines
Figure 2, C and D shows M-A and M-p plots of 267B1 and PC3M as an example of a comparison between cell lines. Fifty-six genes, including 50 genes that have CpG islands within the promoter region, are hybridized more in 267B1 than in PC3M, consistent with more methylation or lower copy number in PC3M. Conversely, 30 genes, including 14 genes that have CpG islands within the promoter region, are significantly hybridized to a greater extent in PC3M (P < .001, ratio > 1.5-fold). It is noteworthy that the method is capable of detecting hybridization changes in any gene that has the appropriate restriction sites, regardless of whether it has a CpG island. Furthermore, it is clear that many genes that do not have CpG islands within the proximal part of the promoter nevertheless showed changes in hybridization, possibly due to methylation changes, but also possibly due to copy number changes or a point mutation in a restriction site. A pairwise comparison of differential hybridization is present for all the cell lines in Table W1 and at http://bioinformatics.skcc.org/mcclelland/methylation.
Association of Some HpaII Fragment Hybridization Differences with Methylation Differences
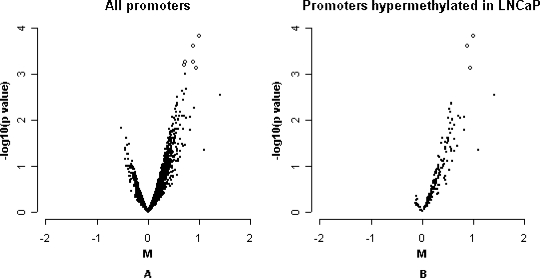
A difference in HpaII fragment hybridization intensity for a promoter between genome samples can occur due to methylation differences, differences in copy number, or restriction site polymorphisms. Global demethylation using DAC [37] was used to determine which differences in hybridization could be partially reversed, indicating methylation as the underlying cause. LNCaP cells were treated with 5-aza-cytidine for 72 hours during which they divided one to two times. Untreated and treated cells were compared with regards to their HpaII fragment profile for all the promoters on the array (Figure 3A). The distribution was consistent with differences in HpaII fragments, detecting a small reduction in DNA methylation for hundreds of genes. Figure 3B plots 191 promoters that had appeared to be most significantly “methylated” in LNCaP relative to at least one of three normal prostate cell lines. Almost all of these genes showed a shift to the right, consistent with demethylation, thus the probability that HpaII fragment amplification differences had correctly measured that these genes are methylated in LNCaP. As a group, the shift of these genes to demethylation was highly significant (P < .001, Mann-Whitney U test).
Figure 3.
Effects of methylation inhibitor (DAC) on methylation status of LNCaP. M-p plots, HpaII fragment hybridization pattern of LNCaP before and after treated with DAC. M: log base 2 ratio of each spot after composite normalization and scale between-array normalization. The P value is from a moderated t-test. The most significant changes are represented by open circles. (A) M-p plot for all promoters. (B) M-p plot for 191 promoters putatively hypermethylated in LNCaP relative to at least one of the three normal prostate cell lines.
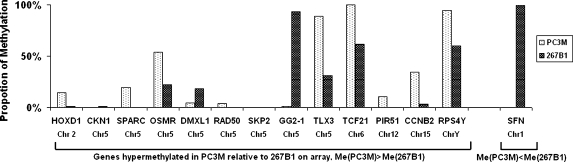
To determine if HpaII fragment hybridization changes for individual genes were due to a methylation change, MSP was performed on 14 genes picked at random, which were putatively differentially methylated between PC3M and 267B1 (Figure 4). Semiquantitative PCR was performed and the proportion of methylation was estimated for each gene and each sample. Eight of 14 were hypermethylated in PC3M relative to 267B1, and one gene was hypermethylated in 267B1, confirming the array data. Of the remaining five genes that showed no changes or changes in the wrong direction, all were located on chromosome 5, which we will show later to be due to a copy number change.
Figure 4.
Detection of DNA methylation changes using methylation-specific semiquantitative PCR. Fourteen promoters that displayed possible differential methylation in the array assay between 267B1 and PC3M (Table 1) were investigated by methylation-specific semiquantitative PCR. The proportion of methylation for each promoter is calculated.
The ratio of HpaII fragment hybridization between two different samples in microarrays is sometimes less than that observed using specific PCR. The reason for this is probably that many spots on the array report an intrinsically relatively high level of hybridization from the complex probe, despite the use of Cot1 subtraction. For example, most of the control human spots that do not have restriction sites still report a level of hybridization greater than nonhuman controls. Thus, the reported difference in hybridization on the microarray is generally a minimal estimate. Encouragingly, however, 18.4% (504/2732) of promoter sequences on the array that had two HpaII sites of the correct size appeared to be differentially hybridized, compared to only 8.0% (28/351) of human DNA that did not have HpaII sites (χ2 = 23.2, P < .001, chi-square analysis). The 28 exceptions have similar sequences elsewhere in the genome, which may cause cross-hybridization.
