Abstract
Streptococcus pyogenes is an important human pathogen that selectively interacts with proteins involved in the humoral defense system, such as immunoglobulins and complement factors. In this report we show that S.pyogenes has the ability to hydrolyze the chitobiose core of the asparagine-linked glycan on immuno globulin G (IgG) when bacteria are grown in the presence of human plasma. This activity is associated with the secretion of a novel 108 kDa protein denoted EndoS. EndoS has endoglycosidase activity on purified soluble IgG as well as IgG bound to the bacterial surface. EndoS is required for the activity on IgG, as an isogenic EndoS mutant could not hydrolyze the glycan on IgG. In addition, we show that the secreted streptococcal cysteine proteinase SpeB cleaves IgG in the hinge region in a papain-like manner. This is the first example of an endoglycosidase produced by a bacterial pathogen that selectively hydrolyzes human IgG, and reveals a novel mechanism which may contribute to S.pyogenes pathogenesis.
Keywords: cysteine proteinase/endo-β-N-acetylglucos aminidase/endoglycosidase/IgG/Streptococcus pyogenes
Introduction
An enzyme expressed by Streptococcus pyogenes capable of releasing the terminal sialic acid residues from glycoproteins such as immunoglobulins was first described by Hayano and Tanaka (1967). Other studies have shown that this release of sialic acid from immunoglobulin G (IgG) is due to a neuraminidase activity produced by S.pyogenes, resulting in autoantigenic and nephritogenic properties in patients with glomerulonephritis (Mosquera et al., 1985). Related endoglycosidases found in Staphylo coccus aureus have been shown to be cluster-dispersing enzymes involved in cell separation (Sugai et al., 1995), and glycosidases in Streptococcus oralis that sequentially deglycosylate N-linked glycoproteins promote growth of the bacterium (Byers et al., 1999). It has also been suggested that a β-N-acetylglucosaminidase from Strepto coccus pneumoniae contributes to pathogenicity, since N-linked sugar residues are common features of several cell-surface proteins in host tissue (Clarke et al., 1995).
Oligosaccharide-cleaving enzymes from other species have been studied in detail, and some are standard tools in glycobiology research. For example, endoglycosidase H from Streptomyces plicatus (Trumbly et al., 1985), and endoglycosidases F1, F2 and F3 from Flavobacterium meningosepticum (Plummer and Tarentino, 1991; Trimble and Tarentino, 1991) are commonly used. These enzymes all belong to family 18 of chitinases (Henrissat, 1991) and catalyze the hydrolysis of the β-1,4-N-acetyl-d-glucos amine linkages in the chitin core of N-linked oligosaccharides in glycoproteins. These enzymes are referred to as true endoglycosidases (Alexander and Elder, 1989; Tarentino and Plummer, 1994). However, a hydrolase isolated from F.meningosepticum, peptide-N-(N-acetyl-β-glucosaminyl)asparagine amidase (PNGase F), has activity on the pentasaccharide core region of asparagine-linked glycans, and is therefore referred to as a deglycosidase (Tarentino et al., 1985).
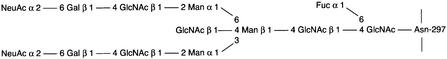
IgG is a 150 kDa glycoprotein that is a key player in the human immune response. It is composed of two identical light chains and two identical heavy chains. The heavy chain comprises one variable domain (VH) and three constant domains (CH1–3), and the light chain consists of one variable domain (VL) and one constant domain (CL). The CH1 and CH2 domains are separated by a protease-sensitive hinge region. Papain, which generates the antigen-binding Fab fragments and the effector function-promoting Fc fragment, can cleave this region. The Fc region is the interaction site for a number of effector function-promoting molecules, including complement factor C1q and Fc receptors on effector cells such as monocytes, macrophages, neutrophils and B cells (for a review see Burton, 1985). In the CH2 domains, there is a conserved N-glycosylation site at Asn297. In human IgG, a complex biantennary oligosaccharide with a bisecting N-acetylglucosamine and a core fucose is attached at this site (Figure 1).
Fig. 1. Structure of the fully substituted complex biantennary type oligosaccharide attached to Asn297 of the IgG γ-chain. GlcNAc, N-acetylglucosamine; Fuc, fucose; Man, mannose; Gal, galactose; NeuAc, sialic acid.
The functional relevance of this oligosaccharide has been implicated in several studies. For instance, murine IgG lacking the oligosaccharide does not activate complement, bind to Fc-receptors on macrophages, or induce antibody-dependent cellular cytotoxicity. Furthermore, such deglycosylated IgG impairs the elimination of antibody–antigen complexes from the circulation (Nose and Wigzell, 1983). It has also been shown that aberrant IgG glycosylation is associated with a number of autoimmune disorders, such as rheumatoid arthritis (RA) and Crohn’s disease (Dube et al., 1990). Agalactosyl IgG autoantibodies have also been demonstrated to be pathogenic in a murine model of collagen-induced arthritis (Rademacher et al., 1994; for a review see Rahman and Isenberg, 1996).
Streptococcus pyogenes is a significant human pathogen that causes infections such as impetigo, scarlatina and pharyngitis, as well as severe invasive diseases such as necrotizing fasciitis and sepsis (Bisno and Stevens, 1996; Cunningham, 2000). Non-suppurative sequelae include glomerulonephritis and acute rheumatic fever with heart complications. Streptococcus pyogenes expresses a number of proteins involved in virulence and manifestations of infection. One important protein is the secreted streptococcal cysteine proteinase, also known as the erythrogenic toxin B SpeB (Gerlach et al., 1983). SpeB degrades several host proteins including vitronectin, fibronectin (Kapur et al., 1993), fibrin and fibrinogen (Elliott, 1945; Matsuka et al., 1999). In addition, SpeB releases pro-inflammatory kinins from kininogens (Herwald et al., 1996). SpeB can also release biologically active fragments from the streptococcal surface, such as M protein, protein H and C5a peptidase (Elliott, 1945; Berge and Björck, 1995). The role of SpeB as an important virulence factor in S.pyogenes pathogenesis has been established using in vivo infection models (Lukomski et al., 1997; Tsai et al., 1998).
Other significant virulence factors of S.pyogenes are the cell wall-anchored M and M-like proteins. These are surface-exposed, dimeric, coiled-coil, α-helical structures associated with the binding of numerous plasma proteins, such as fibrinogen, fibronectin, plasminogen, immunoglobulins and human albumin (Whitnack and Beachey, 1982; Berge and Sjöbring, 1993; Cleary and Retnoningrum, 1994; Retnoningrum et al., 1993; Frick et al., 1995). In addition to expressing M1 protein, the AP1 strain of M1 serotype used in this study expresses the M-like protein H (Gomi et al., 1990). This protein binds with high affinity to the Fc region of human IgG (Åkesson et al., 1990) and has been shown to be important for survival in human blood (Kihlberg et al., 1999).
In this report we describe a novel secreted endoglycosidase in S.pyogenes, EndoS, which specifically hydrolyzes the β-1,4-di-N-acetylchitobiose core of the asparagine-linked glycan of human IgG. Furthermore, we show that the secreted SpeB cleaves the hinge region of IgG in a papain-like manner and thus acts as an IgG protease. This synergistic action of EndoS and SpeB could influence the pathogenicity of S.pyogenes during acute infection as well as in sequelae such as acute post-streptococcal glomerulonephritis and rheumatic fever.
Results
Identification of a secreted protein in S.pyogenes with similarity to an endoglycosidase
When a group A streptococcal strain of M1 serotype, AP1, was grown in C-medium (CM) and the secreted proteins analyzed by SDS–PAGE, a prominent protein was observed in the culture supernatant (Figure 2, lane A). No band at a similar molecular weight was present in the EndoS-mutant strain MC14 (Figure 2, lane B; see below). This protein band was excised and sequenced by Edman degradation, revealing the amino acid sequence EEKTVQVQ. This sequence was used to search the Streptococcal Genome Sequencing Project (SGSP) database (Ferretti et al., 2001; Roe et al., 2001) for similarities using the tBLASTn algorithm (Altschul et al., 1990). A perfect match (8/8) was found in an open reading frame (ORF) of 995 amino acids (Figure 3). The putative protein encoded by this ORF has a predicted size of 108 kDa after removal of a potential signal peptide. This putative protein was then used to search the DDBJ/EMBL/GenBank databases using the BLASTp algorithm, which resulted in similarities with the endo-β-N-acetylglucosamini dase F2 (EndoF2) precursor from F.meningosepticum (Tarentino et al., 1993). The similarity was confined to the first 444 amino acids of the query sequence, where amino acids 227–235 constitute a perfect chitinase family 18 active site, ending with a glutamic acid (Figure 3). The glutamic acid in the active site of chitinases is known to be essential for enzymatic activity (Watanabe et al., 1993).
