Abstract
Chronic gastroesophageal reflux disease is a known risk factor for Barrett's esophagus (BE), that induces oxidative mucosal damage. Glutathione peroxidase-3 (GPx3) is a secretory protein with potent extracellular antioxidant activity. Herein, we have investigated the mRNA and protein expression of GPx3, and explored promoter hypermethylation as an epigenetic mechanism for GPx3 gene inactivation during Barrett's carcinogenesis. Quantitative real-time reverse transcription polymerase chain reaction on 42 Barrett's adenocarcinomas (BAs) revealed consistently reduced levels of GPx3 mRNA in 91% of tumor samples. GPx3 promoter hypermethylation was detected in 62% of Barrett's metaplasia, 82% of dysplasia, and 88% of BA samples. Hypermethylation of both alleles of GPx3 was most frequently seen in BA (P = .001). Immunohistochemical staining of GPx3 in matching tissue sections (normal, BE, Barrett's dysplasia, and BA) revealed a weak-to-absent GPx3 staining in Barrett's dysplasia and adenocarcinoma samples where the promoter was hypermethylated. The degree of loss of immuno-histochemistry correlated with the hypermethylation pattern (monoallelic versus biallelic). The observed high frequency of promoter hypermethylation and progressive loss of GPx3 expression in BA and its associated lesions, together with its known function as a potent antioxidant, suggest that epigenetic inactivation and regulation of glutathione pathway may be critical in the development and progression of BE.
Keywords: Barrett's, dysplasia, cancer, GPx3, ROS
Introduction
Gastroesophageal reflux disease (GERD) is a major public health problem, with a prevalence of 5% to 7% in the general population [1,2]. Approximately 10% of patients with chronic GERD develop a metaplastic condition, where the normal squamous epithelium of the distal esophagus is often replaced by a columnar or intestinalized epithelium with goblet cells, known as Barrett's esophagus (BE). In the setting of continued injury as a result of GERD, BE is a premalignant lesion that can ultimately progress from metaplasia to dysplasia, and subsequent Barrett's adenocarcinoma (BA) [3–6]. BA has one of the fastest-growing incidence rates in the Western world [7–9].
Reactive oxygen species (ROS) play a significant role in the pathogenesis of several diseases of the gastrointestinal (GI) tract, including GERD, BE, and gastritis [10,11]. ROS are potential carcinogens that facilitate mutagenesis, tumor promotion, and progression. Although the extent of the contribution made by oxidative DNA damage has not been well defined, it appears that ROS-induced DNA damage cannot only initiate carcinogenesis, but also facilitate tumor progression [12–15].
Cells have an antioxidant system that controls the balance between production and removal of oxygen radicals, thereby protecting against oxidative damage. Antioxidant defensive mechanisms include the enzymes superoxide dismutase, catalase, and glutathione peroxidase (GPx), as well as non-enzymatic compounds such as α-tocopherol, β-carotene, vitamin C, and glutathione [16,17]. In the GI tract, several peroxide-reducing enzymes are found, including members of the GPx and peroxiredoxin families.
GPx catalyzes the conversion of hydrogen peroxide and lipid peroxides at the expense of glutathione, and is a major scavenger of ROS produced during normal metabolism or after oxidative insult [18]. GPx2 is found in the GI tract and plays a role in colon cancer resistance, whereas plasma glutathione peroxidase-3 (GPx3) is identified in the normal tissues of the esophagus, stomach, small bowel, and colon [19]. GPx3 is an extracellular glycosylated enzyme that can use glutathione, thioredoxin, and glutaredoxin as electron donors, to reduce a broad range of hydroperoxides [20,21]. It reduces hydroperoxides, including fatty acid hydroperoxides and phospholipid hydroperoxides [22,23]. Increased production of mucosal ROS has been demonstrated to occur in BE, suggesting a role for cellular injury in carcino-genesis [24–28]. Antioxidant defenses are therefore critical in the protection of the epithelium against DNA damage and mutation.
The abnormal hypermethylation of CpG sites associated with tumor-suppressor genes can cause transcriptional silencing and is recognized as an important mechanism for gene inactivation in cancer cells [29,30]. Evidence supporting this mechanism of gene inactivation has been observed in BE and many types of human cancer, including esophageal adenocarcinoma [31–33].