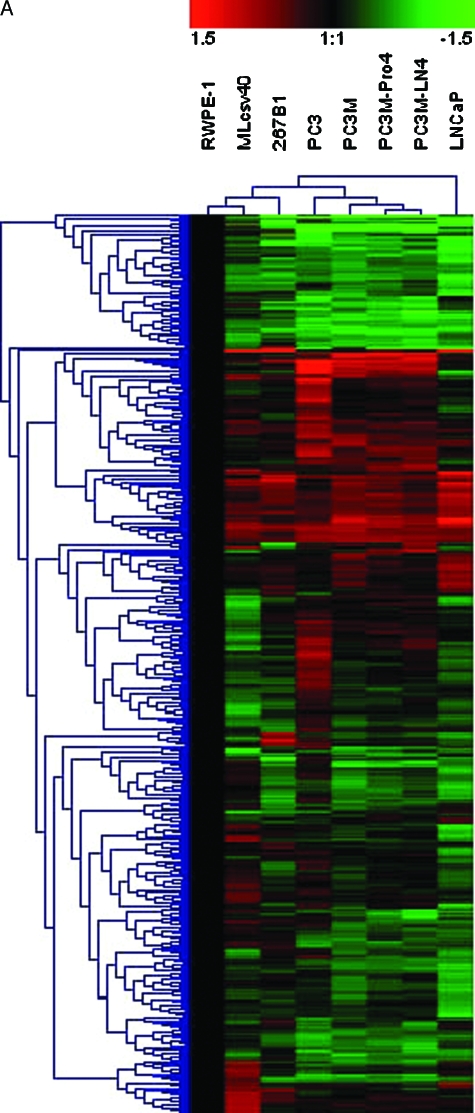
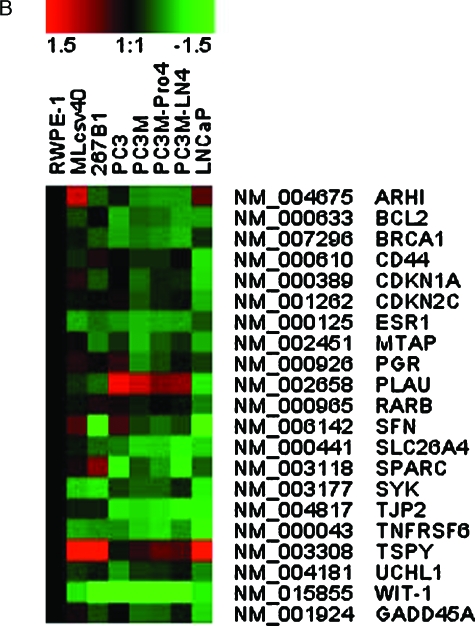
There are 504 promoters that showed statistically significant changes in hybridization among cancer and normal prostate cell lines. Hierarchical clustering of the hybridization patterns of these 504 promoters is displayed in Figure 5A and the corresponding genes are listed in Table W1 in the same order as they appear in the figure. The data are presented as a hybridization difference relative to RWPE-1, a putatively normal transformed prostate epithelia cell from an adult. The clustering of cell lines showed that PC3M-Pro4 and PC3M-LN4 are the most similar. Only one promoter, HAS3, appeared more differentially hybridized between PC3M-Pro4 and PC3M-LN4, possibly being hypermethylated in PC3M-Pro4. PC3M-Pro4 and PC3M-LN4 were clustered with PC3M, then with PC3. These four cell lines were less similar to LNCaP and normal cell lines. This is consistent with the origins of these cell lines. Among these 504 promoters, eight genes are differentially hybridized in prostate cancer cell lines relative to normal lines and are also known as methylation-regulated genes in prostate cancer (CD44, CDKN1A, ESR1, PLAU, RARB, SFN, TNFRSF6, and TSPY) and 13 are known in other cancers (ARHI, bcl-2, BRCA1, CDKN2C, GADD45A, MTAP, PGR, SLC26A4, SPARC, SYK, TJP2, UCHL1, and WIT-1; Table 2 and Figure 5B). The references for these observations are provided in a supplementary version of Table 2. Methylation of SFN and PLAU in LNCaP, but not PC3, has been reported before [38,39], which is consistent with our data.
Figure 5.