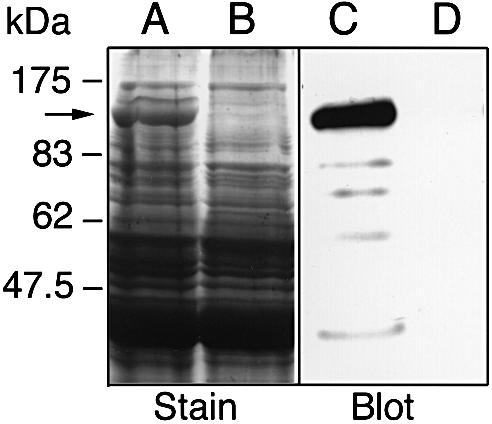
Fig. 2. Identification of a prominent, high molecular weight, secreted protein from S.pyogenes (indicated by an arrow). Proteins present in bacterial culture supernatants were separated by two identical 10% SDS–PAGE, one stained with Coomassie Blue (Stain), the other blotted to a membrane and detected with polyclonal rabbit EndoS antiserum (Blot). Lanes A and C, AP1 (wild type); lanes B and D, MC14 (EndoS–).
Fig. 3. EndoS is similar to EndoF2. Sequence alignment of EndoS from S.pyogenes strain AP1 and EndoF2 from F.meningosepticum. The N-terminal sequence of the protein band isolated from AP1 culture supernatant is underlined (EEKTVQVQ). The chitinase motif is indicated in bold (LDGLDVDVE) and the essential glutamic acid for activity is marked with an asterisk. The consensus sequence is shown under the alignment and similarities are indicated with dots.
Based on this information, the protein was denoted EndoS, for endo-β-N-acetylglucosaminidase of streptococci, and the gene ndoS. The ndoS gene from strain AP1 was cloned and sequenced. It was 99% identical to ndoS in SF370, the strain of M1 serotype used by SGSP. The deduced amino acid sequence was also 99% identical to EndoS from SF370. These sequence data have been submitted to DDBJ/EMBL/GenBank under accession No. AF296340. This suggests that S.pyogenes bacteria are able to secrete a 108 kDa protein with similarities to endo-β-N-acetylglucosaminidases.
Generation of MC14, an isogenic mutant strain lacking EndoS expression
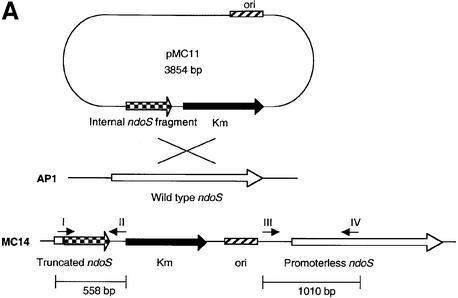
To investigate the properties of secreted EndoS further, an isogenic strain mutated in the ndoS gene of AP1 was constructed using an insertion–duplication strategy. An internal 534 bp fragment from the ndoS gene covering a part of the ORF ending upstream of the putative enzymatic motif was ligated into the streptococcal shuttle vector pFW13 (Podbielski et al., 1996). This resulted in plasmid pMC11. pMC11 was electroporated into AP1 bacteria, and one mutant clone, denoted MC14, was chosen for further characterization (Figure 4A). PCR experiments on chromosomal DNA from AP1 and MC14 bacteria with vector- and ndoS-derived primers were used to verify the integration of pMC11 into the chromosome of MC14. This revealed that both a 558 and a 1010 bp product could be amplified using chromosomal DNA from MC14, but not from wild-type AP1 (Figure 4B), thus confirming disruption of the ndoS gene.
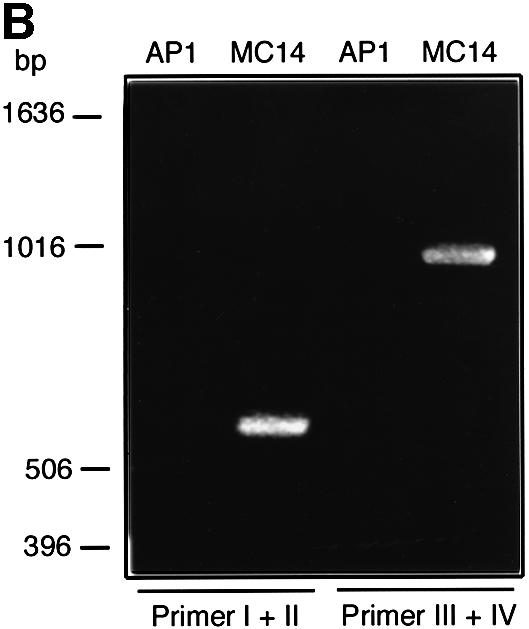
Fig. 4. (A) A schematic drawing of the strategy for mutagenesis of the ndoS gene in AP1 bacteria. An internal 534 bp fragment from the ndoS gene was cloned in the suicide plasmid pFW13, generating pMC11. This plasmid was electroporated into AP1 for homologous recombination. (B) PCR analysis of purified chromosomal DNA from AP1 and MC14. The vector- and ndoS-derived primers used are indicated with roman numerals I–IV in (A) and (B).
The mutation in MC14 did not affect the generation time, and the bacteria reached the same cell density as cultures of wild-type bacteria (data not shown). Secretion of EndoS could also be confirmed using a rabbit anti serum against full-length EndoS purified by preparative SDS–PAGE. Secretion of EndoS was analyzed by western blotting using this EndoS-specific antiserum. Reactivity against a 108 kDa protein and some minor degradation products was apparent in the supernatant from AP1 bacteria (Figure 2, lane C). In contrast, no reactivity was detected in MC14 (Figure 2, lane D). This shows that the disruption of the ndoS gene completely abolished secretion of EndoS.
Streptococcus pyogenes has glycosidase activity on human IgG
In other bacteria, such as S.oralis, it has been shown that endoglycosidases sequentially hydrolyze N-linked glycoproteins for nutritional purposes (Byers et al., 1999). Therefore, we hypothesized that EndoS in S.pyogenes could act in a similar manner and release glycans. Wild-type and EndoS-mutant bacteria were inoculated into poor growth medium, diluted CM (12.5%), which did not promote bacterial growth unless 1% human plasma was added. Bacterial growth was monitored over time, and no significant difference in growth of the two strains was apparent (data not shown). This indicated that EndoS does not play a major role in nutrient acquisition, at least under these experimental conditions.
Since EndoS has similarities to endoglycosidases, we analyzed culture supernatants for degradation products of plasma proteins after bacterial growth. The plasma proteins in culture supernatants were separated by SDS– PAGE. This revealed some apparent differences in the pattern of plasma proteins in the sample from wild-type compared with EndoS-mutant bacteria. A distinct difference was observed in the culture grown in the absence of EndoS (MC14), where a band of ∼31 kDa was detected (Figure 5, lane B). Interestingly, this 31 kDa band could not be detected in the presence of EndoS (AP1), where instead a distinct band of ∼27 kDa was observed (Figure 5, lane A). The 31 kDa protein was isolated and subjected to N-terminal sequencing. The identified sequence, GPS VFLFPP, was 100% identical to amino acids 236–244 in the hinge region of the IgG heavy chain (γ). Neither of these two bands was detected in the medium control (Figure 5, lane C). Interestingly, the cleavage site where papain and SpeB cleave human IgG differs in that papain cleaves at position 224 in human IgG1 (Burton, 1985), while SpeB cleaves between Gly236 and Gly237.