To elucidate the role of GPx3 in Barrett's tumorigenesis, we have investigated the mRNA expression level, promoter methylation status, and protein expression of GPx3 in Barrett's metaplasia, dysplasia, and adenocarcinoma.
Materials and Methods
Tissue Samples
Forty-two frozen tissue samples of Barrett's-related, gastroesophageal junction (GEJ) and lower esophageal adenocarcinomas were collected for the analysis of GPx3 mRNA expression by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR). In addition, 19 normal esophageal and gastric mucosa samples were used as controls for comparison of GPx3 mRNA expression. From the 42 samples with BA, DNA from 12 samples were available for simultaneous analysis for GPx3 promoter methylation. An additional 22 BA tissue samples were processed for DNA extraction to give a total of 34 BA DNA samples. In addition, DNA was purified from 21 BE, 11 BE with dysplasia, and 12 normal esophageal mucosa samples. All DNA samples were analyzed by methylation-specific PCR (MSP). In addition, 55 BA tissues with adjacent BE, dysplasia, and normal tissues were formalin-fixed and embedded in paraffin for immunohistochemical analysis. The histopathology was verified in all tissues (A.R., M.V., and C.A.M.). All clinical tissue samples and histopathologic information were obtained according to approved institutional guidelines at the University of Virginia (Charlottesville, VA) and Otto-von-Guericke University (Magdeburg, Germany). The adenocarcinomas collected ranged from well-differentiated (WD) to poorly differentiated (PD), stages I to IV, with a mix of intestinaltype and diffuse-type tumors.
Quantitative Real-Time RT-PCR
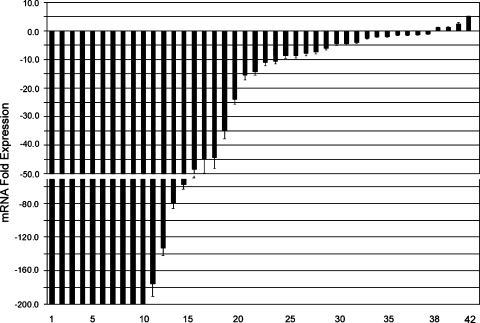
mRNA was isolated using the RNeasy kit (Qiagen, Gmbh, Hilden, Germany), then single-stranded cDNA was subsequently synthesized using the Advantage RT-for-PCR Kit (Clontech, Palo Alto, CA). Quantitative real-time RT-PCR was performed in 42 BA samples and 19 normal gastric mucosal samples, using an iCycler (Bio-Rad, Hercules, CA); threshold cycle number was determined using the iCycler software (version 3.0; BioRad, Hercules, CA), as described earlier [34]. The primers used for real-time RT-PCR were obtained from GeneLink (Hawthorne, NY), and their sequences are available on request. Reactions were performed in triplicate, and threshold cycle numbers were averaged. A single melt curve peak was observed for each sample used in data analysis, thus confirming the purity and specificity of all amplified products. The results for GPx3 were normalized to HPRT1, which had minimal variation in all normal and neoplastic gastric samples tested. The fold expression in tumors was calculated, compared to normal samples, and normalized with HPRT1 values according to the formula: 2(Rt - Et)/2(Rn - En), as described elsewhere [34]. Rt is the threshold cycle number for the reference gene observed in the tumor, Et is the threshold cycle number for the experimental gene observed in the tumor, Rn is the threshold cycle number for the reference gene observed in the normal sample, and En is the threshold cycle number for the experimental gene observed in the normal sample. Rn and En values were taken from the 19 normal mucosa samples that were analyzed. Each tumor sample was compared to the 19 normal samples, and the relative fold expression in tumors, with standard error of the mean (±SEM), is shown in Figure 1.
Figure 1.
Loss of GPx3 expression in BAs. Quantitative real-time RT-PCR was performed on 42 BAs using iCycler (BioRad), in a comparison with 19 normal esophageal and gastric mucosa samples. The horizontal axis shows sample numbers, whereas the fold expression in tumor samples compared to normal is shown in the vertical axis. The expression fold was calculated according to the formula: 2(Rt - Et)/2(Rn - En), as described elsewhere [34]. Each bar represents one tumor sample. Downregulation is shown as negative expression fold values. The displayed mean expression fold for each tumor sample is calculated in comparison with expression in 19 normal samples. The standard error of the mean is shown (±SEM). The expression of GPx3 was normalized to the expression of HPRT1, which showed minimal variation in all normal and neoplastic samples tested. The loss of GPx3 mRNA expression was universal and did not statistically correlate with any of the histopathologic parameters investigated.
