Abstract
Oncolytic adenoviruses are being tested as potential therapies for human malignant tumors, including gliomas. Here we report for the first time that a mutation in the E1A gene results in low levels of E1A protein, conditioning the replication of mutant adenoviruses specifically to cancer cells. In this study, we compared the oncolytic potencies of three mutant adenoviruses encompassing deletions within the CR1 (Delta-39), CR2 (Delta-24) regions, or both regions (Delta-24/39) of the E1A protein. Delta-39 and Delta-24 induced a cytopathic effect with similar efficiency in glioma cells and a comparable capacity for replication. Importantly, the activity of Delta-39 was significantly attenuated compared to Delta-24 in proliferating normal human astrocytes. Direct analyses of the activation of E2F-1 promoter demonstrated the inability of Delta-39 to induce S-phase-related transcriptional activity in normal cells. Interestingly, E1A protein levels in cells infected with Delta-39 were remarkably downmodulated. Furthermore, protein stability studies revealed enhanced degradation of CR1 mutant E1A proteins, and inhibition of the proteasome activity resulted in the striking rescue of E1A levels. We conclude that the level of E1A protein is a critical determinant of oncolytic phenotype and we propose a completely novel strategy for the design and construction of conditionally replicative adenoviruses.
Keywords: glioma, oncolytic, adenovirus, E1A, CR1
Introduction
Oncolytic adenoviruses, engineered to selectively replicate in and lyse cancer cells, hold great potential as efficacious therapeutic agents for primary brain tumors [1–4]. One of the strategies commonly used to design oncolytic adenoviruses is to genetically modify E1A protein binding of key cellular proteins. Comparing E1A protein sequences of various adenovirus serotypes identified three evolutionary conserved regions (CR), pointing to their critical roles in E1A activity [5]. Thus, the CR1 (42–72 aa) and CR2 (115–140 aa) regions in type 5 human adenovirus (Ad5) include E1A binding sites for p300/CBP, Rb, and Rb-related proteins [5,6]. One critical consequence of their ensuing interactions is disrupting pRb binding to E2F. This dysregulation stimulates the transcription of E2F-responsive genes and induces quiescent cells to enter the S-phase [7]. Interactions between E1A and cell cycle modulators would be critical in infected quiescent normal cells in which the G1 checkpoint is tightly regulated by a network of proteins with overlapping functions. The effect of such viral-cellular protein interactions would, however, not be particularly critical in cancer cells where G1/S transition controls are already partially inactivated. This is the underlying rationale that is driving the assessment of E1A mutant adenoviruses as preferentially or exclusively acquiring lytic properties in cancer cells [1,2].
In this work, we compared the antiglioma efficacy and replication selectivity of a CR1 mutant adenovirus (Delta-39) with the CR2 mutant adenovirus, Delta-24 [3]. Importantly, we uncover that cells infected with Delta-39 expressed low levels of E1A protein due to protein instability and enhanced proteosomal-mediated degradation. However, although the low level of E1A did not substantially impair the ability of Delta-39 to replicate in cancer cells, the activity of Delta-39 was significantly attenuated in proliferating normal cells. Delta-39 is thus an excellent potential candidate for further development as an antiglioma therapeutic tool. This is the first report showing that the level of E1A protein is a critical determinant of the selectivity of oncolytic adenoviruses and proposes a completely novel strategy for the design and construction of conditionally replicative adenovirus.
Materials and Methods
Cell Lines
U-87 MG, D-54 MG, U-251 MG, and 293 cells were cultured as previously described [3]. Normal human astrocytes (NHA) were obtained from Clonetics/BioWhittaker (Wakersville, MD) and cultured using the manufacturer's specifications.