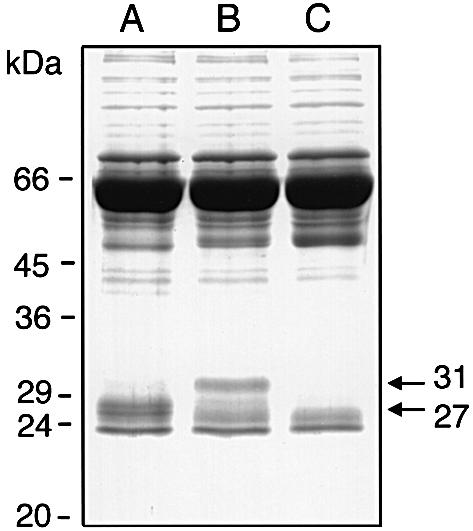
Fig. 5. EndoS has glycosidase activity on IgG in human plasma. Streptococcus pyogenes AP1 and MC14 were grown in diluted CM (12.5%) supplemented with 1% human plasma. After overnight growth, culture supernatants were analyzed by 10% SDS–PAGE. Lane A, proteins from growth with AP1 (wild type); lane B, proteins from growth with MC14 (EndoS–); lane C, medium alone. The 27 and the 31 kDa fragments of the γ-chains are indicated with arrows on the right.
The size difference between the 31 and the 27 kDa bands (Figure 5) most likely represents γ-chains with or without the complex biantennary oligosaccharide that results from inactivation of the ndoS gene in MC14. Plasma IgG contains a range of sugars in the Fc portion (Parekh et al., 1985); the largest attached to the γ-chain at Asn297 is shown in Figure 1. This experiment suggested that the glycan moiety on human IgG present in the growth medium is hydrolyzed by EndoS. Furthermore, a proteolytic cleavage of the hinge region must have occurred in order to generate a Fc fragment. This suggested a protease present in the culture supernatant of both AP1- and MC14-cleaved IgG. A likely candidate for this activity was the streptococcal cysteine proteinase SpeB.
SpeB cleaves IgG in the hinge region and EndoS hydrolyzes the glycan on IgG
Our finding that S.pyogenes bacteria can both hydrolyze the glycan moiety and proteolytically cleave IgG in the γ-chain is novel. Streptococcus pyogenes secretes a cysteine proteinase, SpeB. Recently, the three-dimensional structure of SpeB was solved, and this revealed that it is related in three-dimensional structure to papain (Kagawa et al., 2000). As it is also well established that papain cleaves human IgG into two identical Fab (antigen binding) fragments and an Fc fragment (Porter, 1973), we investigated the ability of SpeB to cleave the hinge region of IgG. Thus, an isogenic mutant strain, AL1, devoid of SpeB expression was constructed.
Purified human IgG was incubated with culture supernatant from wild-type (AP1), EndoS-mutant (MC14) and SpeB-mutant bacteria (AL1), in the absence or presence of the reducing agent dithiothreitol (DTT), and analyzed by SDS–PAGE. The rationale for using a reducing agent is that the activity of SpeB depends on reducing conditions (Liu et al., 1963). Analysis of IgG incubated in the presence of SpeB but in the absence of EndoS under non-reducing conditions (–DTT) revealed a 50 kDa band consistent with fully glycosylated γ-chains and a 25 kDa band representing the light chains (Figure 6, lane B). Incubation with EndoS, both in the presence and absence of SpeB, led to a shift of ∼4 kDa in the size of the γ-chains (Figure 6, lanes A and C). This suggested that the glycan moiety on Asn297 was removed.
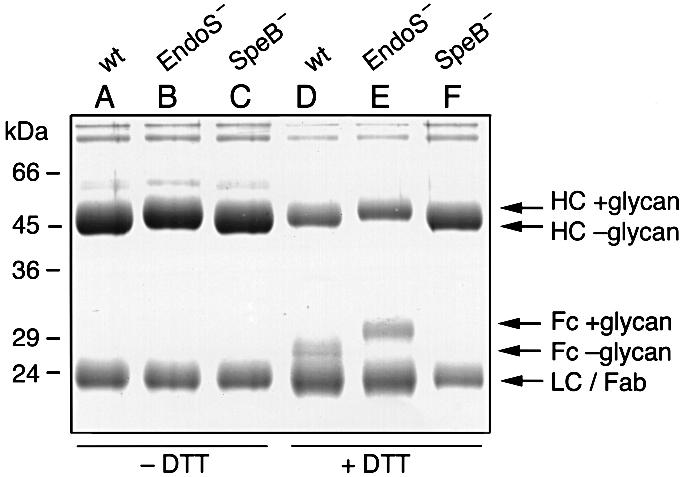
Fig. 6. Removal of Fc glycans by EndoS and cleavage of the hinge region by SpeB. SDS–PAGE analysis of purified human IgG incubated with supernatants from AP1 (wt), MC14 (EndoS–) and AL1 (SpeB–). Samples were incubated under non-reducing (–DTT) or reducing (+DTT) conditions, as indicated. Lanes A and D, wild type (wt); lanes B and E, EndoS–; lanes C and F, SpeB–. Arrows on the right indicate the position of heavy chains with glycan (HC +glycan) or without glycan (HC –glycan), Fc fragment with glycan (Fc +glycan) or without glycan (Fc –glycan), light chains (LC) and Fab fragment of heavy chains (Fab).
However, incubation of purified human IgG with culture supernatant containing both EndoS and SpeB under reducing conditions (+DTT) resulted in partial processing of the γ-chains into a band at 27 kDa (Figure 6, lane D). This corresponds to proteolytically processed γ-chains without the glycan. Furthermore, incubation without EndoS in the presence of SpeB results in a partial processing of the heavy chains into a 31 kDa band (Figure 6, lane E). This most likely represents the Fc fragment of the γ-chains with the glycan still attached. It should also be noted that when incubating in the presence of SpeB under reducing conditions, the generated γ-chain Fab fragment migrated at approximately the same molecular weight as the light chains (Figure 6, lanes D and E). However, in the absence of SpeB (Figure 6, lane F), no proteolytic cleavage of the γ-chains could be detected even under reducing conditions. Together, these data demonstrate that SpeB cleaves IgG in the hinge region and that EndoS hydrolyzes the γ-chains of human IgG.
EndoS cleaves the chitobiose core of the glycan on human IgG
To confirm that the size alteration of IgG was caused by EndoS activity and resulted from removal of the glycan moiety on γ-chains rather than proteolytic degradation, three experiments were set up. In a first approach, a lectin blot analysis was performed. Culture supernatants from wild-type AP1 and EndoS-mutant MC14 bacteria were incubated with purified IgG and separated by SDS–PAGE. Proteins were then analyzed by staining or using a lectin from Galanthus nivalis (GNL) that preferentially recognizes α-1,3 mannose residues found in the biantennary glycan on γ-chains (Figure 1). In the stained gel, a clear difference in the size of the γ-chains was observed in the sample treated with EndoS compared with non-treated IgG (Figure 7A, lanes A and B). Lectin blot analysis of the same samples with the GNL lectin revealed a significantly reduced signal when incubated with EndoS (Figure 7A, lane C). In contrast, the γ-chains were still glycosylated when incubated in the absence of EndoS (Figure 7A, lane D). These data support the proposal that EndoS has the ability to remove structures containing α-1,3 mannose from the γ-chains of human IgG.
