Abstract
DNA sequences (IES elements) eliminated from the developing macronucleus in the ciliate Tetrahymena thermophila are released as linear fragments, which have now been detected and isolated. A PCR-mediated examination of fragment end structures reveals three types of strand scission events, reflecting three steps in the deletion process. New evidence is provided for two steps proposed previously: an initiating double-stranded cleavage, and strand transfer to create a branched deletion intermediate. The fragment ends provide evidence for a previously uncharacterized third step: the branched DNA strand is cleaved at one of several defined sites located within 15–16 nucleotides of the IES boundary, liberating the deleted DNA in a linear form.
Keywords: ciliated protozoa/developmentally programmed genome rearrangements/DNA cleavage/DNA recombination/IES element
Introduction
Developmentally programmed genomic rearrangements have been observed in many organisms. Rearrangements of immunoglobulin genes in vertebrates are one of the best-known examples (Roth and Craig, 1998). Ciliated protozoans also provide some noteworthy examples.
Ciliates have two functionally different nuclei. The polyploid macronucleus is transcriptionally active and is responsible for all gene expression in the cell. The diploid micronucleus is transcriptionally inactive and is an analog of an inactive germ cell nucleus. During sexual reproduction, the existing macronucleus is destroyed and a new one is developed from a copy of the zygotic nucleus (Orias, 1986). Macronuclear development is accompanied by some of the most extensive programmed genomic rearrangements observed in any class of organism. These include fragmentation of micronuclear chromosomes (Yao, 1989). The smaller fragments are then converted into small chromosomes by the addition of telomeres (Blackburn, 1992), and amplified. In addition, a significant fraction of the genome is eliminated in a series of site-specific deletions. The deleted sequences are often referred to as internal eliminated sequences, or IES elements. In Tetrahymena thermophila, ∼15% of the genome is eliminated in the form of ∼6000 individual deletion events (Yao et al., 1984). The deleted DNA consists of moderately repetitive and unique sequences, which range in size from several hundred base pairs to >10 kbp. The chromosomal DNA on either side of the deleted segment comes together to form a macronuclear junction in the mature macronucleus, and the deleted DNA is eventually degraded.
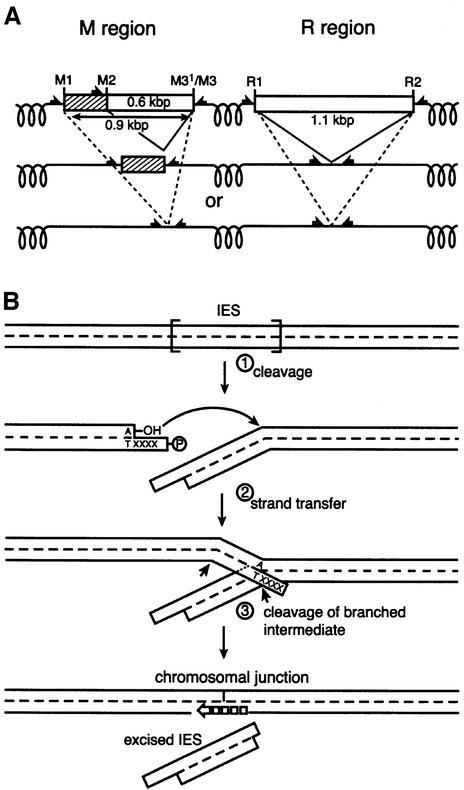
In Tetrahymena, the M and R regions provide the most extensively studied examples of this process (Figure 1A) (Austerberry and Yao, 1987, 1988). The R region is a 1.1 kbp IES that undergoes programmed deletion. In the M region, two deletions of 0.6 and 0.9 kbp occur, both employing the same right boundary. The deletions that occur in these regions are not always identical, but instead display a modest amount of microheterogeneity in their endpoints (Austerberry et al., 1989; Saveliev and Cox, 1994). Deletions in the M region are directed by a cis-acting polypurine tract (A5G5) found 41–54 bp distal to each boundary (Godisca and Yao, 1990, 1993). Cis-acting sequences have also been found in the sequences flanking the R element (Chalker et al., 1999). These direct R region deletion in a similar way, although the identity of the sequences has been less well defined.
Fig. 1. Programmed genomic deletions in T.thermophila. (A) DNA deletions in the M and R regions. Deletions are directed by cis-acting signals located 30–50 bp outside boundaries of the deleted sequences (indicated by black half-arrows). In the M region, there are separate cis-acting signals located at left ends of the 0.6 or 0.9 kbp IES deletions. (B) A model for programmed genomic deletions. (1) The reaction is initiated by a single double-stranded cleavage at one boundary of the IES, generating DNA ends with a 4 nt 5′ extension. There is generally an adenosine residue at the 3′ chromosomal terminus. (2) The 3′ hydroxyl of the adenosine residue serves as the nucleophile in a strand transfer step at a phosphodiester bond at the opposite boundary. This step creates the macronuclear junction on one DNA strand. (3) The other strand must then be cleaved.
The biological significance of DNA elimination in ciliates is unclear. At least some of the eliminated sequences are transposon-like elements (Cherry and Blackburn, 1985; Jaraczewski and Jahn, 1993; Klobutcher et al., 1993; Klobutcher and Herrick, 1995, 1997; Doak et al., 1997; Patil and Karrer, 2000). DNA elimination would lead to removal of the transposons. An alternative idea is that DNA eliminates centromeric sequences (Yao, 1996). During vegetative growth, the macronucleus divides amitotically, and centromeres within the polyploid macronucleus might impose problems with chromosome segregation. These ideas are not mutually exclusive.
Information about the mechanism of programmed genomic deletions in ciliates is limited. Abundant circular forms of eliminated sequences were detected in Euplotes crassus, Oxytricha trifallax and Paramecium tetraurelia (Jaraczewski and Jahn, 1993; Klobutcher et al., 1993; Williams et al., 1993; Betermier et al., 2000). In each organism, the circle junction has a distinct structure, which permits informed speculation about the deletion mechanism. For the deletions in E.crassus, double-strand cleavage at both ends, followed by annealing and repair of the end, provided a good explanation for the observed junctions (Jaraczewski and Jahn, 1993; Klobutcher et al., 1993). A transposition-like mechanism was proposed for the excision of TBE1 element in O.trifallax, initiated with a double-strand cleavage at one end of the element only (Williams et al., 1993). Deletion of eliminated sequences in P.tetraurelia is likely to involve double-strand cuts at both element boundaries (Betermier et al., 2000).
Circular forms of IES elements can be detected in T.thermophila (Saveliev and Cox, 1994; Yao and Yao, 1994), but are so rare that they have been attributed to unusual by-products of the deletion reaction. This suggests that Tetrahymena IES elements are generally deleted in a linear form. An attempt to detect linear forms of the deleted DNA by Southern blot hybridization proved unsuccessful (Austerberry et al., 1984), suggesting that the excised fragments are quickly degraded.
In earlier studies, we detected double-stranded cleavage sites associated with programmed deletions in the M and R regions (Saveliev and Cox, 1995, 1996). The cleavage sites were detected in total nuclear DNA, isolated from conjugating ciliate cells during the period of programmed deletions. The structure of the ends generated by cleavage was identical in the M and R regions: open double-stranded DNA ends with 4 nucleotide (nt) 5′ extensions. An adenosine residue was present at a majority of the recessed 3′ termini.