Hierarchical cluster of hybridized, amplified HpaII fragments for eight cell lines. (A) A total of 504 promoters, which are statistical differentially hybridized between at least one of the five prostate cancer cell lines (LNCaP, PC3, PC3M, PC3M-Pro4, and PC3M-LN4) and at least one of the three relative normal prostate cell lines (RWPE-1, 267B1, and Mlcsv40), is shown. The normalized hybridization ratios of these cell lines relative to RWPE-1 were used for hierarchical clustering. Red indicates higher HpaII fragment hybridization relative to RWPE-1, which usually indicates less methylation or a higher copy number. Green indicates lower hybridization, which usually indicates greater methylation or a lower copy number. (B) Clustering for 21 genes known to be regulated in cancer.
Table 2.
Differential Hybridization of Amplified HpaII Fragments Near Genes Known to be Methylation Targets in Cancer.*
| RefSeq ID | Gene Symbol | Tumor Type |
| NM_004675 | ARHI | Breast cancer |
| NM_000633 | bcl-2 | Colorectal carcinoma |
| Nm_007296 | BRCA1 | Breast cancer |
| Ovarian cancer | ||
| Cervical cancer | ||
| NM_000610 | CD44 | Prostate cancer |
| Colorectal cancer | ||
| Neuroblastoma | ||
| Gastric cancer | ||
| NM_000389 | CDKN1A | Prostate cancer |
| Lymphoma | ||
| Leukemia | ||
| NM_001262 | CDKN2C | Hodgkin's lymphomas |
| NM_000125 | ESR1 | Prostate cancer |
| Colorectal cancer | ||
| Breast cancer | ||
| Lung cancer | ||
| NM_001924 | GADD45A | Breast cancer |
| NM_002451 | MTAP | Malignant melanoma |
| NM_000926 | PGR | Breast cancer |
| Cervical cancer | ||
| NM_002658 | PLAU | Prostate cancer |
| Breast cancer | ||
| NM_000965 | RARB | Prostate cancer |
| Testicular lymphoma | ||
| Cervical cancer | ||
| Breast cancer | ||
| Colorectal cancers | ||
| NM_006142 | SFN | Prostate cancer |
| Ovarian cancer | ||
| Skin cancer | ||
| Lung cancer | ||
| Oral cancer | ||
| Vulval cancer | ||
| Gastric cancer | ||
| Breast cancer | ||
| NM_000441 | SLC26A4 | Thyroid tumorigenesis |
| NM_003118 | SPARC | Pancreatic cancer |
| NM_003177 | SYK | Breast cancer |
| Gastric cancer | ||
| Ovarian cancer | ||
| T-lineage acute lymphoblastic leukemia | ||
| NM_004817 | TJP2 | Pancreatic cancer |
| NM_000043 | TNFRSF6 | Prostate cancer |
| Bladder cancer | ||
| NM_003308 | TSPY | Prostate cancer |
| NM_004181 | UCHL1 | Pancreatic cancer |
| NM_015855 | WIT-1 | Acute myeloid leukemia |
References can be found in (Supplementary Table W2).
That an experiment involving three relatively normal prostate lines and five prostate cancer cell lines pointed to a large number of genes that were previously known to be differentially methylated in cancer, particularly prostate cancer, may indicate that methylation imprinting has been maintained in culture. This provides further support for the recent observation that cell lines and primary tumors generally have similar overall distributions and frequencies of gene methylation [40] and that prostate cancer cell lines may have the same “hypermethylation fingerprint” as primary and metastatic prostate cancers [36].
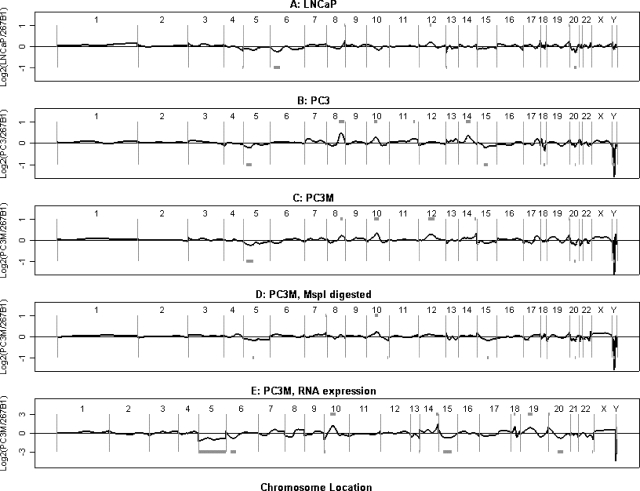
To date, almost all works on sequence-specific methylation have focused on the status of methylation in CpG islands. Interestingly, 17.9% (90/504) of putative differential methylation-regulated promoters in these prostate cell lines were in the parts of promoters that do not have an obvious CpG island. Table W1 includes a column indicating which promoters on the array had CpG islands. A few of the changes in promoter regions without CpG islands were confirmed (Figures 4 and 6). However, it remains possible that copy number changes or restriction site polymorphisms may explain some of those hybridization changes for other genes.