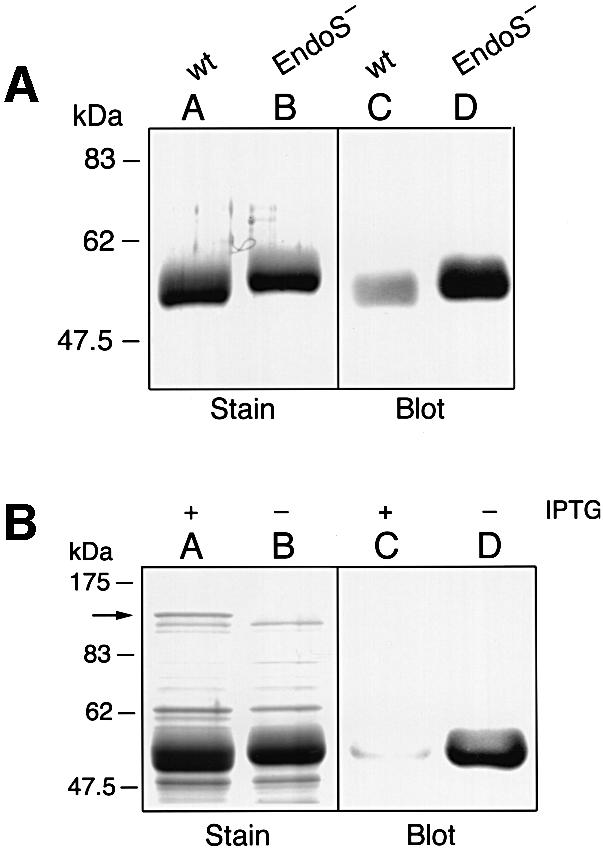
Fig. 7. SDS–PAGE and lectin blot analysis of IgG incubated with and without streptococcal or recombinant EndoS and separated by 10% SDS–PAGE. Gels were analyzed by Coomassie Blue staining (Stain) or by blotting to a membrane that was probed with GNL lectin (Blot). (A) Lanes A and C, IgG incubated with streptococcal EndoS; lanes B and D, IgG incubated without EndoS. (B) IgG incubated with extracts from E.coli expressing EndoS. Lanes A and C, induced cells (+IPTG); lanes B and D, non-induced cells (–IPTG). The arrow on the left indicates the position of recombinant EndoS.
In a second approach, cloning and expressing the gene encoding EndoS in Escherichia coli using pET-30a Xa/LIC expression system provided additional evidence that EndoS has glycosidase activity. Partially purified extracts from isopropyl-β-d-thiogalactopyranoside (IPTG)- induced or non-induced cells were incubated with purified IgG, and analyzed by SDS–PAGE and lectin blotting. This revealed that only the extract prepared from induced cells had the ability to alter the size of the γ-chains (Figure 7B, lanes A and B). Furthermore, analysis using the GNL lectin revealed that only the induced cell preparation had glycosidase activity on IgG (Figure 7B, lanes C and D).
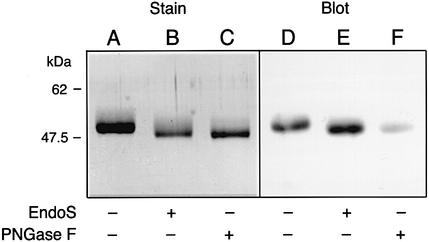
In a third approach, we confirmed that EndoS is an endoglycosidase that cleaves the chitobiose core of human IgG, using a lectin from Ulex europaeus I (UEA-I). UEA-I recognizes α-linked fucose-containing oligosaccharides. Thus, we cleaved human IgG with PNGase F, a highly specific deglycosidase that cleaves the bond between asparagine and the innermost N-acetylglucosamine of the glycan (Tarentino et al., 1985). Purified human IgG was treated with either EndoS or PNGase F, followed by separation by SDS–PAGE, or blotting and probing with the UEA-I lectin in blot analysis. In the stained gel, a clear difference in the size of the γ-chains was observed in the sample treated with EndoS compared with non-treated IgG (Figure 8, lanes A and B). Furthermore, a decrease in size was also apparent after PNGase F treatment (Figure 8, lane C). The lectin blot revealed a signal of equal intensity when the UEA-I lectin bound to untreated or EndoS-treated IgG, suggesting that the fucose group was still attached to the latter (Figure 8, lanes D and E). However, when IgG was incubated with PNGase F, which removes the entire glycan moiety, the UEA-I lectin gave a significantly lower signal (Figure 8, lane F). This suggests that EndoS cleaves the chitobiose core of human IgG, leaving an N-acetylglucosamine with an α-linked fucose. Finally, the α-linked residue’s presence on the glycan moiety did not influence EndoS activity, as analyzed by α-fucosidase treatment prior to incubation with EndoS (data not shown).
Fig. 8. EndoS is an endoglycosidase. Purified human IgG was incubated with either EndoS or PNGase F (PNGase F), separated by 10% SDS–PAGE. Gels were analyzed by Coomassie Blue staining (Stain) or by blotting to a membrane that was probed with UEA-I lectin (Blot). Lanes A and D, untreated IgG; lanes B and E, EndoS-treated IgG; lanes C and F, PNGase F-treated IgG.
Thus, partially purified recombinant EndoS has the capacity to hydrolyze human IgG to the same extent as streptococcal culture supernatants containing EndoS. Taken together, these data clearly show that S.pyogenes is able to secrete an enzyme, EndoS, with endoglycosidase activity on the γ-chains of human IgG.
EndoS has endoglycosidase activity on IgG when bound to the bacterial surface
Wild-type AP1 bacteria express the cell wall-anchored protein H, which binds human IgG with high affinity in the interface between CH2 and CH3 (Åkesson et al., 1990; Frick et al., 1992). We thus addressed whether EndoS affected protein H binding to IgG. This was analyzed by incubating purified IgG in the presence or absence of EndoS, and analyzing the ability of protein H to bind IgG in a ligand blot experiment. Treated IgG was applied to a membrane in serial dilutions and probed with radiolabeled, purified protein H. This revealed that protein H bound IgG to the same extent irrespective of EndoS treatment and indicated that the Fc-binding capacity of protein H is not influenced by EndoS (Figure 8A). This was also the case for staphylococcal protein A, which bound to both intact and EndoS-treated IgG (data not shown), as previously described for aglycosyl mouse IgG (Leatherbarrow and Dwek, 1983; Nose and Wigzell, 1983).
AP1 bacteria have the capacity to absorb IgG on their surface via protein H. We thus asked whether IgG bound to the surface of AP1 could be hydrolyzed by EndoS. Purified IgG was absorbed to the surface of washed AP1 bacteria grown in THY. The rationale for using THY is to suppress SpeB activity (Elliott, 1945), which could otherwise remove protein H from the cell wall (Berge and Björck, 1995). Unabsorbed IgG was washed away, followed by incubation with or without EndoS. After incubation, the bacteria were carefully washed and bound IgG was eluted at low pH. The IgG was then analyzed by 10% SDS– PAGE and by lectin blot analysis. As observed previously with soluble IgG (Figures 5 and 6), the γ-chains eluted from bacteria incubated in the presence of EndoS migrated as a protein band ∼4 kDa smaller than the γ-chains eluted from bacteria incubated in the absence of EndoS (Figure 9B, lanes A and B). Furthermore, lectin blot analysis of the same IgG samples revealed that γ-chains incubated in the presence of EndoS were almost completely hydrolyzed. In contrast, the lectin reacted strongly with the γ-chains incubated in the absence of EndoS (Figure 9B, lanes C and D). Thus, EndoS has endoglycosidase activity on the γ-chains of IgG when bound to the bacterial surface as well as in solution.
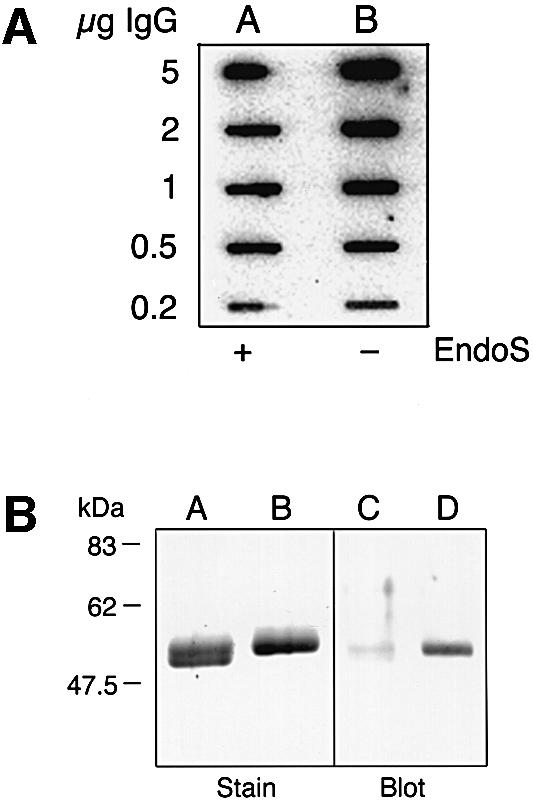
Fig. 9. (A) Ligand blot analysis of protein H binding to glycosylated or EndoS-treated IgG. Purified IgG was incubated with culture supernatant with or without EndoS, applied to a membrane in serial dilutions, and probed with125I-labeled protein H. Lane A, IgG incubated with EndoS; lane B, IgG incubated without EndoS. (B) EndoS has endoglycosidase activity on IgG when bound to the bacterial surface. SDS–PAGE and GNL lectin blot analysis of IgG, bound to the bacterial surface via protein H, treated with or without EndoS prior to elution at low pH. Lanes A and C, surface-bound IgG incubated in the presence of EndoS; lanes B and D, surface-bound IgG incubated without EndoS.