Adenoviruses
Delta-24 characteristics were reported previously [2]. The 39-nucleotide deletion, corresponding to 48 to 60 aa in the CR1 region of the E1A protein, in pXC1 (Microbix Biosystems, Inc., Ontario, Canada) or pXC1-D24 [3]) was accomplished by replacing the BamHI/XbaI fragment, encompassing the 701 to 739 nt region of the Ad5 genome, with two polymerase chain reaction (PCR) fragments flanking this deleted region, resulting in pXC1-D39 (harboring a 701–739 nt deletion in the adenoviral genome) and pXC1-D24/39 (harboring deletions of 701–739 nt and 923–946 nt in the Ad5 genome). XC1-D39 or pXC1-D24/39 was cotransfected with pBHG10 (Microbix Biosystems, Inc.) into 293 cells to generate the Delta-39 and Delta-24/39 adenoviruses, respectively, and then amplified in 293 cells and purified by CsCl gradient centrifugation [8]. To confirm the deletions, PCR amplification of a region of the Ad5 genome, which encompasses the 39- and 24-bp deletions in the E1A region, was followed by XhoII or BstXI enzymatic digestion. Controls include wild-type adenovirus (Adwt) [9], ultraviolet-inactivated adenovirus, and mock infections [3].
Immunoprecipitation and Immunoblotting
Cells infected with Adwt, Delta-24 at 20 MOI, or Delta-24/39 at 50 MOI, or mock-infected were collected 16 hours after infection and suspended in ice-cold PBS plus 0.5% NP40 and a protease inhibitor cocktail (Sigma, St. Louis, MO), and then lysed by sonication. For each sample, 2 µg of anti-E1A antibody and 20 µl of Protein A Agarose beads (Oncogene, Cambridge, MA) were added to 400 µg of cell lysate proteins. After 4 hours of immunoprecipitation at 4°C, the beads were pelleted and washed. The precipitated proteins or cell lysate proteins were dissolved in 1x SDS loading buffer, separated by SDS-PAGE, and probed with antibodies against adenovirus 2 E1A (13 S-5), p300 (C-20), Rb (C-15)-G, or β-actin (Santa Cruz Biotechnology, Santa Cruz, CA). For Western blot experiments, cells were infected with 50 MOI of each adenovirus; however, different viral dosages were used for infection to achieve similar E1A expression levels in cell lysates for immunoprecipitation.
Cell Viability and Viral Replication Assays
Crystal violet, MTT, and tissue culture infection dose50 (TCID50) assays were conducted as described previously [3,10]. Final viral titers were determined as plaque-forming units (pfu/ml) according to the validated method developed by Quantum Biotechnology (Carlsbad, CA).
Luciferase Assay
A total of 3 x 104/well cells was seeded in 24-well plates and grown for 24 hours. Each well was cotransfected with 250 ng of the E2F-1 reporter plasmid [11] and 1 ng of pRL-CMV as control for transfection efficiency (Promega Life Science, Madison, MI) with FuGENE 6 (Roche Molecular Biochemicals, Indianapolis, IN). The cells were infected 2 hours later with 5 MOI of the adenoviruses. Twenty-four hours after transfection, the cells were lysed and luciferase activity was assayed with a luminometer.
E1A Protein Stability Studies
A total of 6 x 104 cells/well was seeded in 24-well plates and infected 24 hours later with adenoviruses at 50 MOI. Sixteen hours later, 100 µM anisomycin (Sigma) was added to the culture to inhibit protein synthesis. Cell lysates were collected in 1x SDS loading buffer (100 µl/well). An equal volume of cell lysates was used for immunoblotting analysis of E1A protein expression.
Proteasomal-Mediated Degradation of E1A
Cells were infected with adenoviruses for 16 hours at 50 MOI and then either mock-treated or treated with 10 µM lactacystin (Calbiochem, La Jolla, CA) for 24 hours. The cell lysates were subjected to immunoblotting analysis.