MSP
DNA was prepared from three to seven 10-µm sections taken from representative paraffin blocks using the Nucleo-Spin Tissue Kit (Machery and Nagel, Duren, Germany). On a mirror-imaged H&E slice, the region of interest was marked, and tissues were manually scraped off to ensure a specific cell population of more than 80% in the preparation. Genomic DNA were extracted from the cell lines by a standard phenol-chloroform procedure. Extracted DNA was bisulfite-modified using the CpGenome DNA modification kit (Intergen, Purchase, NY), as described previously [35]. Briefly, all unmethylated cytosines were deaminated and converted to uracils, whereas 5-methylcytosines remained unaltered. Modified DNA was used as a template for MSP, which was carried out using primers specific for either methylated or modified-unmethylated sequences. CpGenome universal methylated DNA (Intergen) was used as a positive control, whereas DNA from normal lymphocytes was used as a negative control for methylated alleles.
Primers were designed for the CpG-rich region around the start of exon 1 (positions 1984–2201; http://egp.gs.washington.edu), which consists of 17 CpG dinucleotides. The modified DNA was subjected to MSP using the following GPx3-specific primers: for methylated sequences, sense 5′-GGTGGGGAGTTGAGGGTAAGTC-3′ and antisense 5′-CCTACAACAACCGAACCATAACGAAA-3′; and for unmethylated sequences, sense 5′-GGTGGGGAGTTGAGGGTAAGTT-3′ and antisense 5′-CCTACAACAACCAAACCATAACAAAA-3′ (CpGs are given in bold letters). Both these pairs generate PCR products of 219 bp. Methylated PCR products of 12 samples were directly sequenced (sequencing primer: 5′-GGTGGGGAGTTGAGGGTA-3′, which can bind to both methylated and unmethylated sequences) using rhodamine dye terminator chemistry (Big DyeTerminator v1.1 Cycle Sequencing Kit; Applied Biosystems, Foster City, CA) and POP-6 polymer on an ABI PRISM 310 capillary sequencer.
For PCR, 2 µl of bisulfite-modified DNA was amplified in a total volume of 25 µl, containing 1x PCR buffer, 3 mM MgCl2, 12.5 pmol of each primer, 160 µM dNTPs, and 0.5 U of Hot-Goldstar Taq polymerase (Eurogentec, San Diego, CA). PCR conditions were 95°C for 10 minutes, 35 cycles of 95°C for 1 minute, annealing at 60°C (unmethylated) or 64°C (methylated) for 1 minute and 72°C for 1 minute, followed by a final extension step at 72°C for 10 minutes. PCR products were electrophoresed on polyacrylamide gels and visualized by silver staining.
5-Aza-Cytidine Treatment in Cell Lines
MKN45 cells were cultured in RPMI 1640 supplemented with 10% FCS. The MKN45 cells that did not express GPx3 mRNA and for which homozygously (biallelic) methylated promoter status had been confirmed were treated with 5-aza-cytidine (Sigma, St. Louis, MO) that had been dissolved in cold RPMI 1640 immediately prior to use. Cells were grown in a medium containing 1 µM 5-aza-cytidine for 4 days, with the medium and drug being replaced every 48 hours. After 4 days, the drug was removed and the cells were allowed to recover for a further 48 hours (total of 144 hours).
Immunohistochemistry (IHC)
Immunohistochemical analysis of GPx3 protein expression was performed on 55 formalin-fixed, paraffin-embedded tissue samples that included BE, dysplasia, BA, and normal esophageal tissue sections, to determine concordance with results from MSP. Dewaxing and rehydration by descending concentrations of ethanol were followed by antigen retrieval (20 minutes in a microwave, 450 W, 10 mM EDTA, pH 8.0). Blocking was done with 10% goat serum in PBS for 5 minutes. All sections were incubated with antihuman GPx (monoclonal mouse IgG1κ, GPx-347, dilution 1:20, MBL; MoBiTec, Göttingen, Germany) for 1 hour at room temperature, then washed in PBS. For revealing positive immunohistochemical reaction, the Vectastain ABC-AP kit (mouse IgG, vector; Alexis, Gruenberg, Germany) was used as chromogen substrate, and the specimens were counterstained with hematoxylin and mounted with DEPEX. Specificity of immunostaining was checked by omitting single steps in the protocol, and by replacing the primary antibody with nonimmune serum. Slides were examined by two independent reviewers blinded to their identity.