Discussion
In this report we show that the human pathogen S.pyogenes secretes a novel enzyme, EndoS, which removes the glycan moiety from the γ-chains of human IgG. EndoS has similarities to endo-β-N-acetylglucos aminidases that cleave the β1–4 linkage between the two N-acetylglucosamines found in the core of the N-linked glycan of IgG. Furthermore, we show that the streptococcal cysteine proteinase SpeB specifically cleaves in the hinge region of IgG. By utilizing dilute growth medium with small amounts of plasma and an isogenic EndoS-mutant strain, endoglycosidase activities on IgG by EndoS as well as the papain-like cleavage of IgG by SpeB were detected. This probably could not have been achieved by analyzing purified native or recombinantly expressed EndoS. Thus, these two enzymes act synergistically as an IgG endoglycosidase and IgG protease. This is the first described mechanism where S.pyogenes can both cleave and alter the glycan on one of the most significant molecules in the human immune system, IgG.
IgG in human serum appears in different glycoforms. This means that the glycan moiety terminates with different carbohydrate structures, and these various forms are important in the development of disease. For example, in the so-called agalactosyl IgG glycoforms, the glycan moiety terminates with two N-acetylglucosamine residues, instead of the fully substituted oligosaccharide with two galactose residues preceding two terminating sialic acids (Figure 1). This agalactosyl form is often found in patients with rheumatoid arthritis (RA) (Parekh et al., 1985), Crohn’s disease (Dube et al., 1990), mycobacterial infections and systemic lupus erythematosus (SLE) with Sjögren’s syndrome (Parekh et al., 1989). The production of an agalactosyl form of IgG in RA patients, for example, is a result of impaired glycosyltransferase activity in their B cells (Axford et al., 1987). Taken together, numerous observations have shown that the so-called agalactosyl form of IgG plays a role in various human diseases, of which several involve autoimmunity. Aseptic sequelae of S.pyogenes infections are also associated with autoimmunity where the glycosylation state of IgG may be of importance.
Hayano and Tanaka (1967) were the first to describe a neuraminidase that altered immunoglobulins in patients suffering from glomerulonephritis caused by this bacterium. In another report, McIntosh et al. (1972) suggested that a streptococcal neuraminidase activity produced by these strains removed the sialic acid from IgG. This, in turn, led to autoantigenicity and thereby gave rise to nephritogenicity. Thus, the neuraminidase activity augmented the pathological effect seen among patients with glomerulonephritis. Acute post-streptococcal glomerulonephritis is known to be a sequelae to infections by S.pyogenes of several M serotypes, including M1 (Bisno, 1991). This may be a result of delayed clearance of immune complexes formed by deglycosylated IgG from the circulation (Nose and Wigzell, 1983). Thus, EndoS endoglycosidase activity on IgG molecules could possibly enhance the formation of immune complexes that subsequently deposit in the kidney. EndoS can therefore be a factor that contributes to the onset of glomerulonephritis caused by these bacteria.
The AP1 strain used in this study is unique because it expresses protein H, an effective binder of human IgG. AP1 also secretes the cysteine proteinase SpeB, which degrades matrix and plasma proteins (for a review see Musser, 1997). It also releases biologically active fragments of proteins from the bacterial surface, such as protein H (Berge and Björck, 1995). The released protein H can form soluble complexes with IgG that activate complement distant from the bacterium (Berge et al., 1997).
In addition, here we report yet another factor that contributes to the complex pathogenicity of this microorganism, namely the expression of EndoS, an enzyme capable of cleaving the glycan moiety of human IgG. EndoS may function to fine-tune the number of ‘functional’ IgG molecules, both in circulation and when bound in an immune way via the Fab fragment at the bacterial surface. Thus, secreted EndoS may hydrolyze and neutralize IgG molecules that are bound to the bacteria, and impair Fc-mediated effector functions of IgG, such as complement binding and activation or opsonization.
The ability of the streptococcal cysteine proteinase SpeB to cleave IgG γ-chains in the hinge region in a papain-like manner adds a new activity for this enzyme, and to the extensive list of its biological functions. IgG-degrading activity by bacterial enzymes has previously been described for the human pathogen Pseudomonas aeruginosa, where an elastase cleaves both human and mouse IgG (Holder and Wheeler, 1984). In addition, the causative agents of chronic adult periodontitis, Prevotella intermedia and Prevotella nigrescens, have a cysteine proteinase that non-specifically degrades IgG (Jansen et al., 1995). In contrast, specific IgA protease activity is a well established feature of S.pneumoniae and Haemophilus influenzae (Mulks et al., 1980) as well as oral streptococci (Kilian and Holmgren, 1981). However, the specific IgG protease activity of SpeB that mimics papain cleavage, where the γ-chain is cleaved in the hinge region into two distinct fragments, presents an additional example of how a bacterial pathogen has evolved a mechanism to interfere with immune response molecules such as IgG.
The activity of EndoS is specific for IgG in that neither human IgA nor IgM was affected by EndoS (our unpublished observations). In human serum IgM, there are five N-linked glycosylation sites on the µ-chains, but the precise details of the oligosaccharide structure have only been solved for parts of the IgM molecule (Wormald et al., 1991). Human serum IgA1 has two N-linked glycosylation sites, on the α-chain of which the attached oligosaccharides are highly sialylated and galactosylated (Baenziger and Kornfeld, 1974).
The ability of EndoS to act on other plasma or matrix proteins has not yet been fully investigated. However, EndoS has no activity on the highly N-linked plasma glycoprotein α1-acid glycoprotein (data not shown). This is in contrast to the activity described for endoglycosidases of S.oralis (Byers et al., 1999). Furthermore, a cluster-dispersing endoglycosidase from S.aureus affects the growth morphology of the bacteria. However, an electron microscope examination of wild-type and EndoS-mutant bacteria did not reveal any alteration of shape or dispersal of the classical chain-forming growth morphology of the bacteria (data not shown).
In summary, our data reveal the first example of a human pathogen that secretes an enzyme, EndoS, that specifically acts as an endoglycosidase of human IgG both at the bacterial surface and in solution. This, together with the finding that SpeB has IgG-protease activity, adds important new aspects to the molecular pathogenesis of S.pyogenes that could be relevant both during acute infection and in aseptic sequelae.
Materials and methods
Growth conditions
Bacterial strains and plasmids used in this study are described in Table I. For maximal EndoS expression, CM was used. CM consists of 0.5% (w/v) Proteose Peptone No. 2 (Difco, Detroit, MI) and 1.5% (w/v) yeast extract (Oxoid, Basingstoke, UK) dissolved in CM buffer (10 mM K2PO4, 0.4 mM MgSO4, 17 mM NaCl pH 7.5) (Gerlach et al., 1983). To make a poor CM, it was diluted with CM buffer to 12.5% of the original concentration. All culture supernatants were centrifuged at 3800 g for 10 min and sterile-filtered through a 0.22 µm membrane (Millipore, Bedford, MA) prior to use. For absorption experiments, streptococci were cultured in Todd–Hewitt broth (Difco) supplemented with 0.2% yeast extract (THY) at 37°C in a 5% CO2 atmosphere. Escherichia coli strains were cultured in Luria–Bertani (LB) broth at 37°C. When appropriate, 50, 150 or 200 µg/ml kanamycin was added to cultures of E.coli carrying pFW13 and S.pyogenes mutant strains MC14 or AL1, respectively. Preparation of plasmid DNA from E.coli strains was carried out using a standard rapid alkaline extraction method (Birnbiom and Doly, 1979). Chromosomal DNA from S.pyogenes was prepared according to Pitcher et al. (1989) with some minor modifications, including the addition of 1000 U/ml mutanolysin (Sigma, St Louis, MO) in the lysis buffer.