Results
Construction and Characterization of Delta-39 and Delta-24/39 Adenoviruses
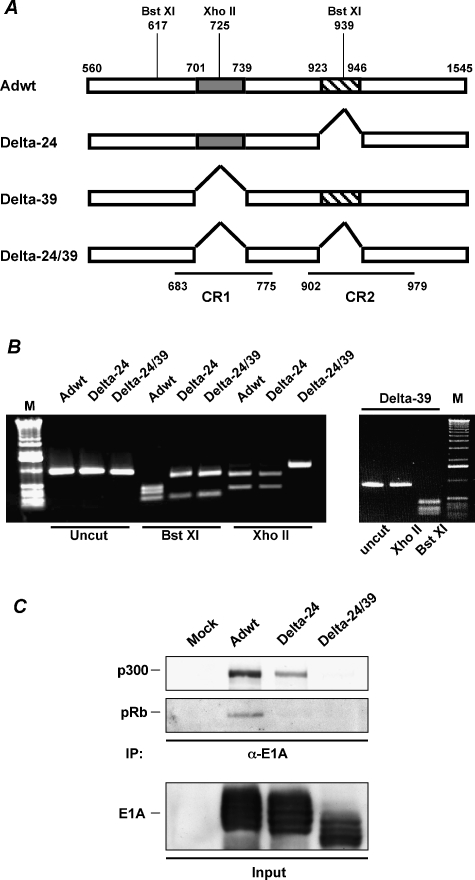
The Delta-24 adenovirus harbors a 24-nucleotide deletion (from 923 to 946 nt in the Ad5 genome) corresponding to 122 to 129 aa in the CR2 region of E1A protein, which is responsible for pRb binding [3]. The Delta-39 adenovirus encompasses a deletion of a 48 to 60 aa region (from 701 to 739 nt in the Ad5 genome) in the CR1 region of the E1A protein, which is responsible for p300 binding [12]. The Delta-24/39 adenovirus was constructed by generating a CR1 mutation similar to the one in Delta-39, on a Delta-24 backbone [3] (Figure 1A). Adenoviral genome deletions were verified through PCR amplification followed by restriction enzyme digestion (Figure 1B), and then confirmed by sequencing (data not shown). As expected, immunoprecipitation analyses of mutant E1A complexes showed that CR1 mutant E1A proteins do not bind p300, and that CR2 mutant proteins do not physically interact with pRb (Figure 1C).
Figure 1.
E1A mutant adenoviruses. (A) Schematic representation of the E1A region of the genome of Adwt, and the E1A mutant adenoviruses Delta-24, Delta-39, and Delta-24/39. The first nucleotide of the start codon and third nucleotide of the stop codon of the E1A gene are indicated. Shown are the 39-bp (gray box) and 24-bp (hatched box) deletions. For clarity, the corresponding DNA segments of the CR1 and CR2 regions in the E1A protein are also indicated. Restriction enzyme sites used to confirm E1A mutations are illustrated. (B) Identification of the deletions through PCR amplification followed by restriction enzyme digestion. The PCR product sizes and enzyme digestion profiles for the tested adenoviruses are as follows: Adwt, a 941-bp PCR fragment was digested into three fragments (207, 322, and 412 bp) with BstXI and into two fragments (315 and 626 bp) with XhoII; Delta-24, a 917-bp PCR fragment was digested into two fragments (207 and 710 bp) with BstXI and two fragments (315 and 602 bp) with XhoII; Delta-39, a 902-bp PCR product, digested into three fragments (207, 283, and 412 bp) with BstXI; Delta-24/39, a 878-bp PCR product, digested into two fragments (207 and 671 bp) with BstXI. Note that the 39-bp deletion (701–739 nt) eliminated a XhoII restriction site. Uncut PCR products are shown for size comparison. Samples were analyzed on 1% agarose gel. M: 1-kb DNA ladder. (C) Disruption of E1A-p300 or E1A-pRb interactions in CR1 or CR2 mutant adenoviruses. U-251 MG cells infected with Adwt, Delta-24, or Delta-24/39, or were mock-infected. The cell lysates were immunoprecipitated with the anti-E1A antibody. Precipitated proteins were separated by 10% SDS-PAGE and immunoblotted for p300 and pRb. Input cell lysates (10% of total) were analyzed for E1A expression.