Figure 6.
DNA copy number changes measured by CGH on promoter array. (A–C) Normalized HpaII fragment hybridization ratios for three different prostate cancer cell lines compared to 267B1 plotted against the relative chromosomal position of each promoter. Candidate copy number changes are represented by grey blocks. (A) LNCaP. (B) PC3. (C) PC3M. (D) PC3M (MspI ligation PCR), compared to 267B1 (MspI ligation PCR), against chromosomal position. (E) PC3M (RNA expression level), compared to 267B1 RNA expression level, against chromosomal position.
The gene TGFBR2 is differentially hybridized in LNCaP relative to the other cell lines. Methylation of the TGFBR2 promoter was confirmed as present in LNCaP, and absent in PC3, PC3M, PC3M-Pro4, PC3M-LN4, 267B1, RWPE-1, and Mlcsv-40 by SQ-PCR across a promoter HpaII site (data not shown). Inactivation of this gene by mutation is common in many cancers, and could play a role in prostate cancer [41]. Given that many genes inactivated by mutation are also targets of methylation as an alternative method of silencing [42,43], it was possible that TGFBR2 may be methylated in some cancers or cancer cell lines. However, differential methylation had not previously been observed for this gene.
In cancer cell lines, relative to normal cell lines, there were fewer genes that showed an increased HpaII fragment hybridization, characteristic of copy number increases or hypomethylation (251 promoters), versus a lower HpaII fragment hybridization, characteristic of copy number decreases or hypermethylation (286 promoters). An example of increased HpaII fragment hybridization (hypomethylation or copy number increase) in cancer lines is the promoter of CTAG1, which is overexpressed in some lung and thyroid cancers [44,45], although this overexpression has not been attributed to hypomethylation or copy number changes.
Other than their dramatic differences in growth properties and metastatic ability [46,47], one of the most striking differences between PC3M and LNCaP is that the latter is almost unique among prostate cancer cell lines in still being androgen-dependent. In this experiment, 29 genes showed loss of hybridization in HpaII fragment in LNCaP and 19 genes showed loss of hybridization in PC3M, consistent with hypermethylation or copy number loss. We looked for differences in hybridization between PC3M and LNCaP among 261 known and suspected androgen-regulated genes present on the array [48]. Among known or suspected androgen receptor-regulated genes that may be methylated or reduced in copy number in LNCaP relative to PC3M were GG2-1 (TNF-induced protein) and GABARAPL2 (GABA(A) receptor-associated protein-like 2). In PC3M, the list included FLJ13782 hypothetical protein, TSPY (testis-specific protein, Y-linked), and RPS4Y (ribosomal protein S4, Y-linked Y isoform).
Expression from the Y chromosome has been of interest in prostate cancer [49,50] and changes in methylation of EIF1AY, MGC26641, PRKY, RPS4Y, SHOX, TSPY, TSPYQ1, and VCY are observed in our experiments, whereas the few other Y chromosome genes on the array act as internal controls for this observation because they are seemingly not differentially methylated.
Correlation between RNA Expression and HpaII Fragment Hybridization
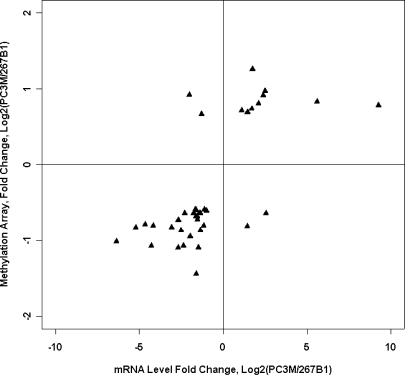
The RNA expression levels of two cell lines, PC3M and 267B1, were obtained using Affymetrix U133A GeneChips. A total of 51.6–53.5% of genes were called as present for these samples. The Affymetrix U133A annotation table was cross-referenced with our human promoter array by RefSeq ID. When methylation differences are plotted against gene expression differences between PC3M and 267B1 for all the genes that showed HpaII fragment hybridization differences and gene expression differences, there is a significant correlation (40 genes, r = 0.68, P < .001; Figure 7 and Table 3). The majority of genes that are differentially hybridized by amplified HpaII fragments in the study are not considered in this section because these genes happen not to be sufficiently expressed, or their gene expression did not change significantly. There are 27 genes, including three genes with no apparent CpG island in the promoter region, that are less hybridized by HpaII fragments (consistent with hypermethylation or copy number loss) in PC3M relative to 267B1 and where gene expression was also downregulated in PC3M, as would be expected if methylation or copy number loss is associated with downregulation of expression. Nine genes, including two genes with no CpG island in the promoter region, were increased by HpaII fragments in hybridization (consistent with hypomethylation or copy number increase) in PC3M relative to 267B1, and the gene expression of these genes is higher in PC3M, also as expected. There were only four genes where the prediction of methylation or copy number loss was associated with an increase in gene expression level. It will be of interest to explore these exceptions further.