Double-strand cleavage sites were detected at all the boundaries of the M and R region deletions, except for one special case. However, the data did not support a cleavage–ligation mechanism in which double-stranded cuts are made at both boundaries of the deleted fragment, followed by ligation of the chromosomal ends. Because the 4 nt extensions are, as a rule, heterologous at the different ends of an individual M or R region IES element, two types of chromosomal junctions would be generated in the ligation step, one type reflecting the sequence of the extensions present at either end. However, at least in some deletion events, we detected only one junction sequence. In addition, at one identified boundary of the 0.6 kbp element in the M region, called M3′, we found only a single 3′ hydroxyl terminus exposed at the end of the deleted DNA, with no corresponding cleavage site in the complementary strand. The data led us to propose a pathway for deletion in which double-stranded cleavage occurs at one boundary only, producing an end with a 4 nt 5′ extension (Saveliev and Cox, 1996). The 3′ hydroxyl terminus generated at the chromosomal (macronuclear retained) end serves as a nucleophile in a strand transfer step, attacking a phosphodiester bond at the opposite boundary. This creates a chromosomal junction on one strand as well as the 3′ hydroxyl terminus at the end of the IES element (such as that observed at M3′). It also leaves the IES element as a branch connected to the chromosomal DNA via the other strand. This pathway is outlined in Figure 1B.
Additional work to test this proposed pathway is needed. The total nuclear DNA used in the earlier studies contained a significant background of non-specific breaks. Double-stranded cleavage at the M3′ boundary might, in principle, elude detection. In addition, the model predicted that the non-spliced strand would be cleaved to promote a final release of the eliminated DNA from the chromosome. However, our examination of total nuclear DNA did not provide any information about these putative late steps in the deletion process.
In the present study, our focus shifts from total nuclear DNA to the deleted DNA fragments themselves. Reasoning that the deleted fragment ends should provide some clues as to the nature of the final steps in deletion, we applied gel purification along with sensitive anchor-based PCR techniques to successfully detect linear forms of DNA fragments deleted in the M and R regions. We found that the M and R elements are excised in double-stranded, linear forms. Analysis of these deleted DNA segments provided further evidence consistent with the pathway of Figure 1B, and also provided new information about the late steps in the pathway.
Results
M and R region IES elements are excised in a linear form
Because of the reported failure of Southern blot hybridization to detect linear deleted DNA fragments from the M and R regions (Austerberry et al., 1984), we applied PCR-based techniques to provide greater sensitivity. Total nuclear DNA, isolated from conjugating cells 15 h after conjugation commenced, was loaded onto an agarose gel. The 15 h time point corresponds to the period of programmed genomic deletions in conjugating Tetra hymena cells. After electrophoresis was complete, we cut the agarose gel lane into pieces containing DNA fragments of different sizes. We then extracted DNA from the gel pieces and searched for the deleted DNA using PCR. Note that by the mechanism of Figure 1B, three kinds of fragment endpoints might be expected. First, an endpoint might be generated by the staggered double-strand cleavage that is proposed to initiate the deletion process (step 1, Figure 1B). One end of a deleted fragment should thus have the resulting 4 nt 5′ extension structure. At the other end of the deleted fragment, the 3′ end is generated in the strand transfer step (step 2), and the 5′ end must be generated in subsequent cleavage steps to eliminate the branched intermediate from the chromosome (step 3). Since the deletion can be initiated at either end of the deleted DNA, both ends of the deleted fragments may exhibit evidence of all three deletion steps in different members of the DNA fragment population.
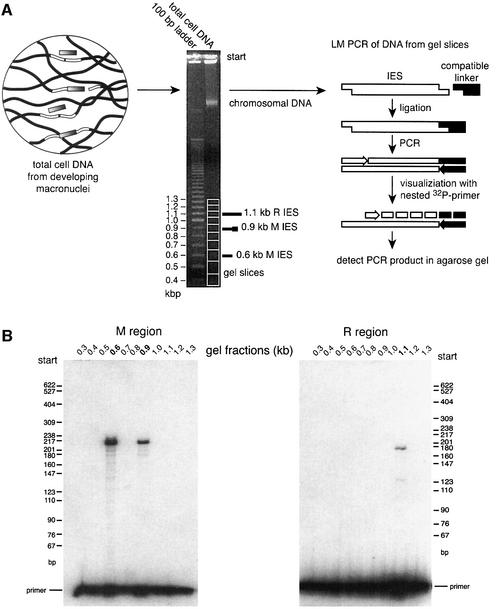
We used ligation-mediated (LM)-PCR to detect linear forms of the eliminated sequences. The initiating double-strand cleavage events, creating open DNA ends with 4 nt 5′ extensions, were characterized in earlier studies (Saveliev and Cox, 1996). Designed compatible linkers will ligate at the end of the linear fragment if the fragment is generated by means of the expected initiation cleavage event. The ligated end can be detected by LM-PCR with a high degree of specificity, as shown in Figure 2A.
Fig. 2. Purification and detection of linear forms of IES elements excised from the M and R regions. (A) Technique to purify a linear IES. After electrophoresis, DNA is isolated from gel slices corresponding to fragments of different sizes. The 100 bp ladder of markers was required to follow the migration of linear forms of IESs, which are invisible because of their low abundance. (B) Linear forms of IES elements excised from the M region were detected in gel fractions corresponding to DNAs of 600 and 900 ± 50 bp in length, Linear IES elements from the R region were detected only in fractions containing DNA of ∼1.1 kbp.
We detected M region-containing linear fragments in gel fractions corresponding to 0.6 and 0.9 kbp DNA fragments, as well as R region-containing linear fragments in fractions containing 1.1 kbp fragments (Figure 2B). The size of the fragments corresponded to the length of sequences deleted from the M and R regions (see Figure 1A). This suggested that the deleted DNA is excised as a linear molecule, and that deletion in this form is a characteristic of at least two Tetrahymena IES elements. We expected that the deleted DNA fragments might undergo a gradual degradation. However, we were unable to detect M and R eliminated sequences in gel fractions containing shorter DNA fragments, even with a very sensitive PCR procedure using nested primers (data not shown). The low concentration of the deleted DNA fragments in the genomic DNA preparations is thus not readily explained by gradual degradation. Rather, these DNA fragments appear to exist in an intact form for some period of time prior to rapid digestion to nucleotides.
Two 3′ termini and one 4 nt staggered double-stranded cleavage are generated at the right end of the 0.6 kbp M linear fragment
The fact that the deleted sequences could be detected in an intact linear form gave us a unique opportunity to study the late steps of the DNA deletion reaction. These steps must logically be reflected in the structure of the ends of the linear fragments. If deletion involves two double-stranded cleavage events, followed by ligation, the linear product fragments will all have two open double-stranded ends, both with 4 nt 5′ extensions. If the mechanism follows the model outlined in the Introduction, we would find a different pattern of end structures. The fractions containing the linear fragments appear to be essentially free of randomly broken chromosomal fragments, which would otherwise produce a background of non-specific signals in our experiments. Fragments generated by random breakage in the chromosomal M and R regions are generally much longer than the linear IES deletion fragments, and they reside in a fraction of chromosomal DNA near the top of the gel.