Delta-39 and Delta-24 Induced Comparable Oncolytic Effects in Glioma Cells
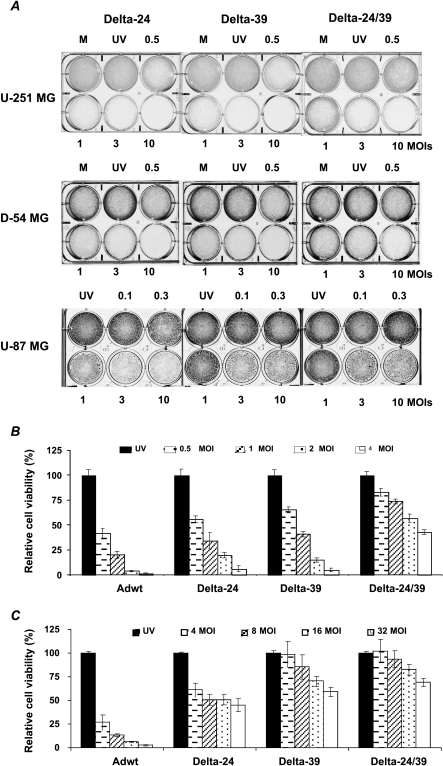
We first studied the in vitro antiglioma activity of the Delta-24, Delta-39, and Delta-24/39 adenoviruses in three glioma cell lines: U-251 MG, D-54 MG, and U-87 MG. Crystal violet assays showed that all three E1A mutant adenoviruses displayed antiglioma activity and that there was no marked difference between the oncolytic effects of Delta-24 and Delta-39 (Figure 2A). The antiglioma activity of Delta-24/39 was inferior to the other two mutant adenoviruses (Figure 2A). A quantitative assessment of cell viability by MTT demonstrated a consistent adenovirus-mediated cytopathic effect in U-251 MG glioma cells (Figure 2B). It showed that the viral dose required to induce a 50% decrease in cell viability (IC50) was 0.5 MOI for Adwt, 1 MOI for Delta-24 and Delta-39, and 4 MOI for Delta-24/39 (Figure 2B). As expected, the cytotoxicity of all three E1A mutant adenoviruses was attenuated with respect to wild-type adenovirus in NHA (Figure 2C). The IC50 was less than 4 MOI for Adwt and about 32 MOI for Delta-24. Importantly, with the maximum tested dose, more than 65% and 75% of the Delta-39- and Delta-24/39-infected cells, respectively, were still viable (Figure 2C). We conclude that, whereas the antiglioma efficiencies of Delta-24 and Delta-39 were similar, the double mutant E1A adenovirus, Delta-24/39, produced a relatively attenuated anticancer effect. Additionally, both CR1 mutant adenoviruses exhibited an attenuated oncolytic profile, compared to Delta-24, in proliferating normal astrocytes.
Figure 2.
In vitro antiglioma activity of the E1A mutant adenoviruses. (A) Adenoviral-mediated CPE was assayed by crystal violet staining. Glioma cells were infected with Delta-24, Delta-39, or Delta-24/39 adenoviruses at the indicated doses. Experiments were completed when one of the adenovirus induced a CPE of 80% or more at 10 MOI. Thus, the staining was performed in U-251 MG, D-54 MG, and U-87 MG cells at the fifth, fourth, and ninth days of the viral infection, respectively. M, mock; UV, UV-inactivated adenovirus (10 MOI). (B and C) Cell viability was assessed by MTT analysis. U-251 MG (B) or NHA (C). Cells were infected at the indicated MOI of Adwt, Delta-24, Delta-39, and Delta-24/39, or at the highest dose of UV-inactivated Adwt. Cell viability was assessed 6 days after infection. Data are mean ± SD of eight determinations, and are represented as cell viability relative to UV-inactivated adenovirus-treated cells (equal to 100%); UV, UV-inactivated adenovirus.
Delta-39 Showed Broader Therapeutic Index than Delta-24
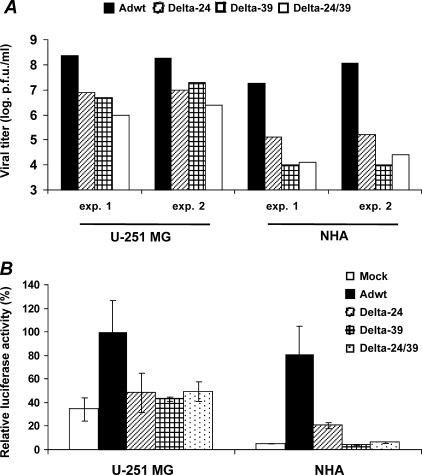
To determine whether the CPE was related to adenoviral replication, we used a TCID50 assay. Delta-24 and Delta-39 replicated with a similar potency in glioma cells, whereas the activity of Delta-24/39 was relatively attenuated (Figure 3A). Notably, these experiments showed that the final titers of Delta-39 and Delta-24/39 were slightly inferior to the initial dose (5 x 104 pfu), highlighting the substantially diminished ability of CR1 mutant adenoviruses to replicate in a dividing NHA population (Figure 3A). In summary, Delta-24 and Delta-24/39 replicated approximately 100 times better in cancer cells than in normal cells, but Delta-39 replicated approximately 1000 times more efficiently in glioma cells than in dividing NHA. Collectively, cell viability and viral replication experiments indicate that Delta-39 exhibited an improved therapeutic index when compared to Delta-24 or Delta-24/39.