Results
Reduced Levels of GPx3 mRNA Were Found in BAs Compared with Normal Mucosa
We detected comparably high mRNA expression levels of GPx3 mRNA in all 19 normal mucosae. A quantitative real-time RT-PCR analysis of 42 Barrett's-related adenocarcinomas included all stages of development (TNM stages I–IV), histopathology (WD to PD, intestinal type and diffuse type), and location (lower esophageal to GEJ). This analysis revealed consistently low levels of GPx3 expression in 90.5% (38 of 42) of BA samples compared with 19 normal mucosal samples. There was a dramatic reduction in levels of GPx3 in more than half of the cases, with a 10-fold reduction compared with the normal samples (Figure 1). The loss of GPx3 was universal and did not statistically correlate with any of the histopathologic parameters investigated.
High Frequency of GPx3 Promoter Methylation Observed in BAs
MSP revealed frequent monoallelic (one band for methylated product and one band for unmethylated product) and biallelic (one band for methylated product) GPx3 promoter methylation in tumor samples and preneoplastic dysplasia. However, only 2 of 12 normal esophageal mucosa samples that were studied with MSP showed GPx3 promoter hypermethylation, which was always monoallelic (Figures 2–4). Furthermore, these two samples were taken from patients with adjacent BE, one of whom had biallelic GPx3 promoter hypermethylation and the other monoallelic GPx3 promoter hypermethylation (Figure 2, B11 and B12). Thus, methylation in normal samples may be due to a contaminant cell population that was not identified histologically. However, GPx3 promoter hypermethylation was detected in 61.9% (13 of 21) of Barrett's metaplasia, 81.8% (9 of 11) of dysplasia, and 88.2% (30 of 34) of BA samples (Table 1 and Figure 2). Among the samples with GPx3 promoter hypermethylation, 6 BE, 1 dysplastic, and 16 BA samples showed biallelic hypermethylation. These data clearly demonstrate a significant correlation between histomorphologic diagnosis and GPx3 promoter hypermethylation (P = .001).
Figure 2.
GPx3 promoter methylation status. BE, Barrett's esophagus; BA, Barrett's adenocarcinoma. Black boxes indicate biallelic methylation (M+/M+) and gray boxes indicate monoallelic methylation (M+/M-) as detected in MSP, whereas blank boxes indicate samples that are only unmethylated (M-/M-). GPx3 promoter hypermethylation was detected in 30 of 34 (88.2%) adenocarcinoma samples, with 16 samples showing biallelic methylation. Of the dysplasia samples, 9 of 11 (81.8%) were methylated, with only one sample showing biallelic methylation. In Barrett's metaplasia, methylation was only detectable in 13 of 21 samples (61.9%). Ten of 12 normal samples were unmethylated, with only monoallelic methylation in the remaining two normal samples. Among 12 samples available for the comparison of mRNA expression level and methylation status, nine samples had downregulation of GPx3 mRNA levels together with hypermethylation.
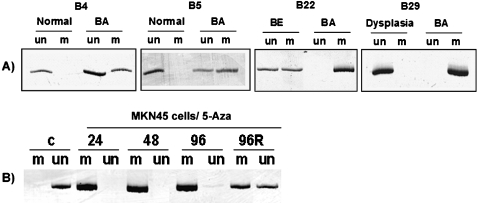
Figure 3.
MSP analysis of the GPx3 promoter region. (A) This figure shows two cases of matched normal mucosa and BA (B4 and B5), one case of matched BE and BA (B22), and one sample of matched dysplasia and BA (B29). Unmethylated (un) and methylated (m) PCR products are shown. (B) MSP of the GPx3 promoter after the treatment of MKN45 cells with 5-aza-cytidine for 24, 48, and 96 hours. A slight unmethylated band can be recognized already after 96 hours of treatment with 5-aza-cytidine. After an additional 48 hours of recovery from the drug (96R), the unmethylated band further increases and the methylated and unmethylated PCR bands reach similar signal intensities.