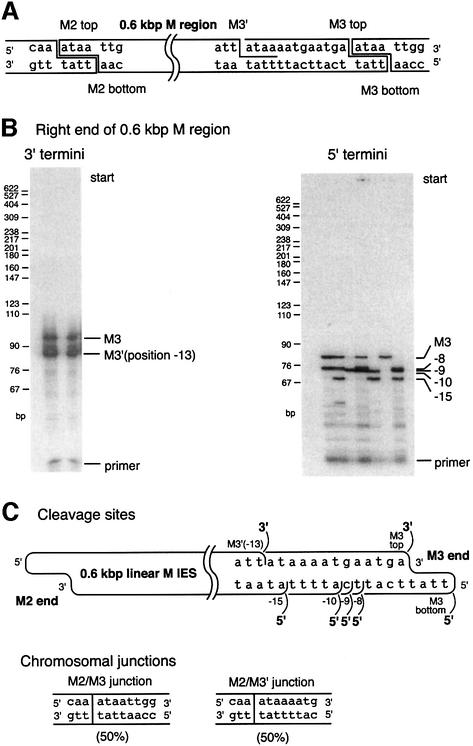
The 0.6 kbp deletion in the M region was a primary focus of our study. The deletion generates two chromosomal junctions, occurring at equal frequency in Tetrahymena (Saveliev and Cox, 1994). The deletion events share the same left boundary (M2), whereas the right boundaries (M3′ and M3) are shifted by 13 bp relative to each other. For the deletions bounded by M2 and M3, deletion initiation can occur at either site and thus the 4 nt staggered cleavage events can be detected at both M2 and M3 (Saveliev and Cox, 1996; Figure 3A). However, the deletions bounded by M2 and M3′ appear to be initiated only at M2, as discussed above. No double-strand cleavage has been detected at M3′. Using the more sensitive techniques in the current experiments, we wished to investigate the possibility that a double-stranded cleavage at the M3′ site eludes detection because of the background of random double-stranded breaks in total nuclear DNA. If we could confirm the absence of a double-stranded cleavage at this junction, it would tend to support the original model. It would also provide an excellent opportunity to examine the fate of the other DNA strand at the ‘receiving’ end of the strand transfer step (step 2 in Figure 1B) at a location, M3′, where no deletion initiations occurred to interfere with the analysis.
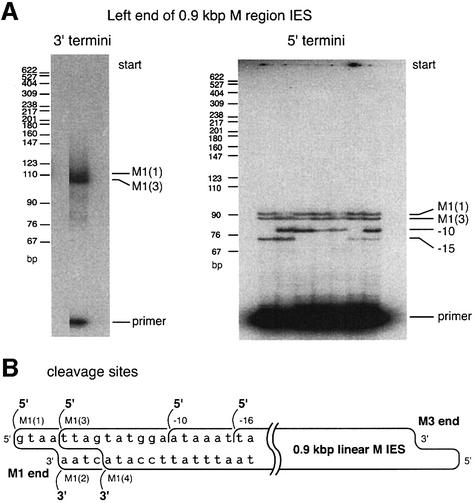
Fig. 3. DNA end structure at the M3 end of the isolated 0.6 kbp linear M IES elements. (A) The structure of DNA cleavage events detected for the 0.6 kbp M region IES in an earlier study (Saveliev and Cox, 1994). A double-stranded cleavage is detected at the M3 site, while the M3′ site features a cleavage detectable in only the upper strand. (B) Characterization of the 3′ and 5′ termini at the M3 end of the isolated 0.6 kbp linear IES elements. For the 3′ termini, two separate trials are shown. For the 5′ termini, seven separate trials are shown. In total, two 3′ and five 5′ termini were detected. The origin of a third band generated during study of 3′ termini (seen just above M3′ product) is unclear. It does not generate any PCR product during re-amplification for sequencing purposes. Most likely, it is a non-specific PCR product. (C) Map of all termini detected in this and previous work. All the termini shown in (B) were sequenced to pinpoint their exact positions. The 3′ and 5′ termini at the far right are generated during an initiating double-stranded cleavage. All other termini appear to represent single-strand breaks. The known chromosomal junctions show that macronucleus-retained flanking sequences join at positions (called macronuclear or chromosomal junctions) corresponding to the 3′ termini, but not the 5′ termini.
First, the locations of the 3′ termini in the upper strand were confirmed. By the mechanism of Figure 1B, all 3′ termini should correspond to known macronuclear junctions. Terminal transferase-based PCR was applied to detect 3′ termini at the right end of the 0.6 kbp M linear fragments. PCR generated two products present at equal levels (Figure 3B), indicating the presence of two classes of fragments with different ends. Sequences of the PCR products confirmed that the 3′ termini were generated at the M3 and M3′ positions (Figure 3C), consistent with earlier work (Saveliev and Cox, 1996).
If double-stranded cleavage occurs at both M3 and M3′, the flanking sequences are such that both should generate DNA ends with identical 5′ extensions (see sequences in Figure 3A). However, we detected only one double-stranded cleavage at this end (Figure 2B, 0.6 kbp fraction). The sequence of the corresponding PCR product showed that the cleavage occurs at the M3 site. Thus, the 3′ terminus on the top strand at the M3′ site is not accompanied by a cognate staggered cleavage on the lower strand detectable even with the more sensitive approaches used here.
Several 5′ termini are scattered within a 15 nt range at the right end of the 0.6 kbp M linear fragment
Although no 4 nt staggered double-stranded cleavage occurs at the M3′ site, strand transfer to join the chromosomal DNA ends (step 2, Figure 1B) does occur here to generate a chromosomal junction on the upper strand to produce the 3′ end seen in Figure 3B. Cleavage of the bottom strand must logically occur somewhere near this site during M2/M3′ deletion to release the 0.6 kbp DNA fragment. Such cleavage steps will generate 5′ termini at the right end of the 0.6 kbp M linear fragment, on the strand complementary to the one where the strand transfer occurred. To detect these termini, we applied yet another PCR protocol, a modified version of LM-PCR described earlier (Saveliev and Cox, 1995). As usual, any observed PCR product was sequenced multiple times to map the precise position of ends.
Somewhat surprisingly, we detected five distinct 5′ termini at the end of the linear fragment (Figure 3B). The termini were highly reproducible, with all of them or subsets found in nine separate trials. The distal 5′ terminus is coincident with the site of the expected 4 nt staggered position for double-strand cleavage to initiate deletion at the M3 site (Figure 3C). The other four termini were never detected in previous studies. They exhibit two important features. First, in contrast to the cleavage sites generating the 3′ ends on the opposite strand, none of the 5′ ends map to a point corresponding to the known macronuclear boundary of an eliminated sequence. Secondly, none of them occurs at a position that is staggered by 4 nt relative to the position of a known cleavage site on the complementary strand.
The data support the idea that the M2/M3′ junction is generated when chromosomal DNA boundaries are joined by strand transfer on the upper strand. This reaction liberates a free 3′ terminus at the M3′ end of the deleted IES element on the upper strand. The intact complementary strand is then cleaved at one of four optional positions in close proximity to the junction, finally liberating the M3′ end from the chromosomal DNA.