Figure 3.
Replication potency of the E1A mutant adenoviruses in glioma cells and proliferating normal astrocytes. (A) Viral replication profile in U-251 MG and NHA cells. The input dose was 1 MOI (5 x 104 pfu totally), and the final titer of the new adenoviral progeny was quantified 3 days after the infection using the TCID50 method. Viral titer is expressed as logarithmic scale of plaque-forming units per milliliter. Shown are two independent experiments for cancer and normal cells. (B) Modulation of the E2F-1 promoter activity in U-251 MG and NHA cells. After adenoviral infection, U-251 MG and NHA cells were cotransfected with an E2F-1 promoter reporter construct and the control vector, pRL-CMV. Then the cultures were infected with the indicated viruses. Luciferase activity was defined as firefly luciferase activity (reporter) normalized by renilla luciferase activity (internal control), and expressed relative to the values obtained in the Delta-24-treated U-251 MG cells (equal to 100%). Each experiment was performed in triplicate. Data are shown as mean ± SD.
We next asked whether Delta-39 triggered the cellular DNA replication machinery differently in cancer versus normal cells. To stimulate adenoviral replication, E1A protein binds pRb and releases E2F1 transcriptional activity [7]. Because E2F-1 can be autoactivated [11], E1A-mediated release of E2F-1 transcriptional activation is an indicator of the adenovirus' ability to activate the DNA replication machinery of the host cell. Hence, we directly analyzed the E1A-mediated activation of an E2F-1 promoter coupled to a luciferase gene. Cancer cells infected with any of the three E1A mutant adenoviruses showed a similar E2F-1 promoter activity, which was not significantly different from the activity in mock-treated cells (P > 0.1, double-sided t-test) (Figure 3B), underscoring the inefficient repression of Rb on E2F-1 in these glioma cells [13]. However, when NHA were infected with Delta-24, E2F-1 promoter activity was upregulated compared to mock-treated cells (P < .01, double-sided t-test). In contrast, infection with Delta-39 or Delta-24/39 did not significantly modify E2F-1 promoter activity compared to control cultures (P > .1, double-sided t-test) (Figure 3B), which is not surprising because the CR1 deletion in Delta-39 also weakens the binding of E1A to pRb and p107, in addition to abrogating the binding to p300 protein [12,14,15]. These results supported the replication assay findings, and suggested that Delta-39 and Delta-24 adenoviruses fundamentally differed in their abilities to trigger unscheduled S-phase in normal cells.
Enhanced Proteasomal-Mediated Degradation Resulted in Low Levels of CR1 Mutant E1A Protein
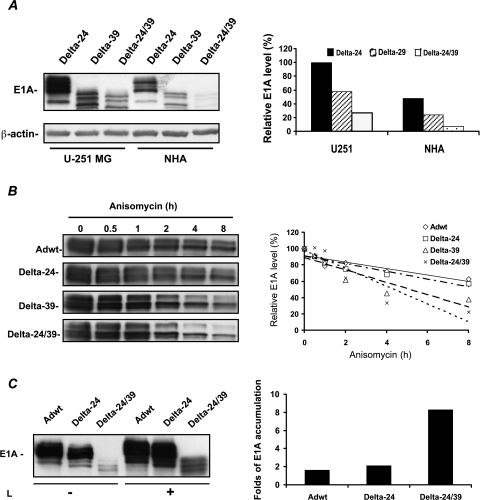
To examine how the CR1 deletion affects E1A protein levels, we analyzed E1A protein expression in glioma cells and NHA after infection with the E1A mutant adenoviruses. Intriguingly, we found that the E1A protein was expressed at markedly lower levels in U-251 MG and NHA treated with Delta-39 or Delta-24/39 than in both Delta-24-infected cultures. E1A protein expression in glioma cultures treated with Delta-39 and Delta-24/39 was approximately 40% and 75% lower, respectively, than in cultures treated with Delta-24 (Figure 4A). In NHA cells infected with Delta-39 and Delta-24/39, the level of expression of E1A protein was similarly decreased (Figure 4A).