Figure 7.
Comparison of amplified HpaII fragment data to Affymetrix RNA expression data. Decreases in signal from HpaII fragments (which is usually due to an increase in DNA methylation) between cell lines are generally associated with a decrease in RNA expression. The upper right quadrant contains genes with higher RNA expressions in PC3M than in 267B1 and higher yields of HpaII fragments, in PC3M. The lower left quadrant contains genes with lower RNA expressions in PC3M than in 267B1 and also lower yields of HpaII fragment hybridization in PC3M.
Table 3.
Promoters That Show Large Differences in Both HpaII Fragment Amplification and RNA Expression, between PC3M and 267B1.
| RefSeq ID | Gene Symbol | Gene Full Name | Expression Ratio* |
| Genes with reduced HpaII fragment hybridization in PC3M (candidate hypermethylated genes) | |||
| NM_000138 | FBN1 | Fibrillin 1 | -5.21 |
| NM_000546 | TP53 | Tumor protein p53 | -4.66 |
| NM_000985 | RPL17 | Ribosomal protein L17 | -1.17 |
| NM_001008 | RPS4Y† | Ribosomal protein S4, Y-linked Y isoform | -6.36 |
| NM_001790 | CDC25C | Cell division cycle 25C protein, isoform a | -2.29 |
| NM_002082 | GPRK6 | G protein-coupled receptor kinase 6 | -1.76 |
| NM_002749 | MAPK7 | Mitogen-activated protein kinase 7 isoform 1 | -1.62 |
| NM_003118 | SPARC† | Secreted protein, acidic, cysteine-rich (osteonectin) | -4.28 |
| NM_003714 | STC2 | Stanniocalcin 2 | -1.42 |
| NM_003999 | OSMR | Oncostatin M receptor | -2.52 |
| NM_004472 | FOXD1† | Forkhead box D1 | -2.68 |
| NM_004663 | RAB11A | RAB11A, member RAS oncogene family | -1.13 |
| NM_004701 | CCNB2 | Cyclin B2 | -1.78 |
| NM_005509 | DMXL1 | Dmx-like 1 | -1.37 |
| NM_005732 | RAD50 | RAD50 homolog isoform 1 | -2.69 |
| NM_005983 | SKP2 | S-phase kinase-associated protein 2 isoform 1 | -1.47 |
| NM_006282 | STK4 | Serine/threonine kinase 4 | -1.36 |
| NM_006479 | PIR51 | RAD51-interacting protein | -1.97 |
| NM_012382 | OSRF | Osmosis-responsive factor | -2.36 |
| NM_014621 | HOXD4 | Homeo box D4 | -3.07 |
| NM_018163 | FLJ10634 | Hypothetical protein FLJ10634 | -1.66 |
| NM_018268 | FLJ10904 | Hypothetical protein FLJ10904 | -4.17 |
| NM_024501 | HOXD1 | Homeo box D1 | -1.61 |
| NM_024558 | C14orf138 | Hypothetical protein FLJ13920 | -1.56 |
| NM_024796 | FLJ22639 | Hypothetical protein FLJ22639 | -1.00 |
| NM_033028 | BBS4 | Bardet-Biedl syndrome 4 | -1.54 |
| NM_133338 | RAD17 | RAD17 homolog isoform 1 | -1.63 |
| NM_006194 | PAX9‡ | Paired box gene 9 | 2.55 |
| NM_053001 | OSR2‡ | Odd-skipped-related 2A protein | 1.43 |
| Genes with increased HpaII fragment hybridization in PC3M (candidate hypomethylated genes) | |||
| NM_001123 | ADK | Adenosine kinase isoform a | 1.69 |
| NM_002290 | LAMA4 | Laminin, alpha 4 precursor | 2.11 |
| NM_002467 | MYC | v-myc myelocytomatosis viral oncogene homolog | 2.51 |
| NM_002658 | PLAU | Plasminogen activator, urokinase | 1.75 |
| NM_004693 | K6HF† | Cytokeratin type II | 9.27 |
| NM_005555 | KRT6B† | Keratin 6B | 1.10 |
| NM_006142 | SFN | Stratifin | 5.60 |
| NM_030759 | NRBF-2 | Nuclear receptor binding factor-2 | 1.45 |
| NM_032804 | FLJ14547 | Hypothetical protein FLJ14547 | 2.39 |
| NM_018649 | H2AFY2‡ | Core histone macroH2A2.2 | -2.02 |
| NM_020177 | FEM1C‡ | Feminization 1 homolog a | -1.30 |
Ratios: log2(PC3M/267B1).