Fragment end structure is similar at the right ends of the 0.6 and 0.9 kbp M linear fragments
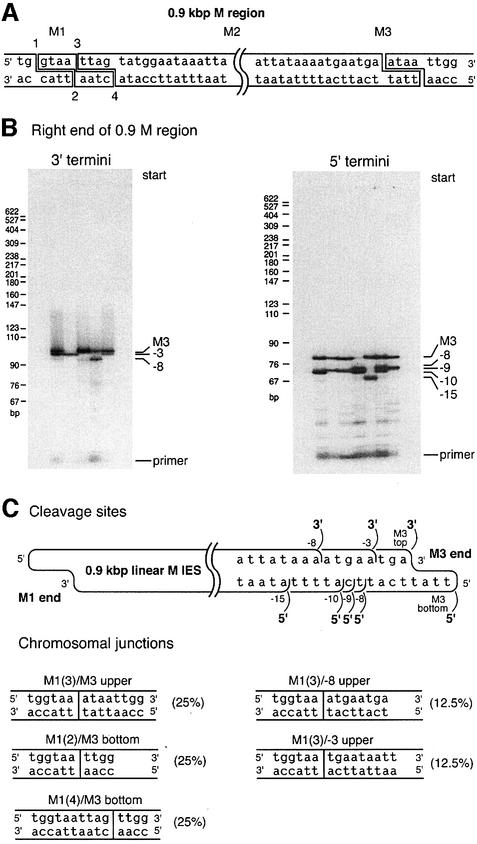
To determine whether the pattern of end structures documented in Figure 3 is applicable to other deleted IES elements in Tetrahymena, we switched our attention from the 0.6 kbp IES deletion in the M region to the 0.9 kbp M region deletion. We thus examined the right ends of the DNA fragments extracted from the gel section corresponding to 0.9 kbp linear DNAs.
We first characterized the 3′ termini at the right end, which again should correspond to known chromosomal junctions. In total nuclear DNA, we previously detected only one 3′ terminus at the right end (Saveliev and Cox, 1996; Figure 4A). In the present study, using the gel-fractionated DNA fragments, we unexpectedly detected three 3′ termini at the end of 0.9 kbp M linear IES (Figure 4B). The outermost 3′ terminus (the one furthest to the right as shown) was the one detected in previous work (Saveliev and Cox, 1996). The two additional 3′ termini were found at the –8 and –3 positions (Figure 4C).
Fig. 4. DNA ends at the M3 end of the 0.9 kbp linear M IES. (A) The structure of DNA cleavage events detected for the 0.9 kbp M region IES in an earlier study (Saveliev and Cox, 1994). Double-stranded cleavage at the left boundary of the 0.9 kbp M region can occur at either of two sites shifted by 4 bp relative to each other. Joining of macronucleus-retained DNA ends at these sites generates the three most common chromosomal junctions (Saveliev and Cox, 1994). (B) Characterization of the 3′ and 5′ termini at the M3 end of the isolated 0.9 kbp linear IES element. In total, three 3′ and five 5′ termini were detected. Each lane of each gel corresponds to an individual trial. (C) Map of the DNA termini and macronuclear junctions. All the termini shown in (B) were sequenced to pinpoint their exact positions. As in the 0.6 kbp M region, the distal 3′ and 5′ termini at the M3 end of the 0.9 kbp M region are generated during initiating double-stranded cleavage, whereas all other termini appear to be single-stranded breaks. Two new chromosomal junctions generated during the 0.9 kbp deletion in the M region are also shown. Junctions with the ‘upper’ designation were generated by an initiating DNA cleavage on the left side of the element, followed by strand transfer on the upper strand as drawn. Those with the ‘bottom’ designation underwent an initiating cleavage on the right side of the element, with strand transfer involving the bottom strand. The absence of an M1(1)M3upper junction has been tentatively attributed to the unique presence of a 3′ terminal G residue at M1, which may not function in strand transfer (Saveliev and Cox, 1996). As for the termini observed in Figure 3, each of the detected 3′ termini corresponds to defined macronuclear junctions, whereas some of the detected 5′ termini do not.
We characterized the new ends further. Additional 3′ ends in the vicinity of M3 could be generated in several different ways. The simplest explanation was that these are additional strand transfer sites, and that there are additional deletion endpoints in the 0.9 kbp M region that were not identified earlier. The new fragment endpoints could be introduced into the 0.9 kbp deleted M region sequence during these uncharacterized deletions.
We therefore went back to total nuclear DNA (without gel fractionation) and amplified chromosomal junctions generated by the 0.9 kbp deletion. PCR products were precisely resolved in 4% polyacrylamide gel, and sequenced. We found that the 0.9 kbp deletion actually creates five different chromosomal junctions rather than the three detected previously (Figure 4C). The three major junctions, each accounting for ∼25% of all junctions, were those described previously and include one that corresponds to the known 3′ end in Figure 5 (Austerberry et al., 1989; Saveliev and Cox, 1994). Two new minor junctions, each accounting for ∼12% of all junctions, were also present [Figure 4C, junctions M1(3)/–8 and M1(3)/–3] that apparently eluded detection in the earlier studies.
Fig. 5. Characterization of DNA ends at the M1 end of the 0.9 kbp linear M IES. (A) As determined by DNA sequencing, the slightly smeared single band represents two 3′ termini with ends only 4 bp apart. Four 5′ termini were detected and easily resolved in the gel. (B) Map of the DNA ends. All the termini were sequenced and placed on the map. Two new 5′ termini detected in this study (–10 and –16) appear to represent single-strand breaks. Each gel lane represents an individual trial.
Thus, after comparison of chromosomal junctions found in mature macronuclei with the 3′ termini detected at the right end of the 0.9 kbp linear fragment during macronuclear development, we found that all the 3′ termini identified map to demonstrable boundaries generated in the normal deletion process. The M1 (left) chromosomal end joins with the right boundary of the deleted DNA at positions –8 and –3 on the upper strand to create junctions M1(3)/–8 and M1(3)/–3, respectively.
We then turned our attention to 5′ termini on the complementary strand at the same right end of the 0.9 kbp M region IES. This is where the final cleavage events needed to liberate the linear fragment from the chromosomal DNA should be found. Five 5′ termini were detected (Figure 4B). The pattern of the breaks was identical to that of breaks on the same strand at the right end of the 0.6 kbp fragment (compare Figures 3C and 4C), with 5′ termini found at defined positions within a 15 nt range. As in the 0.6 kbp linear fragment, none of the 5′ termini, except for the outermost one, maps to the observed chromosomal boundary of an M region deletion.
Both ends of the 0.9 kbp M linear fragments exhibit similar end structures
Our analysis of the structure of the right ends of the 0.6 and 0.9 kbp linear fragments showed that each fragment is actually a defined mixture of several fragments that have nearly the same size, but slightly different end structures. Within the population of fragments, the observed right ends reflect all three steps of the deletion reaction. In principle, the left end of these same fragments should exhibit a very similar pattern. The 0.9 kbp M element generates the highest variety of chromosomal junctions. Therefore, study of this element should give us the most comprehensive information about the deletion reaction.