Figure 4.
Expression and stability of E1A proteins. (A) Expression of E1A proteins in glioma cells and normal astrocytes. Immunoblotting analysis of the expression of E1A protein in U-251 MG and NHA (left panel). Cells were collected 16 hours after being infected with the indicated viruses at 50 MOI. Proteins in the cell lysates were separated by 10% SDS-PAGE and immunoblotted with anti-E1A and anti-β-actin antibodies. The E1A signals were quantified by densitometry followed by normalization to β-actin levels (right panel). Data are represented as relative to E1A expression level displayed by Delta-24 in U-251 MG cells (equal to 100%). (B) Stability of E1A proteins. U-251 MG cells were infected with the indicated adenoviruses and then treated with the protein synthesis inhibitor, anisomycin. Cell lysates were collected at the indicated time points after anisomycin treatment, separated by 10% SDS-PAGE, and immunoblotted with the anti-E1A antibody (left panel). Quantification of the E1A level was performed by densitometry analysis. Protein levels were represented as relative to E1A protein levels present at 0 hour of the addition of anisomycin (equal to 100%). For clarity, data are presented as a linear regression graph (right panel). (C) Proteasomal-mediated degradation of E1A. U-251 MG cells were infected with the indicated adenoviral constructs and then treated with lactacystein. Proteins were immunoblotted for E1A protein (left panel). Protein levels were quantified by densitometry analysis. Shown are the ratios of E1A expressed by lactacystein-treated cultures versus untreated cultures (right panel). L: lactacystin.
To determine whether the CR1 mutation affected the stability of E1A protein, we monitored the temporal profile of E1A protein degradation by infecting U-251 MG cells with Adwt, CR1 mutant, or CR2 mutant adenoviruses and by treating infected cells with the protein synthesis inhibitor, anysomicin. Analyses of the E1A protein levels after the addition of anisomycin showed that the stability of the CR2 mutant E1A protein was similar to the wild-type E1A protein (Figure 4B). However, the degradation of CR1 mutant proteins was drastically accelerated (Figure 4B). As expected, the stability of E1A protein was even more compromised in the double CR1–CR2 mutant E1A adenovirus (Figure 4B). These results suggest that the 39-bp deletion in the E1A CR1 region severely destabilizes the E1A protein. Because E1A is a substrate for proteasomal-mediated degradation [16], we were interested in ascertaining whether proteosomal degradation was affecting mutant E1A levels. To more precisely gauge the contribution of the proteasome to CR1 mutant E1A degradation, U-251 MG cells infected with Delta-24/39 were treated with the proteasomal inhibitor, lactacystein. The CR1 mutant E1A species were increased around eight-fold after prolonged lactacystin treatment, whereas lactacystin treatment of cells infected with Delta-24 or Adwt resulted in about two-fold increase in E1A protein (Figure 4C). Collectively, these data showed that the CR1 mutation produced a mutant form of E1A protein whose stability was compromised by enhanced proteasomal-mediated degradation.
Discussion
This is the first study to show that a mutation in the CR1 region of the E1A protein results in reduced protein stability and a low level of E1A protein expression due to proteosomal-mediated degradation. Importantly, a low level of E1A protein does not significantly attenuate the capability of CR1 mutant adenovirus to activate S-phase-associated genes and replicate in glioma cells, but critically diminishes its activity in normal astrocytes. These data indicate that the level of E1A protein could be manipulated to construct mutant adenoviruses with replicative capacity restricted to cancer cells.