Promoter does not have a CpG island within the amplified promoter region.
Promoter where HpaII fragment hybridization was not correlated with RNA expression level.
CGH
The protocol (Figure 1) reports polymorphisms, changes in DNA copy number, or DNA methylation status. This feature is also true of other array-based protocols that measure methylation at multiple locations, simultaneously [15–19], and at 2D gels (RLGS) [51]. Figure 6, A–C presents an analysis of the HpaII ligation PCR data for three cell lines, plotted in genome order, and essentially uses the data in the same way as in the original representational analysis method [9]. The best candidate chromosome regions for widespread methylation or aneuploidy are highlighted. Note the dramatic apparent aneuploidy of chromosome 5 in PC3M, which is responsible for the identification, earlier in this manuscript, of genes that appeared to be differentially methylated, but which are, in fact, altered in copy number. Among the aneuploidies seen here that have been previously reported are changes in chromosome 6 in LNCaP, in chromosomes 8, 10, and 14 in PC3 [28], and many sporadic changes previously observed in prostate cancer [52]. The same analysis was performed for each pair of cell lines; Table W1, which contains 504 genes that are differentially hybridized among the cell lines, was annotated to indicate cases where the difference in hybridization could be due to ploidy differences.
We also measured copy number changes in PC3M relative to 267B1 using MspI ligation PCR. MspI is an enzyme that cuts at the same CCGG site as HpaII but which is insensitive to methylation at most sites. The normalized ratio (PC3M/267B1) was plotted against the chromosomal position of each promoter (Figure 6D). This is a simple variation on the CGH method [53]. Finally, the ratios of PC3M expression data relative to 267B1 were plotted against chromosome position in Figure 6E. Perhaps surprisingly, there are detectable global effects of aneuploidy on averaged RNA expression along the chromosomes.
Summary
Overall, we identified many candidate genes that were rarely, if ever, previously associated with gene regulation by methylation changes in cancer. These genes include PAK6 (p21-activated protein kinase 6), RAD50 (RAD50 homolog isoform 1), TLX3 (T-cell leukemia, homeo box 3), PIR51 (RAD51-interacting protein), MAP2K5 (mitogen-activated protein kinase kinase 5 isoform B), INSR (insulin receptor), FBN1 (fibrillin 1), and GG2-1 (TNF-induced protein). Many genes have exciting functions that could be plausibly associated with cancer progression, but we will not speculate further on those roles here. These genes will be worthy of assessment in tumors by methods that assay individual genes, such as bisulfite sequencing.
Relying on cleavage by enzymes that detect methylation [15–19,51] has limitations, including the need to parse out copy number and SNPs at a subsequent step. In addition, tumor samples are usually mixtures of various cell types, in addition to tumor cells. However, the protocol can use very little DNA. It is a future goal to demonstrate that enriched tumor samples can be assayed by this class of methods.
Acknowledgements
We thank Fred Long and Shalu Mittal for expert bioinformatics assistance; Kemal Korkmaz and Jun Hayakawa for helpful discussions; and Sidney Kimmel and Ira Lechner, without whose consistent support this work would not have been possible.
Footnotes
This work was supported, in part, by grant DAMD17-03-1-0022 and NIH grants R01CA68822, R01CA84107, and U01CA84998. Y.W. received the AACR-Gary J. Miller Scholar-in-Training Award for this work.
This article refers to supplementary material, which is designated by “W” (ie, Table W1, Figure W1) and is available online at www.bcdecker.com.