On the bottom strand at the left end of the isolated 0.9 kbp linear fragments, the data show that the 3′ termini are identical to those detected earlier in total nuclear DNA (Saveliev and Cox, 1996; Figure 5A and B). These termini should be generated either by initiating double-strand cleavage events or by strand transfer, and both 3′ termini map precisely to previously identified chromosomal junctions left behind after deletion. A different result was obtained in the study of the 5′ termini on the upper strand. We found four 5′ termini at the left end of the fragment (Figure 5A). Two of the termini correspond to double-strand-initiating cleavage events as well as to previously characterized deletion boundaries (Figure 5B). However, the other two termini were not detected in earlier studies. In both cases, there are no cognate cleavage points staggered by 4 nt on the other strand and neither of these two 5′ ends maps to a point corresponding to the known macronuclear boundary of an eliminated sequence. The 5′ termini are all found within a 16 bp region at the end of the IES element, similar to the 15 nt range observed at the opposite end. Thus, the observed pattern of cleavage sites is similar at both ends of the deleted 0.9 kbp linear fragments.
M region IES excision is initiated only with double-strand breaks
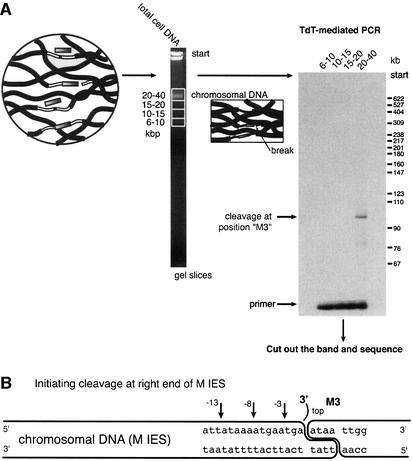
While hypothesizing that the new 5′ endpoints illustrated in Figures 3–5 reflect cleavage events in the last stages of the deletion process, we wished to explore further the possibility that they could appear at earlier stages of the reaction. We therefore examined gel fractions with much larger chromosomal DNA fragments (average size >20 kbp). Any strand scissions reproducibly seen in this DNA at or near the ends of the M region could be associated with early stages of the deletion reaction, before the linear fragments are released from the chromosome.
We used terminal transferase-based PCR to detect 3′ termini generated by initiating cleavage events. The right end of the M element was analyzed, and a DNA cleavage site was identified (Figure 6A). The signal is weaker than that generated by the right ends of the isolated 0.6 or 0.9 kbp linear fragments. We reasoned that the DNA break at this site was transient. Sequencing of the PCR products showed that the initiating cleavage occurs at the M3 position only (Figure 6B). This position corresponds to a previously characterized double-stranded cleavage at the boundary. No 3′ termini reflecting DNA strand breaks were detected at the positions corresponding to the M3′, –8 or –3 positions in 10 subsequent trials, even when the X-ray films were heavily overexposed. In general, the data are consistent with a model in which the new termini are generated in the late stages of the IES deletion process.
Fig. 6. Initiating cleavage events at M3 in chromosomal DNA. (A) Protocol illustration. Chromosomal DNA was purified in 1.5% agarose gel to remove linear IES elements. The 3′ termini generated by an initiating cleavage at the M3 end were detected by TdT-mediated PCR. Only one 3′ terminus was detected, and this only in the fraction containing the chromosomal DNA fragments (DNA fragment sizes in kbp are indicated at the top of each gel lane). (B) Map of the initiating cleavage with other 3′ ends also shown. The PCR product shown in (A) was sequenced multiple times and shown to coincide with the known double-stranded cleavage at the M3 boundary. The positions of detected single-strand breaks (–3, –8, –13) are indicated to stress that an initiating cleavage occurs only at M3.
Discussion
During programmed genomic deletions of the M region in T.thermophila, deleted DNA sequences are released in a linear, double-stranded form. The linear fragments do not show any evident signs of degradation. The fragments are generated by excision between the known boundaries of the M region deletions and they do not contain any extra sequences derived from the flanking chromosomal DNA. They are, however, not abundant. A very sensitive technique like PCR is required to detect the linear fragments, suggesting that they are short-lived. The ends of these fragments provide information about the final stages of the deletion process.
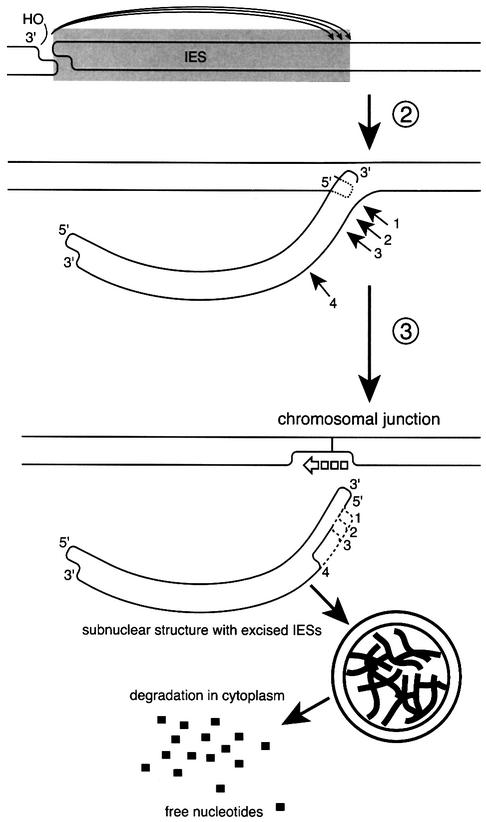
Figure 7 presents some details of the final steps of the proposed pathway of programmed genomic deletions in Tetrahymena, and explains the three types of DNA ends observed in this and previous studies. The first type is an end that is part of a double-stranded cleavage staggered to produce open DNA ends with 4 nt 5′ extensions. We have associated these with the initiating event in the deletion process (Saveliev and Cox, 1996).
Fig. 7. A model for the late stages of programmed deletions in the M and R regions of T.thermophila. Circled step numbers correspond to those in Figure 1. The strand transfer step shown in Figure 1B creates the macronuclear junction on one strand, and also creates a new 3′ terminus on the displaced strand of the IES element. The IES element is still joined as a branch of the other strand. The branched DNA strand is then cleaved at any of several defined sites located within 15–16 nt of the boundary. This cleavage (step 3) would remove the IES element, leaving short single-strand branches (which can not anneal with the sequences on the chromosomal junction strand) in the chromosomal DNA. Subsequently, these branches would have to be removed by nucleases (a step we have no evidence for), and the resulting gap filled in by DNA polymerase. As a result of the various cleavage events, one end of the excised IES has a 4 nt 5′ extension, whereas the opposite end has a 3′ extension of up to 12 nt. The excised IES may be quickly packaged into subnuclear structures together with thousands of other deleted DNA fragments to avoid their re-integration into chromosomal DNA. These could be transported outside of the developing macronucleus and digested to nucleotides.
The second type is a 3′ terminus on the deleted fragment, generated during the postulated strand transfer step (step 2, Figure 1B). The 3′ terminus observed at M3′ in the deletion of 0.6 kbp fragments in the M region appears to provide an unambiguous example of this type of DNA end. Multiple attempts to find a cognate staggered cleavage on the complementary strand at M3′ in chromosomal DNA have been unsuccessful. In addition, two new 3′ ends of this same type were identified in the present study. These are positioned at –3 and –8 near M3, and are observed only during the 0.9 kbp M region deletion events. Although these positions did not correspond to chromosomal junctions seen in earlier studies (as they should according to the model), closer examination of chromosomal DNA revealed that they did indeed correspond to a minor population of chromosomal junctions for this deletion process.