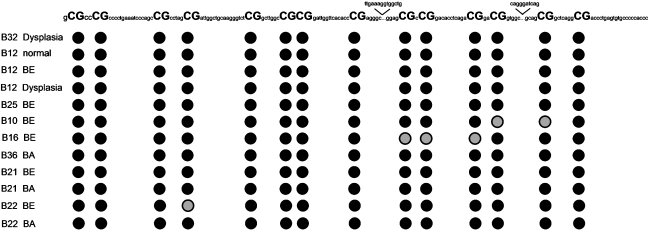
Figure 4.
Sequencing analysis of the GPx3 promoter region. Twelve samples showing a methylated band were randomly chosen for bisulfite sequencing. The figure shows the methylation pattern of a partial GPx3 promoter region that was used for MSP analysis, consisting of 14 CpG sites. Three additional CpG sites were already covered by the primer regions. Gray circles indicate the presence of a C and a T at the same position, reflecting a partial methylation, whereas closed circles indicate methylated sites (only a C was seen). These data demonstrate a dense methylation of CpG sites in this GPx3 promoter region.
Table 1.
Frequency of GPx3 Promoter Hypermethylation According to Histomorphologic Diagnosis.
| GPx3 Methylation Status | Histologic Diagnosis | |||
| Normal (n = 10) | BE (n = 21) | Dysplasia (n = 11) | BA (n = 34) | |
| No methylation (M-) | 10 (83%) | 8 (38%) | 2 (18%) | 4 (12%) |
| Monoallelic (M+/M-) | 2 (17%) | 7 (33%) | 8 (73%) | 14 (41%) |
| Biallelic (M+/M+) | 0 | 6 (29%) | 1 (9%) | 16 (47%)* |
M, methylation; (-) negative; (+) positive. Monallelic methylated samples displayed one band for methylated product (M+) and one band for unmethylated product (M-). In biallelic methylation, only one band that represents the MSP product was visualized (M+/M+).
Biallelic methylation was significantly more frequent in BA (P < .01).
Treatment with 5-Aza Demethylates GPx3 Promoter and Restores mRNA Expression in MKN45 Cells
The MSP following 5-aza-cytidine treatment of the MKN cell line revealed an intense unmethylated band, whereas the methylated band nearly disappeared. RT-PCR demonstrated a significant increase in the level of GPx3 mRNA expression after 96 hours of treatment followed by 48 hours of recovery from the drug (Figure 3). Thus, these results confirmed that promoter methylation plays an essential role in the silencing of GPx3 expression.
GPx3 Promoter Methylation Correlates with Loss of GPx3 mRNA and Protein Expression
Of the 34 BA samples that were subjected to methylation analysis, 12 samples were further analyzed using quantitative real-time RT-PCR to measure GPx3 mRNA expression. All but two revealed low GPx3 expression and nine of them corresponded to promoter hypermethylation (Figure 2). One sample had no detectable GPx3 promoter methylation and showed a minimal reduction of gene expression (B44). Figure 3 demonstrates a representative gel electrophoresis image for MSP of matched normal mucosa and BA, BE, or dysplasia. Bisulfite sequencing of 12 methylated samples identified the CpG sites, within the GPx3 promoter region that we studied, that were vulnerable to methylation (Figure 4). Immunohistochemical analysis of GPx3 protein expression was performed on all samples that were subjected to methylation analysis in this study. High expression levels, determined by a strong, diffuse staining of the cytoplasm, was found in all normal esophageal and Barrett's epithelia tissues that had unmethylated promoter regions. In contrast, BA and nearby dysplasia, which were determined to be methylated, had weak to absent immunostaining for GPx3 protein (Figure 5).
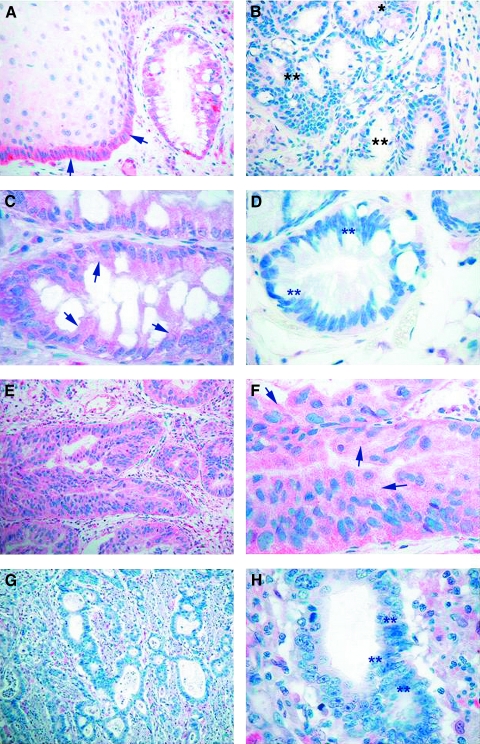
Figure 5.