References
- 1.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 4.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002;33:632–632. doi: 10.2144/02333rv01. (pp. 634, 636–649) [DOI] [PubMed] [Google Scholar]
- 5.Liu ZJ, Maekawa M. Polymerase chain reaction-based methods of DNA methylation analysis. Anal Biochem. 2003;317:259–265. doi: 10.1016/s0003-2697(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 6.Rush LJ, Plass C. Restriction landmark genomic scanning for DNA methylation in cancer: past, present, and future applications. Anal Biochem. 2002;307:191–201. doi: 10.1016/s0003-2697(02)00033-7. [DOI] [PubMed] [Google Scholar]
- 7.Smiraglia DJ, Plass C. The study of aberrant methylation in cancer via restriction landmark genomic scanning. Oncogene. 2002;21:5414–5426. doi: 10.1038/sj.onc.1205608. [DOI] [PubMed] [Google Scholar]
- 8.Hirotsune S, Hatada I, Komatsubara H, Nagai H, Kuma K, Kobayakawa K, Kawara T, Nakagawara A, Fujii K, Mukai T, et al. New approach for detection of amplification in cancer DNA using restriction landmark genomic scanning. Cancer Res. 1992;52:3642–3647. [PubMed] [Google Scholar]
- 9.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trenkle T, Mathieu-Daude F, Welsh J, McClelland M. Reduced complexity probes for DNA arrays. Methods Enzymol. 1999;303:380–392. doi: 10.1016/s0076-6879(99)03023-2. [DOI] [PubMed] [Google Scholar]
- 11.Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, Cigudosa JC, Huang TH, Esteller M. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 13.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 14.Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 15.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 16.Shi H, Wei SH, Leu YW, Rahmatpanah F, Liu JC, Yan PS, Nephew KP, Huang TH. Triple analysis of the cancer epigenome: an integrated microarray system for assessing gene expression, DNA methylation, and histone acetylation. Cancer Res. 2003;63:2164–2171. [PubMed] [Google Scholar]
- 17.Shi H, Maier S, Nimmrich I, Yan PS, Caldwell CW, Olek A, Huang TH. Oligonucleotide-based microarray for DNA methylation analysis: principles and applications. J Cell Biochem. 2003;88:138–143. doi: 10.1002/jcb.10313. [DOI] [PubMed] [Google Scholar]
- 18.Tompa R, McCallum CM, Delrow J, Henikoff JG, van Steensel B, Henikoff S. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr Biol. 2002;12:65–68. doi: 10.1016/s0960-9822(01)00622-4. [DOI] [PubMed] [Google Scholar]
- 19.Hatada I, Kato A, Morita S, Obata Y, Nagaoka K, Sakurada A, Sato M, Horii A, Tsujimoto A, Matsubara K. A microarray-based method for detecting methylated loci. J Hum Genet. 2002;47:448–451. doi: 10.1007/s100380200063. [DOI] [PubMed] [Google Scholar]
- 20.Lippman Z, Gendrel A, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 21.Hayakawa J, Mittal S, Wang Y, Korkmaz K, Adamson E, English C, Omichi M, McClelland M, Mercola D. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation by genotoxic stress. Mol Cell. 2004;16:521–535. doi: 10.1016/j.molcel.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Adamson E, De Belle I, Mittal S, Wang Y, Hayakawa J, Korkmaz K, O'Hagan D, McClelland M, Mercola D. Egr1 signaling in prostate cancer. Cancer Biol Ther. 2003;2:610–616. [PubMed] [Google Scholar]
- 23.Craig JM, Kraus J, Cremer T. Removal of repetitive sequences from FISH probes using PCR-assisted affinity chromatography. Hum Genet. 1997;100:472–476. doi: 10.1007/s004390050536. [DOI] [PubMed] [Google Scholar]
- 24.Ihaka I, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 25.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. (Article 3) [DOI] [PubMed] [Google Scholar]
- 27.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 28.Clark J, Edwards S, Feber A, Flohr P, John M, Giddings I, Crossland S, Stratton MR, Wooster R, Campbell C, et al. Genome-wide screening for complete genetic loss in prostate cancer by comparative hybridization onto cDNA microarrays. Oncogene. 2003;22:1247–1252. doi: 10.1038/sj.onc.1206247. [DOI] [PubMed] [Google Scholar]
- 29.Daruwala RS, Rudra A, Ostrer H, Lucito R, Wigler M, Mishra B. A versatile statistical analysis algorithm to detect genome copy number variation. Proc Natl Acad Sci USA. 2004;101:16292–16297. doi: 10.1073/pnas.0407247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 31.Chaib H, Rubin MA, Mucci NR, Li L, Taylor JMG, Day ML, Rhim JS, Macoska JA. Activated in prostate cancer: a PDZ domain-containing protein highly expressed in human primary prostate tumors. Cancer Res. 2001;61:2390–2394. [PubMed] [Google Scholar]