Table I. Bacteria and plasmids used in this study.
| Strain or plasmid | Relevant genotype | Characteristics | References |
|---|---|---|---|
| Bacteria | |||
| Streptococcus pyogenes | |||
| AP1 | wild type | M1 serotype, IgG-binding via protein H | WHO Prague collectiona |
| MC14 | ndoS::Km | ndoS inactivated | this study |
| AL1 | speB::Km | speB inactivated | this study |
| Escherichia coli | |||
| DH5α | recA1 endA1 hsdr17 | cloning strain | BRLb |
| Bl21(DE3)pLysS | F– ompT hsdSB | cloning strain | Novagenc |
| Plasmids | |||
| pFW13 | kanR | streptococcal shuttle vector | Podbielski et al. (1996) |
| pMC11 | kanR | plasmid construct to inactivate ndoS | this study |
| pJRS233 | ermR | temperature-sensitive shuttle vector | Perez-Casal et al. (1993) |
| pJRS233-kana-speB | kanR, ermR, speB::Km | plasmid construct to inactivate speB | Svensson et al. (2000) |
| pET-30a Xa/LIC | kanR | ligation-independent expression vector | Novagenc |
| pETndoS | kanR | recombinant EndoS clone | this study |
aWHO Collaborating Center for Reference and Research on Streptococci, Institute of Hygiene and Epidemiology, Prague, Czech Republic.
bGibco-Bethesda Research Laboratories Inc., Gaithersburg, MD.
cNovagen, Madison, WI.
Generation of an EndoS-specific antiserum
A culture supernatant from AP1 grown in CM was precipitated with 5% trichloroacetic acid and separated by 10% preparative SDS–PAGE. The 108 kDa EndoS band (Figure 1) was excised from the gel and used for immunization of rabbits according to standard protocols (Harlow and Lane, 1988).
SDS–PAGE and N-terminal sequencing
To identify secreted proteins, bacterial culture supernatants were precipitated with a final concentration of 5% trichloroacetic acid, separated by 10% SDS–PAGE and stained with Coomassie Blue. Protein samples for N-terminal sequencing were separated by 10% SDS– PAGE and electroblotted to Immobilon-P™ (Millipore) as described (Matsudaira, 1987). Bands of interest were excised and subjected to Edman degradation (Edman and Begg, 1967).
For growth-promotion experiments and the identification of plasma protein substrates for EndoS, AP1 and MC14 bacteria were cultured for 24 h at 37°C in 12.5% CM containing 1% heparinized human plasma. After growth, bacteria were pelleted by centrifugation at 15 000 g for 5 min and 1 µl of each supernatant was separated by 10% SDS–PAGE. For analysis of EndoS and SpeB activity on purified IgG, culture supernatants from AP1, MC14 and AL1 bacteria were incubated for 2 h at 37°C with an equal amount of purified human IgG [2 mg/ml in phosphate-buffered saline (PBS)] in the absence or presence of a final concentration of 10 mM DTT. Two microliters of the samples were separated by 10% SDS–PAGE, followed by staining or blotting to membranes and analysis with a lectin as described below.
Western- and lectin-blotting experiments
For western blotting experiments, proteins were blotted to an Immobilon-P™ PVDF membrane (Millipore) according to Towbin et al. (1979). After blotting, membranes were blocked in PBS containing 0.1% Tween-20 and 5% skim milk (PBSTS) for 20 min at room temperature. After addition of the primary EndoS antibodies, diluted 1:1000 in PBSTS, membranes were incubated for an additional 30 min at 37°C. Membranes were washed three times for 5 min each in PBST, incubated for 30 min at 37°C with peroxidase-labeled protein A (Sigma), diluted 1:5000 in PBSTS, and washed as above. After incubation, immunoreactive proteins were detected by the Immunoprint method (Nesbitt and Horton, 1992) and exposed on Cronex X-ray film (Sterling Diagnostic Imaging, Newark, DE).
For lectin analysis, treated or untreated IgG preparations were separated by 10% SDS–PAGE and electroblotted to PVDF membranes as above. Membranes were blocked with Tris-buffered saline (TBS) with 0.1% Tween-20 (TBST), and incubated with 0.5 µg/ml biotinylated GNL or UEA-I lectin (Vector Laboratories, Burlingame, CA). After washing in TBST, the membranes were incubated with 1 µg/ml peroxidase-labeled streptavidin (Vector Laboratories). Membranes were washed in TBST and developed with the Immunoprint method as described above.
Mutagenesis of ndoS and speB in AP1
For mutagenesis of the ndoS gene in AP1, plasmid pFW13, which does not replicate in streptococci, was used for genomic integration of a truncated ndoS gene. A PCR product that covers bp 305–3232 of the ndoS gene was generated from AP1 chromosomal DNA using oligonucleotide primers 5′-CTGTACATATGGAGGAGAAGACTG-3′ and 5′-TTAATC TCGAGGTTGCTATCTAAG-3′. This product was cloned using the pGEM™-T TA cloning vector system (Promega, Madison, WI) according to the manufacturer’s instructions. One TA plasmid was digested with SpeI, which cleaves in the pGEM-T vector, and HindIII, which cleaves in the ndoS gene, to generate a fragment representing bp 305–839 of the ndoS gene (DDBJ/EMBL/GenBank accession No. AF296340). This fragment was ligated into XbaI- and HindIII-digested pFW13. Escherichia coli strain DH5α was transformed using standard protocols (Sambrook et al., 1989) and transformants were selected for kanamycin resistance. One clone, pMC11/DH5α, was isolated for further analysis. Purified plasmid pMC11 was electroporated into competent AP1 as described (Schalén et al., 1995). Recombinants were selected on THY plates containing 150 µg/ml kanamycin. Mutants were analyzed using PCR with vector- and ndoS-specific oligonucleotide primers, and one mutant, designated MC14, was chosen for further analysis.
An isogenic speB mutant was generated using the temperature-sensitive shuttle vector pJRS233 (Perez-Casal et al., 1993) containing a cloned speB fragment interrupted by a kanamycin resistance gene, generating the plasmid pJRS233-kana-speB (Svensson et al., 2000). This plasmid was electroporated into AP1 strain according to Hanski et al. (1995), generating a mutant strain designated AL1. Lack of cysteine proteinase activity was confirmed using an assay with azocasein as the substrate, as previously described (Collin and Olsén, 2000).
α-fucosidase and PNGase F treatment of human IgG
Human IgG (10 µg) was incubated either with 0.01 U of α-fucosidase from bovine kidney (Calbiochem, Darmstadt, Germany) in 50 mM sodium phosphate buffer pH 5.0 for 18 h, or with 1 U of PNGase F from Chryseobacterium meningosepticum (Calbiochem) in 50 mM sodium phosphate buffer pH 7.5 for 18 h. After incubation, the hydrolyzed IgG was separated, blotted and probed with UEA-I lectin as described above.
Ligand blot experiments
Purified human IgG (Sigma) was incubated for 2 h with culture supernatants from AP1 and MC14 bacteria grown in CM. After incubation, 0.2–5 µg of the treated IgG were applied to a PVDF membrane using a MilliBlot™ apparatus (Millipore). The membrane was blocked in PBST with 0.25% bovine serum albumin, washed with PBST and probed with 4 × 105 c.p.m./ml recombinant protein H labeled with 125I according to Bolton and Hunter (1973). After incubation, the membranes were washed extensively with PBST and scanned using a Bio-Imaging Analyzer BAS-2000 (Fuji Photo Films Co. Ltd, Japan).
Bacterial absorption experiments
AP1 bacteria were grown in THY for 16 h and harvested by centrifugation. Bacteria were washed three times with PBST containing 0.02% azide (PBSAT). Approximately 2 × 108 bacteria in 100 ml of PBSAT were incubated with 100 ml of purified human IgG (2 mg/ml in PBS) for 30 min at room temperature. Bacteria were washed as above, resuspended in 200 ml of PBSAT and incubated for 2 h at 37°C with 100 ml of culture supernatants from AP1 (EndoS+) or MC14 (EndoS–) bacteria. After washing in PBSAT, bound IgG was eluted with 50 ml of 0.2 M glycine pH 2.0 and neutralized with Tris base prior to analysis of 10 ml by 10% SDS–PAGE.