After our first report on the E1A mutant oncolytic Delta-24 adenovirus, it became one of the best characterized oncolytic adenoviruses to have been developed using functional E1A deletions [3,17,18]. A modification of Delta-24, Delta-24-RGD, enhanced the anticancer effect in glioma cells in vitro and in vivo during preclinical characterization assessments [4]. Current efforts are focused on improving the therapeutic index of Delta-24 and its ability to utilize deregulated E2F activity to preferentially replicate in cancer cells [3,19,20]. Previously, mutations in the CR1 region were shown to abrogate or decrease the ability of E1A to interact with p300/CBP, as well as Rb and Rb-related proteins [5]. Moreover, the CR1 and CR2 regions are apparently required to activate viral and cellular genes through independent, redundant pathways, as only E1A mutants that fail to bind to p300 and pRb simultaneously are completely defective. Therefore, CR1 and CR2 mutations would be expected to weaken E1A-mediated disruption of the tightly regulated cell cycle checkpoints to force normal cells into S-phase. This tenet is supported by demonstrating improved replicative selectivity from a double CR1/CR2 mutant adenovirus over a CR2 mutant adenovirus in models with proliferating cells, such as organotypic keratinocyte cultures [23]. Our data are consistent with these observations and suggest that the CR1 deletion is primarily responsible for the attenuated replication pattern demonstrated by double CR1/CR2 mutant adenoviruses [23].
In addition to modification of the well-studied E1A interactions with cell proteins [5], deletions in E1A protein may have other effects. A novel finding in this study is that the CR1 deletion in the Delta-39 construct markedly reduces the stability of E1A protein. Interestingly, this observation is consistent with previous work reporting on the reduction of E1A autoactivation of an E1A mutant with a similar CR1 deletion [24]. Our data and previous data suggest that there is a stoichiometric relationship between the level of expression of E1A and its ability to modulate the activity of cellular and adenoviral proteins. If this hypothesis were true, cancer cells that already have impaired cell cycle checkpoint activity would be permissive to the replication of E1A mutant adenoviruses expressing low E1A protein levels. Low levels of E1A expression would, however, be disadvantageous in the setting of multiple, extremely redundant, and overlapping mechanisms of cell cycle control in normal cells. Although it would be difficult to pinpoint the exact contribution of a given level of E1A expression to the selective replication of CR1 mutant adenoviruses in cancer cells, the deletion in the Delta-39 construct apparently decreases E1A protein to a level lower than the hypothetically required threshold needed to activate DNA replication in normal cells. Our data showed that, together with disruptions in p300/CBP and pRb binding, a low level of E1A could further detriment the ability of E1A to properly inactivate cell cycle regulators in normal cells; none of these two conditions, however, modified the ability of the mutant adenovirus to replicate in cancer cells. Our results indicate that further in vivo analyses need to be performed to determine the impact of our mechanistic studies on the full translation of Delta-39 to the clinical setting.
In summary, we report for the first time that a mutation in the E1A gene results in low levels of E1A protein, conditioning the replication of the mutant adenoviruses specifically to cancer cells. Thus, Delta-39 has the potential to be developed further into an efficacious antiglioma therapeutic tool.
Acknowledgements
We greatly appreciate the assistance of Joann Aaron (Scientific Editor, Department of Neuro-Oncology, M. D. Anderson Cancer Center) in the edition and preparation of the manuscript.
Abbreviations
- CR
conserved regions
- CPE
cytopathic effect
- MOI
multiplicity of infection
- PFU
plaque-forming units
Footnotes
The work was supported by National Institutes of Health grant RO1CA90879 (J.F.) and M.D. Anderson Cancer Center Institutional Research grant 43721231 (H.J.).