The third and final end type also occurs in only one strand, but can not be detected in chromosomal DNA fractions. These are the new 5′ ends characterized in the present study and seen only in the isolated linear deletion products. Several of these are generally found within a 15–16 bp region near each boundary of the deleted DNA (with the boundary defined by the chromosomal junctions left behind after deletion). We propose that these 5′ ends are associated with the final cleavage events needed to release the deleted DNA from the chromosome, as shown in Figure 7.
Following the release of the deleted IES element, one strand of the chromosomal DNA left behind must be repaired. We do not have any information about these steps, but they presumably involve nucleolytic removal of any unpaired DNA strand ends, DNA synthesis to fill in any gaps, and ligation. Similar mechanisms are often employed in the final steps of chemically related processes such as transposition (Haren et al., 1999) and intron retrohoming (Belfort and Perlman, 1995). We note that all cleavage events detected in these studies generate 3′ hydroxyl termini and phosphorylated 5′ termini, as required by the PCR protocols used to detect them. These end structures are those most commonly used as substrates by enzymes of DNA metabolism.
In principle, the deletion event could be initiated at either end of the IES element, with strand transfer and processing occurring at the opposite end. Both ends of the element should thus exhibit a set of end structures reflecting all three steps of the deletion process. This is generally observed, with observed 5′ ends reflecting both the initiating double-strand cleavage and the new 5′ ends we associate with the processing steps being found on every set of fragment ends examined. The unique 3′ ends associated with the strand transfer step are not always evident on the fragment ends, since some strand transfer events presumably occur at sites coincident with the initiating cleavage events which may occur (in other deletion events in the population) at the same end. However, unambiguous examples can be seen at the M3′ site for 0.6 kbp deletions, and at the –3 and –8 positions near the right end of the 0.9 kbp IES near M3, as described above.
A number of other possible mechanisms for the deletion of the M and R region IES elements have been considered in the past (Saveliev and Cox, 1994, 1995, 1996; Yao and Yao, 1994; Yao, 1996). The most prominent alternative is a mechanism in which both ends of the element are cleaved, and the chromosomal ends later re-ligated. The element itself would be deleted as a linear DNA fragment, as we observe here, or the ends could be ligated to circularize the deleted DNA. Previous examination of the cleavage sites at the ends of the M and R regions revealed elements that had been cleaved at one end and not the other, and a precise single-strand break at M3′, as already described (Saveliev and Cox, 1995, 1996). This evidence was consistent with the model of Figures 1 and 7, but difficult to reconcile with a mechanism in which both ends were cleaved. Circular forms of the deleted DNA can be detected with very sensitive PCR protocols (Saveliev and Cox, 1994; Yao and Yao, 1994), but they are so rare that they are likely to be anomalous reaction by-products rather than primary reaction intermediates. The present work also addresses this alternative mechanism. Two new isolated single-strand breaks were found in the present study, as noted in the preceding paragraph (the breaks at –8 and –3 in Figure 4). Also, the population of 3′ right ends in the deleted fragments is distinct for the 0.9 and 0.6 kbp M region deletion processes (in both cases corresponding to the known macronuclear junctions that are the products of each deletion event). However, the population of 5′ ends is the same for the two deletion events on this same right side of the released fragments. Non-identity in the 3′ ends, but identity in the 5′ ends for these two deletion events, is completely consistent with the model of Figures 1 and 7. If a double-strand cleavage occurred instead on both sides of the 0.6 and 0.9 kbp IES elements, the enzyme(s) would have to cleave in different places on the top strand on the right side while selecting the same sites on the bottom strand for the two deletions. In general, we can not rigorously eliminate the alternative mechanism, but the results are much more easily reconciled by the proposal in Figures 1 and 7.
The formation of covalently closed hairpins at some or all of the cleaved DNA ends is also a mechanistic possibility. Such hairpins are intermediates in VDJ recombination and in the reactions of some transposons (Roth and Craig, 1998; Bhasin et al., 1999; Fugmann et al., 2000). These intermediates allow both strands of a duplex DNA to be cleaved within a single enzyme active site. We can not eliminate the possibility that hairpin intermediates occur in the pathway for M or R region excision (particularly in the initiating double-strand cleavage step), but currently have no evidence for them. They would not be detected by any of the PCR protocols used in this or previous studies. If they exist, they would be readily accommodated as extra steps within the proposed pathway.
Allis and colleagues (Madireddi et al., 1996) demonstrated that the IES DNA sequences in Tetrahymena were packaged into subnuclear protein structures immediately after excision, suggesting the following scenario. The linear DNA deletion fragments could be quickly packaged into such structures, protecting them from nuclease digestion until they were transported outside of the developing macronucleus. The sequences could then be quickly digested to nucleotides in the cytoplasm without the generation of detectable partially fragmented forms. This scenario would explain the apparent lack of gradual degradation of the linear forms of M and R deleted sequences as observed in this study. The very low level of circular forms of excised DNA fragments, detected in earlier studies (Saveliev and Cox, 1994; Yao and Yao, 1994), could arise by rare ligation of the ends of the linear forms.
In summary, this study expands upon the model shown in Figure 1B for the deletion of the M and R region IES elements, and adds to the evidence supporting it. The model should provide a useful basis for further work on the deletion mechanism both in vivo and in vitro.
Materials and methods
Strains and culture conditions
Tetrahymena thermophila strains were CU427.2VI and CU428.1VII, obtained from P.Bruns, Cornell University (Ithaca, NY). Cell cultivation and mating were as described previously (Bruns and Brussard, 1974). Microscopic analysis showed that mating efficiency was >90% in all trials.
Preparation of cell DNA
Conjugating cells were collected 15 h after mating was initiated. This time corresponds to the period of programmed genomic deletion in our experiments (Saveliev and Cox, 1995, 1996). Cells in 500 ml of cell culture were collected by centrifugation and lysed in 10 ml of SDS–pronase buffer [1% SDS, 0.5 M EDTA, 10 mM Tris–HCl final pH 9.5, 0.2 mg/ml pronase (Austerberry and Yao, 1987)] at 60°C for 4 h. The lysate was first dialyzed against TE (10 mM Tris pH 7.5, 1 mM EDTA) at room temperature for several hours, then dialyzed again at 4°C overnight in fresh buffer. The dialyzed lysate was deproteinized twice with 3 vols of chloroform:isoamyl ethanol, and dialyzed again at 4°C overnight to remove traces of chloroform. Finally, DNA was divided into five equal aliquots. To each aliquot was added ammonium acetate to 2 M. DNA was then precipitated with 2.5 vols of ethanol and stored in 80% ethanol until use.