Recombinant expression of EndoS in E.coli
The ndoS gene was cloned in E.coli using the pET-30a Xa/LIC ligation-independent cloning vector system according to the manufacturer’s instructions (Novagen, Madison, WI). A PCR product covering bp 316–3195 of the intact ndoS gene was generated using oligonucleotide primers 5′-GGTATTGAGGGTCGCGAGGAGAAGACTGTT-3′ and 5′-AGAGGAGAGTTAGAGCCTTATTTTTTTAGCAG-3′ (vector compatible overhangs are underlined). This fragment was annealed to digested pET-30a vector generating pETndoS. Escherichia coli strain BL21(DE3)pLysS was transformed with pETndoS using standard protocols (Sambrook et al., 1989). Lysates from cells induced with 1 mM IPTG (Sigma) and non-induced cells were prepared by freezing of the bacterial pellets at –70°C, followed by resuspension in PBS. Lysates were cleared of cell debris by centrifugation at 15 000 g for 10 min. Supernatants were incubated with purified IgG and separated by 10% SDS–PAGE for analysis of the activity of recombinantly expressed EndoS.
Acknowledgments
Acknowledgements
Ulla Johannesson is acknowledged for excellent technical assistance, Mikael Svensson for plasmid pJRS233-kana-speB, Dr Inga-Maria Frick for purified protein H, and the SGSP funded by USPHS/NIH grant #AI38406 for making genomic sequence available. Grants from the Swedish Medical Research Council (project 13062), the Foundations of Bergvall, Crafoord, Kock, Nilson, Royal Physiografic Society and Österlund, and the Medical Faculty of Lund University supported this work.
References
- Åkesson P., Cooney,J., Kishimoto,F. and Björck,L. (1990) Protein H—a novel IgG binding bacterial protein. Mol. Immunol., 27, 523–531. [DOI] [PubMed] [Google Scholar]
- Alexander S. and Elder,J.H. (1989) Endoglycosidases from Flavo bacterium meningosepticum: application to biological problems. Methods Enzymol., 179, 505–518. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Axford J.S., Mackenzie,L., Lydyard,P.M., Hay,F.C., Isenberg,D.A. and Roitt,I.M. (1987) Reduced B-cell galactosyltransferase activity in rheumatoid arthritis. Lancet, 2, 1486–1488. [DOI] [PubMed] [Google Scholar]
- Baenziger J. and Kornfeld,S. (1974) Structure of the carbohydrate units of IgA1 immunoglobulin. I. Composition, glycopeptide isolation and structure of the asparagine-linked oligosaccharide units. J. Biol. Chem., 249, 7260–7269. [PubMed] [Google Scholar]
- Berge A. and Björck,L. (1995) Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem., 270, 9862–9867. [DOI] [PubMed] [Google Scholar]
- Berge A. and Sjöbring,U. (1993) PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem., 268, 25417–25424. [PubMed] [Google Scholar]
- Berge A., Kihlberg,B.-M., Sjöholm,A.G. and Björck,L. (1997) Streptococcal protein H forms soluble complement-activating complexes with IgG, but inhibits complement activation by IgG-coated targets. J. Biol. Chem., 272, 20774–20781. [DOI] [PubMed] [Google Scholar]
- Birnbiom H.C. and Doly,J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno A.L. (1991) Group A streptococcal infections and acute rheumatic fever. N. Engl. J. Med., 325, 783–793. [DOI] [PubMed] [Google Scholar]
- Bisno A.L. and Stevens,D.L. (1996) Streptococcal infection of skin and soft tissue. N. Engl. J. Med., 334, 240–245. [DOI] [PubMed] [Google Scholar]
- Bolton A.E. and Hunter,W.M. (1973) The labelling of proteins to high specific radioactivities by conjugation to a 125I-containing acylating agent: application to the radioimmunoassay. Biochem. J., 133, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R. (1985) Immunoglobulin G: functional sites. Mol. Immunol., 22, 161–206. [DOI] [PubMed] [Google Scholar]
- Byers H.L., Tarelli,E., Homer,K.A. and Beighton,D. (1999) Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human α1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology, 9, 469–479. [DOI] [PubMed] [Google Scholar]
- Clarke V.A., Platt,N. and Butters,T.D. (1995) Cloning and expression of the β-N-acetylglucosaminidase gene from Streptococcus pneumoniae. Generation of truncated enzymes with modified aglycon specificity. J. Biol. Chem., 270, 8805–8814. [DOI] [PubMed] [Google Scholar]
- Cleary P. and Retnoningrum,D. (1994) Group A streptococcal immunoglobulin-binding proteins: adhesins, molecular mimicry or sensory proteins? Trends Microbiol., 2, 131–136. [DOI] [PubMed] [Google Scholar]
- Collin M. and Olsén,A. (2000) Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol., 36, 1306–1318. [DOI] [PubMed] [Google Scholar]
- Cunningham M.W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev., 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube R., Rook,G.A., Steele,J., Brealey,R., Dwek,R., Rademacher,T. and Lennard-Jones,J. (1990) Agalactosyl IgG in inflammatory bowel disease: correlation with C-reactive protein. Gut, 31, 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman P. and Begg,G. (1967) A protein sequenator. Eur. J. Biochem., 1, 80–91. [DOI] [PubMed] [Google Scholar]
- Elliott S.D. (1945) A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J. Exp. Med., 81, 573–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti J.J. et al. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl Acad. Sci. USA, 98, 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick I.-M., Wikström,M., Forsén,S., Drakenberg,T., Gomi,H., Sjöbring,U. and Björck,L. (1992) Convergent evolution among immunoglobulin G-binding bacterial proteins. Proc. Natl Acad. Sci. USA, 89, 8532–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick I.-M., Crossin,K.L., Edelman,G.E. and Björck,L. (1995) Protein H—a bacterial surface protein with affinity for both immunoglobulin and fibronectin type III domains. EMBO J., 14, 1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach D., Knöll,H., Köhler,W., Ozegowski,J.-H. and Hribalova,V. (1983) Isolation and characterization of erythrogenic toxins V. Communication: identity of erythrogenic toxin type B and streptococcal proteinase precursor. Zentralbl. Bakteriol. Mikrobiol. Hyg. A, 255, 221–233. [PubMed] [Google Scholar]
- Gomi H., Hozumi,T., Hattori,S., Tagawa,C., Kishimoto,F. and Björck,L. (1990) The gene sequence and some properties of protein H—a novel IgG binding protein. J. Immunol., 144, 4046–4052. [PubMed] [Google Scholar]
- Hanski E., Fogg,G., Tovi,A., Okada,N., Burstein,I. and Caparon,M. (1995) Molecular analysis of Streptococcus pyogenes adhesion. Methods Enzymol., 253, 269–305. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hayano S. and Tanaka,A. (1967) Streptococcal sialidase. I. Isolation and properties of sialidase produced by group K Streptococcus. J. Bacteriol., 93, 1753–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J., 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwald H., Collin,M., Müller-Esterl,W. and Björck,L. (1996) Streptococcal cysteine proteinase releases kinins: a novel virulence mechanism. J. Exp. Med., 184, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder I.A. and Wheeler,R. (1984) Experimental studies of the pathogenesis of infections owing to Pseudomonas aeruginosa: elastase, an IgG protease. Can. J. Microbiol., 30, 1118–1124. [DOI] [PubMed] [Google Scholar]
- Jansen H.J., Grenier,D. and Van der Hoeven,J.S. (1995) Characterization of immunoglobulin G-degrading proteases of Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immunol., 10, 138–145. [DOI] [PubMed] [Google Scholar]
- Kagawa T.F., Cooney,J.C., Baker,H.M., McSweeney,S., Liu,M., Gubba,S., Musser,J.M. and Baker,E.N. (2000) Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease. Proc. Natl Acad. Sci. USA, 97, 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur V., Topouzis,S., Majesky,M.W., Li,L.-L., Hamrick,M.R., Hamill,R.J., Patti,J.M. and Musser,J.M. (1993) A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog., 15, 327–346. [DOI] [PubMed] [Google Scholar]