Immunohistochemical analysis for human GPx. Immunohistochemical staining was performed using the GPx-347 monoclonal mouse IgG1j antibody. (A) Normal esophageal squamous epithelium and columnar Barrett's epithelium, determined as unmethylated by MSP, show strong, diffuse cytoplasmic staining (red color) representing a normal expression of GPx (original magnification, x200) (indicated by arrows). (B) Barrett's epithelium with monoallelic methylation of GPx shows weak protein expression (indicated by *) and demonstrates loss of GPx immunoreactivity in dysplastic epithelia (indicated by **) with determined biallelic methylation (original magnification, x100). (C) Barrett's epithelium, determined as unmethylated, shows moderate to strong, diffuse cytoplasmic staining (original magnification, x400) (indicated by arrows). (D) Barrett's epithelium with biallelic methylation shows complete loss of immunoreactivity (original magnification, x200) (indicated by **). (E and F) In BA lacking methylation of the GPx promoter, a strong, diffuse cytoplasmic staining was observed (original magnification, x100 and x200) (indicated by arrows). (G and H) BA with biallelic methylation demonstrates loss of immunoreactivity for a GPx antibody (original magnification, x100 and x400) (indicated by **).
Discussion
In this study, we have demonstrated that loss of the GPx3 mRNA and protein expression is a consistent and progressive molecular alteration during Barrett's tumorigenesis that occurs due to hypermethylation of the GPx3 promoter. A monoallelic methylation pattern associated with partial loss of GPx3 expression was seen as early as metaplasia. However, a more frequent and consistent biallelic methylation, together with a dramatic loss of GPx3 expression, was demonstrated in almost all BA samples. This observation indicates that methylation of one allele starts as early as metaplasia, whereas for cancer samples, both alleles become methylated, with a dramatic loss of expression of GPx3. This finding is particularly interesting because Barrett's tumors arise following a characteristic sequence of events that is initiated by chronic GERD. Several reports have demonstrated increased mucosal ROS levels in reflux esophagitis and BE [24–27]. ROS are mainly produced by phagocytic cells, neutrophils, and epithelial cells as a consequence of inflammatory tissue damage [36], and have been implicated as important factors for both tissue and DNA damage [13–15]. Additionally, it has previously been demonstrated that the levels of glutathione are markedly decreased in Barrett's epithelium [37], suggesting that the antioxidant defense mechanism is impaired in this premalignant lesion. GPx is considered to be the first line of defense for the protection of epithelial cells against this oxidative damage process. GPx3 is a secreted isoform of GPx and is an efficient extracellular antioxidant [38,39].
Mörk et al. [19] demonstrated that both plasma GPx3 and GI GPx1 and GPx2 are expressed in the normal mucosa of the esophagus. GPx3 mRNA expression was markedly high in the squamous epithelium of the esophagus, which may represent a characteristic and physiological feature of squamous esophageal mucosa. In particular, the high expression levels may contribute to adequate extracellular neutralization of ROS [19]. We observed a constantly low level of expression of GPx3 mRNA in mucosal samples from BE and BA, with the adenocarcinomas showing a particularly dramatic loss of GPx3 expression. This finding was confirmed immunohistochemically, where a remarkable loss of GPx3 immunostaining in all studied Barrett's dysplasia and adenocarcinoma samples was contrasted by a strong, diffuse cytoplasmic staining in the normal esophageal mucosa samples. These findings suggest that GPx3 function is impaired in BE, a consequence of which is likely to be an increased amount of hydrogen peroxide and other ROS. This would induce DNA damage, driving the carcinogenic process to the final stages of BA.