Purification of linear forms of deleted IES elements in an agarose gel
An aliquot of ethanol-precipitated cell DNA derived from 100 ml of conjugating cell culture was redissolved in 1 ml of TE. Preliminary studies showed that the preparation contains RNA that will impact DNA mobility if a highly concentrated DNA solution is loaded onto a gel. Therefore, the solution was treated with 100 µl/ml RNase A at 37°C for 2 h. RNase was removed by treatment of the solution with 3 vols of chloroform:isoamyl ethanol. DNA was precipitated from the aqueous phase with ethanol and ammonium acetate as described above. DNA was rinsed with 80% ethanol to remove salt from the DNA pellet and redissolved in 150 µl of TE. Cell DNA was then loaded onto a 1.5% agarose gel. When electrophoresis was complete, the gel was stained with ethidium bromide to visualize DNA markers. The gel lanes containing fractionated Tetrahymena DNA were then cut into slices containing DNA fragments of a particular size. DNA was extracted from gel slices with the use of QIAquick columns (Qiagen). DNA from each gel fraction was eluted in 300 µl of TE.
Detection of DNA ends
Double-stranded DNA ends were detected by LM-PCR as described earlier (Saveliev and Cox, 1996), with modifications. Preliminary experiments showed that double-stranded DNA ends of linear IES elements were easy to amplify. Therefore, we used only 0.5 µl of a solution of gel-purified DNA in a detection protocol. Ligation was carried out at 16°C for 4 h in 15 µl of mixture containing 0.5 µl of a solution of gel-purified DNA, 0.1 Weiss unit of T4 DNA ligase (Promega), 0.3 µM of the appropriate linker, 30 mM Tris–HCl pH 7.8, 10 mM MgCl2, 10 mM dithiothreitol (DTT) and 1 mM ATP. A 5 µl aliquot of the ligated mixture was used in the amplification step. Amplification was carried out as described earlier (Saveliev and Cox, 1996), except that PCR was run in an MJ Research PTC-200 Peltier Thermal Cycler for 25 cycles (this cycler was used in every protocol involving denaturation or amplification in this study). Each cycle consisted of 10 s at 94°C, 45 s at 55°C and 1 min at 72°C. Cycling was concluded with a final extension at 72°C for 7 min. PCR products were visualized as described earlier (Saveliev and Cox, 1996).
To detect open double-stranded ends at the M3 side of the 0.6 and 0.9 kbp linear M IESs, we ligated the DNA to a linker consisting of the annealed oligonucleotides I and JM3MIC. The ligated end was then amplified using oligonucleotides I and M3MIC as primers. The PCR product was visualized using a 32P end-labeled M3MICfirst oligonucleotide as a primer for run-off synthesis. To detect open double-stranded ends at the R1 side of the linear R IES, we ligated DNA with a linker consisting of annealed oligonucleotides I and JR1MIC. The ligated end was then amplified using oligonucleotides I and R1MICfirst as primers. The PCR product was visualized with 32P end-labeled R1MICvis oligonucleotide.
5′ termini at the ends of linear IESs were detected with a different application of a LM-PCR (Saveliev and Cox, 1995), with modifications. A 5 µl aliquot of a solution of gel-purified DNA was mixed with 0.1 pmol of M3MIC (in studies involving the M3 end) or M1MICfirst (in studies involving the M1 end) oligonucleotides. The reaction volume was then brought to 50 µl by adding water, MgCl2 to 2.5 mM, the dNTPs dGTP, dATP, dCTP and dTTP, each to 0.2 mM, KCl to 50 mM, Tris–HCl pH 9.0 to 10 mM, Triton X-100 to 0.1% and glycerol to 10%. The linear DNA fragments were denatured by incubation of the mixture at 94°C for 2 min, and the oligonucleotide was annealed with a linear IES at 55°C for 20 min. One unit of sequencing grade Taq polymerase was added and an extension step at 72°C for 7 min carried out. Taq polymerase was removed by treatment of the mixture with 3 vols of chloroform:isoamyl ethanol (twice). DNA was precipitated with 3 vols of ethanol in the presence of 2 M ammonium acetate and 200 µg/ml glycogen at –70°C for 2 h. The DNA was blunt-end ligated to a duplex fragment (annealed oligonucleotides I and J) as described (Saveliev and Cox, 1995). The amplification step was as in the protocol above for detection of double-stranded DNA ends, except that the number of cycles was increased to 35. PCR products were visualized with 32P end-labeled M3MICvis or M1MICvis to detect amplified 5′ termini at M3 and M1 ends, respectively, via run-off synthesis as described (Saveliev and Cox, 1995).
3′ termini at the ends of linear IESs as well as in chromosome-integrated IESs were recovered with an anchor-based PCR protocol similar to that described earlier (Saveliev and Cox, 1996), with modifications. A 5 µl aliquot of a solution of gel-purified IES DNA was used in each detection experiment. First, the aliquot was mixed with 60 µl of TE. The DNA was then denatured by incubation of the mixture at 94°C for 3 min. Our preliminary experiments showed that QIAquick-purified DNA contains a contaminant of an unknown nature that inhibits terminal deoxynucleotidyl transferase (TdT) used on the next step of the detection protocol. Therefore, we additionally purified the denatured DNA by treatment with chloroform:isoamyl ethanol before TdT treatment, and precipitated it as we did after the extension step in the 5′ termini detection protocol above. The 3′ termini were then labeled with poly(dG) tails in a 100 µl reaction mixture containing 100 mM cacodylate buffer pH 6.8, 1 mM CaCl2, 0.1 mM DTT, 0.01 mM dGTP, 0.2 U/µl TdT (Promega), incubating at 37°C for 30 min. DNA was precipitated by adding ammonium acetate to 2 M, followed by 3 vols of ethanol, and leaving the solution at –70°C for 2 h. The precipitate was collected by microcentrifugation for 15 min at 4°C and redissolved in 50 µl of PCR mixture containing 0.2 mM each of dGTP, dATP, dTTP and dCTP, 2.5 mM MgCl2, 10 mM Tris–HCl pH 9.0, 0.1% Triton X-100, 10% glycerol and 0.002 µM oligo A(C). The mixture was incubated at 94°C for 2 min and then 50°C for 30 min. All other steps were as described (Saveliev and Cox, 1996), except that each PCR cycle consisted of 15 s at 94°C, 45 s at 55°C and 1 min at 72°C. M3MIC and M3MICfirst oligonucleotides were used to detect 3′ termini at the M3 end (in the first and nested PCR reactions, respectively) in combination with oligo A. PCR products were visualized with 32P end-labeled M3MICvis oligonucleotide as a primer for run-off synthesis. M1MICfirst and M1MICsecond oligonucleotides were used to amplify 3′ termini at the M1 end (in the first and nested PCR reactions, respectively) in combination with oligo A. PCR products were visualized with 32P end-labeled M1MICvis oligonucleotide.
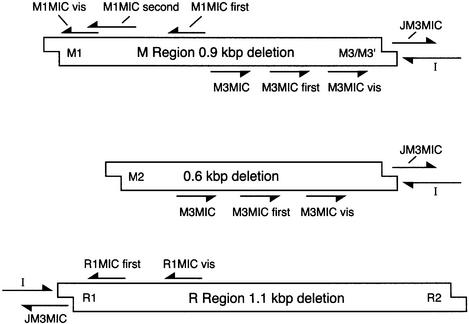
PCR primers and oligonucleotides are summarized in Figure 8, and their sequences are given in Table I.