References
- 1.Chiocca EA. Oncolytic viruses. Nat Rev Cancer. 2002;2:938–950. doi: 10.1038/nrc948. [DOI] [PubMed] [Google Scholar]
- 2.Jiang H, Conrad C, Fueyo J, Gomez-Manzano C, Liu TJ. Oncolytic adenoviruses for malignant glioma therapy. Front Biosci. 2003;8:d558–d577. doi: 10.2741/923. [DOI] [PubMed] [Google Scholar]
- 3.Fueyo J, Gomez-Manzano C, Alemany R, Lee PS, McDonnell TJ, Mitlianga P, Shi YX, Levin VA, Yung WK, Kyritsis AP. A mutant oncolytic adenovirus targeting the Rb pathway produces antiglioma effect in vivo. Oncogene. 2000;19:2–12. doi: 10.1038/sj.onc.1203251. [DOI] [PubMed] [Google Scholar]
- 4.Fueyo J, Alemany R, Gomez-Manzano C, Fuller GN, Khan A, Conrad CA, Liu TJ, Jiang H, Lemoine MG, Suzuki K, et al. Preclinical characterization of the antiglioma activity of a tropism-enhanced adenovirus targeted to the retinoblastoma pathway. J Natl Cancer Inst. 2003;95:652–660. doi: 10.1093/jnci/95.9.652. [DOI] [PubMed] [Google Scholar]
- 5.Frisch SM, Mymryk JS. Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 6.Avvakumov N, Wheeler R, D'Halluin JC, Mymryk JS. Comparative sequence analysis of the largest E1A proteins of human and simian adenoviruses. J Virol. 2002;76:7968–7975. doi: 10.1128/JVI.76.16.7968-7975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shenk T. Adenoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Virology. Philadelphia, PA: Lippincott-Raven Publishers; 1996. pp. 2111–2148. [Google Scholar]
- 8.Graham FL, Prevec L. Manipulation of adenovirus vectors. Methods Mol Biol. 1991;7:109–128. doi: 10.1385/0-89603-178-0:109. [DOI] [PubMed] [Google Scholar]
- 9.Jones N, Shenk T. Isolation of deletion and substitution mutants of adenovirus type 5. Cell. 1978;13:181–188. doi: 10.1016/0092-8674(78)90148-4. [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Manzano, Balague C, Alemany R, Lemoine MG, Mitlianga P, Jiang H, Khan A, Alonso M, Lang FF, Conrad CA, et al. A novel E1A–E1B mutant adenovirus induces glioma regression in vivo. Oncogene. 2004;23:1821–1828. doi: 10.1038/sj.onc.1207321. [DOI] [PubMed] [Google Scholar]
- 11.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 12.Egan C, Jelsma TN, Howe JA, Bayley ST, Ferguson B, Branton PE. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol Cell Biol. 1988;8:3955–3959. doi: 10.1128/mcb.8.9.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fueyo J, Gomez-Manzano C, Yung WK, Clayman GL, Liu TJ, Bruner J, Levin VA, Kyritsis AP. Adenovirus-mediated p16/CDKN2 gene transfer induces growth arrest and modifies the transformed phenotype of glioma cells. Oncogene. 1996;12:103–110. [PubMed] [Google Scholar]
- 14.Fattaey AR, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda MA, Nevins JR. Identification of distinct roles for separate E1A domains in disruption of E2F complexes. Mol Cell Biol. 1993;13:7029–7035. doi: 10.1128/mcb.13.11.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnell AS, Grand RJ, Gorbea C, Zhang X, Wang W, Mymryk JS, Gallimore PH. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 2000;19:4759–4773. doi: 10.1093/emboj/19.17.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, Ng L, Nye JA, Sampson-Johannes A, Fattaey A, et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 18.Heise C, Hermiston T, Johnson L, Brooks G, Sampson-Johannes A, Williams A, Hawkins L, Kirn D. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med. 2000;6:1134–1139. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- 19.Whyte P, Ruley HE, Harlow E. Two regions of the adenovirus early region 1A proteins are required for transformation. J Virol. 1988;62:257–265. doi: 10.1128/jvi.62.1.257-265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whyte P, Williamson NM, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 21.Lee BH, Liu M, Mathews MB. Regulation of the human proliferating cell nuclear antigen promoter by the adenovirus E1A-associated protein p107. J Virol. 1998;72:1138–1145. doi: 10.1128/jvi.72.2.1138-1145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mymryk JS, Bayley ST. Induction of gene expression by exon 2 of the major E1A proteins of adenovirus type 5. J Virol. 1993;67:6922–6928. doi: 10.1128/jvi.67.12.6922-6928.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balague C, Noya F, Alemany R, Chow LT, Curiel DT. Human papillomavirus E6E7-mediated adenovirus cell killing: selectivity of mutant adenovirus replication in organotypic cultures of human keratinocytes. J Virol. 2001;75:7602–7611. doi: 10.1128/JVI.75.16.7602-7611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida K, Higashino F, Fujinaga K. Transcriptional regulation of the adenovirus E1A gene. Curr Top Microbiol Immunol. 1995;199:113–130. doi: 10.1007/978-3-642-79586-2_6. [DOI] [PubMed] [Google Scholar]