- 32.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 33.Stephenson RA, Dinney CP, Gohji K, Ordonez NG, Killion JJ, Fidler IJ. Metastatic model for human prostate cancer using orthotopic implantation in nude mice. J Natl Cancer Inst. 1992;84:951–957. doi: 10.1093/jnci/84.12.951. [DOI] [PubMed] [Google Scholar]
- 34.Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2:1627–1636. [PubMed] [Google Scholar]
- 35.Horoszewicz JS, Leong SS, Chu TM, Wajsman ZL, Friedman M, Papsidero L, Kim U, Chai LS, Kakati S, Arya SK, et al. The LNCaP cell line—a new model for studies on human prostatic carcinoma. Prog Clin Biol Res. 1980;37:115–132. [PubMed] [Google Scholar]
- 36.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Huper G, Guo Y, Murphy SK, Olson JA, Marks JR. Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene. 2005;24:2705–2714. doi: 10.1038/sj.onc.1208464. [DOI] [PubMed] [Google Scholar]
- 38.Urano T, Takahashi S, Suzuki T, Fujimura T, Fujita M, Kumagai J, Horie-Inoue K, Sasano H, Kitamura T, Ouchi Y, et al. 14-3-3 Sigma is down-regulated in human prostate cancer. Biochem Biophys Res Commun. 2004;319:795–800. doi: 10.1016/j.bbrc.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 39.Pakneshan P, Xing RH, Rabbani SA. Methylation status of uPA promoter as a molecular mechanism regulating prostate cancer invasion and growth in vitro and in vivo. FASEB J. 2003;17:1081–1088. doi: 10.1096/fj.02-0973com. [DOI] [PubMed] [Google Scholar]
- 40.Lind GE, Thorstensen L, Lovig T, Meling GI, Hamelin R, Rognum TO, Esteller M, Lothe RA. A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol Cancer. 2004;3:28. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu WH, Thomas TZ, Masumori N, Bhowmick NA, Gorska AE, Shyr Y, Kasper S, Case T, Roberts RL, Shappell SB, et al. The loss of TGF-beta signaling promotes prostate cancer metastasis. Neoplasia. 2003;5:267–277. doi: 10.1016/S1476-5586(03)80058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plass C, Soloway PD. DNA methylation, imprinting and cancer. Eur J Hum Genet. 2002;10:6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 43.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 44.Maio M, Coral S, Sigalotti L, Elisei R, Romei C, Rossi G, Cortini E, Colizzi F, Fenzi G, Altomonte M, et al. Analysis of cancer/testis antigens in sporadic medullary thyroid carcinoma: expression and humoral response to NY-ESO-1. J Clin Endocrinol Metab. 2003;88:748–754. doi: 10.1210/jc.2002-020830. [DOI] [PubMed] [Google Scholar]
- 45.Sugita M, Geraci M, Gao B, Powell RL, Hirsch FR, Johnson G, Lapadat R, Gabrielson E, Bremnes R, Bunn PA. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62:3971–3979. [PubMed] [Google Scholar]
- 46.Blagosklonny MV, Dixon SC, Robey R, Figg WD. Resistance to growth inhibitory and apoptotic effects of phorbol ester and UCN-01 in aggressive cancer cell lines. Int J Oncol. 2001;18:697–704. doi: 10.3892/ijo.18.4.697. [DOI] [PubMed] [Google Scholar]
- 47.Blagosklonny MV, Darzynkiewicz Z, Figg WD. Flavopiridol inversely affects p21(WAF1/CIP1) and p53 and protects p21-sensitive cells from paclitaxel. Cancer Biol Ther. 2002;1:420–425. doi: 10.4161/cbt.1.4.21. [DOI] [PubMed] [Google Scholar]
- 48.DePrimo SE, Diehn M, Nelson JB, Reiter RE, Matese J, Fero M, Tibshirani R, Brown PO, Brooks JD. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0032. (RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dasari VK, Deng D, Perinchery G, Yeh CC, Dahiya R. DNA methylation regulates the expression of Y chromosome specific genes in prostate cancer. J Urol. 2002;167:335–338. [PubMed] [Google Scholar]
- 50.Lau YF, Lau HW, Komuves LG. Expression pattern of a gonadoblastoma candidate gene suggests a role of the Y chromosome in prostate cancer. Cytogenet Genome Res. 2003;101:250–260. doi: 10.1159/000074345. [DOI] [PubMed] [Google Scholar]
- 51.Hayashizaki Y, Hirotsune S, Okazaki Y, Hatada I, Shibata H, Kawai J, Hirose K, Watanabe S, Fushiki S, Wada S, et al. Restriction landmark genomic scanning method and its various applications. Electrophoresis. 1993;14:251–258. doi: 10.1002/elps.1150140145. [DOI] [PubMed] [Google Scholar]
- 52.Luo JH, Yu YP. Genetic factors underlying prostate cancer. Expert Rev Mol Med. 2003;5:1–26. doi: 10.1017/S1462399403006057. [DOI] [PubMed] [Google Scholar]
- 53.Matsuta M, Nishiya I. Comparative genomic hybridization (CGH) Hum Cell. 1993;6:231–236. [PubMed] [Google Scholar]