- Kihlberg B.-M., Collin,M., Olsén,A. and Björck,L. (1999) Protein H, an antiphagocytic surface protein in Streptococcus pyogenes. Infect. Immun., 67, 1708–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian M. and Holmgren,K. (1981) Ecology and nature of immuno globulin A1 protease-producing streptococci in the human oral cavity and pharynx. Infect. Immun., 31, 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherbarrow R.J. and Dwek,R.A. (1983) The effect of aglycosylation on the binding of mouse IgG to staphylococcal protein A. FEBS Lett., 164, 227–230. [DOI] [PubMed] [Google Scholar]
- Liu T.-Y., Neumann,N.P., Elliott,S.D., Moore,S. and Stein,W.H. (1963) Chemical properties of streptococcal proteinase and its zymogen. J. Biol. Chem., 238, 251–256. [PubMed] [Google Scholar]
- Lukomski S., Sreevatsan,S., Amberg,A., Reichardt,W., Woischnik,M., Podbielski,A. and Musser,J.M. (1997) Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J. Clin. Invest., 99, 2574–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem., 262, 10035–10038. [PubMed] [Google Scholar]
- Matsuka Y.V., Pillai,S., Gubba,S., Musser,J.M. and Olmsted,S.B. (1999) Fibrinogen cleavage by the Streptococcus pyogenes extracellular cysteine protease and generation of antibodies that inhibit enzyme proteolytic activity. Infect. Immun., 67, 4326–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh R.M., Kaufman,D.B., McIntosh,J.R. and Griswold,W. (1972) Glomerular lesions produced by autologous serum and autologous IgG modified by treatment with a culture of α-haemolytic streptococcus. J. Med. Microbiol., 5, 1–7. [DOI] [PubMed] [Google Scholar]
- Mosquera J.A., Katiyar,V.N., Coello,J. and Rodriguez-Iturbe,B. (1985) Neuraminidase production by streptococci from patients with glomerulonephritis. J. Infect. Dis., 151, 259–263. [DOI] [PubMed] [Google Scholar]
- Mulks M.H., Kornfeld,S.J. and Plaut,A.G. (1980) Specific proteolysis of human IgA by Streptococcus pneumoniae and Haemophilus influenzae. J. Infect. Dis., 141, 450–456. [DOI] [PubMed] [Google Scholar]
- Musser J.M. (1997) Streptococcal superantigen, mitogenic factor and pyrogenic exotoxin B expressed by Streptococcus pyogenes. Structure and function. Prep. Biochem. Biotechnol., 27, 143–172. [DOI] [PubMed] [Google Scholar]
- Nesbitt S.A. and Horton,M.A. (1992) A non radioactive biochemical characterization of membrane proteins using enhanced chemi luminescence. Anal. Biochem., 206, 267–272. [DOI] [PubMed] [Google Scholar]
- Nose M. and Wigzell,H. (1983) Biological significance of carbohydrate chains on monoclonal antibodies. Proc. Natl Acad. Sci. USA, 80, 6632–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh R.B. et al. (1985) Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature, 316, 452–457. [DOI] [PubMed] [Google Scholar]
- Parekh R., Isenberg,D., Rook,G., Roitt,I., Dwek,R. and Rademacher,T. (1989) A comparative analysis of disease-associated changes in the galactosylation of serum IgG [published erratum appears in J. Autoimmun., 1989, 2, 307]. J. Autoimmun., 2, 101–114. [DOI] [PubMed] [Google Scholar]
- Perez-Casal J., Price,J.A., Maguin,E. and Scott,J.R. (1993) An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequence to the chromosome of S. pyogenes. Mol. Microbiol., 8, 809–819. [DOI] [PubMed] [Google Scholar]
- Pitcher D.G., Saunders,N.A. and Owen,R.J. (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol., 8, 151–156. [Google Scholar]
- Plummer T.H. Jr and Tarentino,A.L. (1991) Purification of the oligosaccharide-cleaving enzymes of Flavobacterium meningo septicum. Glycobiology, 1, 257–263. [DOI] [PubMed] [Google Scholar]
- Podbielski A., Spellerberg,B., Woischnik,M., Pohl,B. and Lütticken,R. (1996) Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene, 177, 137–147. [DOI] [PubMed] [Google Scholar]
- Porter R.R. (1973) Structural studies of immunoglobulins. Science, 180, 713–716. [DOI] [PubMed] [Google Scholar]
- Rademacher T.W., Williams,P. and Dwek,R.A. (1994) Agalactosyl glycoforms of IgG autoantibodies are pathogenic. Proc. Natl Acad. Sci. USA, 91, 6123–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.A.A. and Isenberg,D.A. (1996) Glycosylation of IgG in rheumatic disease. In Isenberg,D.A. and Rademacher,T.W. (eds), Abnormalities of IgG Glycosylation and Immunological Disorders. John Wiley & Sons, Chichester, UK, pp. 101–118.
- Retnoningrum D.S., Podbielski,A. and Cleary,P.P. (1993) Type M12 protein from Streptococcus pyogenes is a receptor for IgG3. J. Immunol., 150, 2332–2340. [PubMed] [Google Scholar]
- Roe B.A., Linn,S.P., Song,L., Yuan,X., Clifton,S., McLaughlin,R.E., McShan,M. and Ferretti,J. (2001) Streptococcus pyogenes genome sequencing—strain M1 GAS. http://www.genome.ou.edu/strep.html, November 2, 2000 (access date).
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schalén C., Gebreselassie,D. and Ståhl,S. (1995) Characterization of an erythromycin resistance (erm) plasmid in Streptococcus pyogenes. APMIS, 103, 59–68. [DOI] [PubMed] [Google Scholar]
- Sugai M., Komatsuzawa,H., Akiyama,T., Hong,Y.M., Oshida,T., Miyake,Y., Yamaguchi,T. and Suginaka,H. (1995) Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J. Bacteriol., 177, 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson M.D., Scaramuzzino,D.A., Sjöbring,U., Olsén,A., Frank,C. and Bessen,D.E. (2000) Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol. Microbiol., 38, 242–253. [DOI] [PubMed] [Google Scholar]
- Tarentino A.L. and Plummer,T.H.,Jr (1994) Enzymatic deglycosylation of asparagine-linked glycans: purification, properties and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningo septicum. Methods Enzymol., 230, 44–57. [DOI] [PubMed] [Google Scholar]
- Tarentino A.L., Gomez,C.M. and Plummer,T.H.,Jr (1985) Deglycosyl ation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry, 24, 4665–4671. [DOI] [PubMed] [Google Scholar]
- Tarentino A.L., Quinones,G., Changchien,L.M. and Plummer,T.H.,Jr (1993) Multiple endoglycosidase F activities expressed by Flavo bacterium meningosepticum endoglycosidases F2 and F3. Molecular cloning, primary sequence and enzyme expression. J. Biol. Chem., 268, 9702–9708. [PubMed] [Google Scholar]
- Towbin H., Staehelin,T. and Gordon,J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble R.B. and Tarentino,A.L. (1991) Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningo septicum: endo F1, endo F2 and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J. Biol. Chem., 266, 1646–1651. [PubMed] [Google Scholar]
- Trumbly R.J., Robbins,P.W., Belfort,M., Ziegler,F.D., Maley,F. and Trimble,R.B. (1985) Amplified expression of Streptomyces endo-β-N-acetylglucosaminidase H in Escherichia coli and characterization of the enzyme product. J. Biol. Chem., 260, 5683–5690. [PubMed] [Google Scholar]
- Tsai P.-J., Kuo,C.-F., Lin,K.-Y., Lin,Y.-S., Lei,H.-Y., Chen,F.-F., Wang,J.-R. and Wu,J.-J. (1998) Effect of group A streptococcal cysteine proteinase on invasion of epithelial cells. Infect. Immun., 66, 1460–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Kobori,K., Miyashita,K., Fujii,T., Sakai,H., Uchida,M. and Tanaka,H. (1993) Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J. Biol. Chem., 268, 18567–18572. [PubMed] [Google Scholar]
- Whitnack E. and Beachey,E.H. (1982) Antiopsonic activity of fibrinogen bound to M protein on the surface of group A streptococci. J. Clin. Invest., 69, 1042–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormald M.R., Wooten,E.W., Bazzo,R., Edge,C.J., Feinstein,A., Rademacher,T.W. and Dwek,R.A. (1991) The conformational effects of N-glycosylation on the tailpiece from serum IgM. Eur. J. Biochem., 198, 131–139. [DOI] [PubMed] [Google Scholar]