Fig. 8. Map of the oligonucleotides used in this work. Sequences are listed in ITable I. Arrows indicate 3′ ends. Maps are not drawn to scale, and are intended only to indicate the relative linear order of the sequences to which the oligos pair. Some oligos used in specialized procedures at DNA ends, and which do not pair with any sequence in the IES elements [A, A(C) and J], are not shown.
Table I. Sequences of PCR primers and oligonucleotides.
| Sequence | |
|---|---|
| I | 5′-GGCTCGGACCGTGGCTAGCATTAGT |
| J | 5′-ACTAATGCTAG’ |
| JM3MIC | 5′-ATAAACTAATGCTAG |
| JR1MIC | 5′-TCACACTAATGCTAG |
| M1MICfirst | 5′-GGAGATTTTCTTTAAGTCAAGGATGGAAAC |
| M1MICsecond | 5′-CTCTATCTATACAAACACAGTTGATGGT |
| M1MICvis | 5′-TGATGGTATTTTAATTTCAGGATTAGCAAT |
| M3MIC | 5′-GTGTGGTACAATAGGTTGTCGTAGATTTTG |
| M3MICfirst | 5′-AAAGCAAGAAGGCTACTTAGCTTTCAAATT |
| M3MICvis | 5′-GCATTAATCACAATTTTGTTTCGGATTTTC |
| R1MICfirst | 5′-TGCTTAGAGTATCTTATTAATGAGATATTG |
| R1MICvis | 5′-CGGAAATACTTCGTTCATATTTATTTGTAT |
| A(C) | 5′-GATCTCATGCTGGAGTTCTTCGCCAAGT(C)16 |
| A | 5′-GATCTCATGCTGGAGTTCTTCGCC |
Acknowledgments
Acknowledgements
This work was supported by grant MCB-9808381 from the United States National Science Foundation.
References
- Austerberry C.F. and Yao,M.-C. (1987) Nucleotide sequence structure and consistency of a developmentally regulated DNA deletion in Tetrahymena thermophila. Mol. Cell. Biol., 7, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C.F. and Yao,M.-C. (1988) Sequence structures of two developmentally regulated, alternative DNA deletion junctions in Tetrahymena thermophila. Mol. Cell. Biol., 8, 3947–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C.F., Allis,C.D. and Yao,M.-C. (1984) Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc. Natl Acad. Sci. USA, 81, 7383–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C.F., Snyder,R.O. and Yao,M.-C. (1989) Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic Acids Res., 17, 7263–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M. and Perlman,P.S. (1995) Mechanisms of intron mobility. J. Biol. Chem., 270, 30237–30240. [DOI] [PubMed] [Google Scholar]
- Betermier M., Duharcourt,S., Seitz,H. and Meyer,E. (2000) Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol. Cell. Biol., 20, 1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin A., Goryshin,I.Y. and Reznikoff,W.S. (1999) Hairpin formation in Tn5 transposition. J. Biol. Chem., 274, 37021–37029. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (1992) Telomerases. Annu. Rev. Biochem., 61, 113–129. [DOI] [PubMed] [Google Scholar]
- Bruns P.J. and Brussard,T.B. (1974) Pair formation in Tetrahymena pyriformis, an inducible developmental system. J. Exp. Zool., 188, 337–344. [DOI] [PubMed] [Google Scholar]
- Chalker D.L., La Terza,A., Wilson,A., Kroenke,C.D. and Yao,M.-C. (1999) Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement. Mol. Cell. Biol., 19, 5631–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J.M. and Blackburn,E.H. (1985) The internally located telomeric sequences in the germ-line chromosomes of Tetrahymena are at the ends of transposon-like elements. Cell, 43, 747–758. [DOI] [PubMed] [Google Scholar]
- Doak T.G., Witherspoon,D.J., Doerder,F.P., Williams,K. and Herrick,G. (1997) Conserved features of TBE1 transposons in ciliated protozoa. Genetica, 101, 75–86. [DOI] [PubMed] [Google Scholar]
- Fugmann S.D., Lee,A.I., Shockett,P.E., Villey,I.J. and Schatz,D.G. (2000) The RAG proteins and V(D)J recombination: complexes, ends and transposition. Annu. Rev. Immunol., 18, 495–527. [DOI] [PubMed] [Google Scholar]
- Godisca R. and Yao,M.-C. (1990) A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell, 61, 1237–1246. [DOI] [PubMed] [Google Scholar]
- Godisca R. and Yao,M.-C. (1993) A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev., 7, 2357–2365. [DOI] [PubMed] [Google Scholar]
- Haren L., Ton-Hoang,B. and Chandler,M. (1999) Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol., 53, 245–281. [DOI] [PubMed] [Google Scholar]
- Jaraczewski J.W. and Jahn,C.L. (1993) Elimination of Tec elements involves a novel excision process. Genes Dev., 7, 95–105. [DOI] [PubMed] [Google Scholar]
- Klobutcher L.A. and Herrick,G. (1995) Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res., 23, 2006–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher L.A. and Herrick,G. (1997) Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol., 56, 1–62. [DOI] [PubMed] [Google Scholar]
- Klobutcher L.A., Turner,L.R. and LaPlante,J. (1993) Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev., 7, 84–94. [DOI] [PubMed] [Google Scholar]
- Madireddi M.T., Coyne,R.S., Smothers,J.F., Mickey,K.M., Yao,M.-C. and Allis,C.D. (1996) Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell, 87, 75–84. [DOI] [PubMed] [Google Scholar]
- Orias E. (1986) Ciliate conjugation. In Gall,J.G. (ed.), The Molecular Biology of Ciliated Protozoa. Academic Press, Orlando, FL, pp. 45–84.
- Patil N.S. and Karrer,K.M. (2000) A developmentally regulated deletion element with long terminal repeats has cis-acting sequences in the flanking DNA. Nucleic Acids Res., 28, 1465–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.B. and Craig,N.L. (1998) VDJ recombination: a transposase goes to work. Cell, 94, 411–414. [DOI] [PubMed] [Google Scholar]
- Saveliev S.V. and Cox,M.M. (1994) The fate of deleted DNA produced during programmed genomic deletion events in Tetrahymena thermophila. Nucleic Acids Res., 22, 5695–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev S.V. and Cox,M.M. (1995) Transient DNA breaks associated with with programmed genomic deletions in conjugating cells of Tetrahymena thermophila. Genes Dev., 9, 248–255. [DOI] [PubMed] [Google Scholar]
- Saveliev S.V. and Cox,M.M. (1996) Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J., 15, 2858–2869. [PMC free article] [PubMed] [Google Scholar]
- Williams K., Doak,T.G. and Herrick,G. (1993) Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J., 12, 4593–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.-C. (1989) Site-specific chromosomal breakage and DNA deletions in ciliates. In Berg,D.E. and Howe,M.M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 715–734.
- Yao M.-C. (1996) Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet., 12, 26–30. [DOI] [PubMed] [Google Scholar]
- Yao M.-C. and Yao,C.H. (1994) Detection of circular excised DNA deletion elements in Tetrahymena thermophila during development. Nucleic Acids Res., 22, 5702–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.-C., Choi,S.J., Yokoyama,S., Austerberry,C.F. and Yao,C.H. (1984) DNA elimination in Tetrahymena: a developmental proccess involving extensive breakage and rejoining of DNA at defined sites. Cell, 36, 433–440. [DOI] [PubMed] [Google Scholar]