In this study, we investigated GPx3 promoter hypermethylation as a mechanism of GPx3 gene silencing. We have demonstrated a significant correlation of GPx3 promoter hypermethylation and the appearance of BE and adenocarcinoma. In addition, significant GPx3 promoter hypermethylation was detected in the majority of BE, dysplasia, and BAs, but not in normal esophageal mucosa. Moreover, there was a strong concordance between the promoter hypermethylation of GPx3 and the data obtained during mRNA and immunohistochemical analyses. Promoter hypermethylation resulted in the downregulation of mRNA levels in 9 of 11 samples and reduced or loss of protein expression in samples with monoallelic or biallelic methylation, respectively. These data provide strong evidence that GPx3 promoter hypermethylation is a main mechanism involved in GPx3 gene inactivation, resulting in impaired GPx3 function. This impairment could be an important step in the neoplastic transformation of BE.
In addition to the hypermethylation seen in pathologic lesions, we detected monoallelic GPx3 promoter hypermethylation in 2 of 12 normal esophageal mucosal samples. Interestingly, these two samples were adjacent to BE and/or dysplasia that had GPx3 promoter hypermethylation, suggesting that GPx3 promoter hypermethylation may be an early epigenetic event during the multistep process of neoplastic progression in BE. If this were the case, then these samples may represent submicroscopic lesions that were beyond histologic evaluation, but were susceptible to the development of BE. Similar observations were made for the tumor-suppressor gene CDKN2/p16, with respect to promoter hypermethylation in BE [33].
In summary, we have demonstrated constantly low mRNA and protein expression levels of GPx3 in BAs. Promoter hypermethylation of GPx3 started as early as BE and was often seen as monoallelic. However, GPx3 biallelic hypermethylation and inactivation increased significantly with progression toward neoplasia. Therefore, epigenetic inactivation of GPx3 may play an important role in the development of BE and its carcinogenesis cascade.
Acknowledgements
We would like to thank Manfred Stolte and Sarah de la Rue for their assistance during this study.
Abbreviations
- BE
Barrett's esophagus
- BA
Barrett's adenocarcinoma
- GEJ
gastroesophageal junction
Footnotes
This work was supported by grant awards from the National Cancer Institute (NCI; R01CA106176) and the Medical Faculty Magdeburg (Spitzenbonus). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the NCI or the University of Virginia.
References
- 1.Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett's esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002;122(1):26–33. doi: 10.1053/gast.2002.30297. [DOI] [PubMed] [Google Scholar]
- 2.Cameron AJ, Lagergren J, Henriksson C, Nyren O, Locke GR, III, Pedersen NL. Gastroesophageal reflux disease in monozygotic and dizygotic twins. Gastroenterology. 2002;122(1):55–59. doi: 10.1053/gast.2002.30301. [DOI] [PubMed] [Google Scholar]
- 3.Rana PS, Johnston DA. Incidence of adenocarcinoma and mortality in patients with Barrett's oesophagus diagnosed between 1976 and 1986: implications for endoscopic surveillance. Dis Esophagus. 2000;13(1):28–31. doi: 10.1046/j.1442-2050.2000.00069.x. [DOI] [PubMed] [Google Scholar]
- 4.O'Connor JB, Falk GW, Richter JE. The incidence of adenocarcinoma and dysplasia in Barrett's esophagus: report on the Cleveland Clinic Barrett's Esophagus Registry. Am J Gastroenterol. 1999;94(8):2037–2042. doi: 10.1111/j.1572-0241.1999.01275.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferraris R, Bonelli L, Conio M, Fracchia M, Lapertosa G, Aste H. Incidence of Barrett's adenocarcinoma in an Italian population: an endoscopic surveillance programme. Gruppo Operativo per lo Studio delle Precancerosi Esofagee (GOSPE) Eur J Gastroenterol Hepatol. 1997;9(9):881–885. doi: 10.1097/00042737-199709000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Haggitt RC. Barrett's esophagus, dysplasia, and adenocarcinoma. Hum Pathol. 1994;25(10):982–993. doi: 10.1016/0046-8177(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton SR, Smith RR, Cameron JL. Prevalence and characteristics of Barrett esophagus in patients with adenocarcinoma of the esophagus or esophagogastric junction. Hum Pathol. 1988;19(8):942–948. doi: 10.1016/s0046-8177(88)80010-8. [DOI] [PubMed] [Google Scholar]
- 8.Phillips RW, Wong RK. Barrett's esophagus. Natural history, incidence, etiology, and complications. Gastroenterol Clin North Am. 1991;20(4):791–816. [PubMed] [Google Scholar]
- 9.Blot WJ, Devesa SS, Fraumeni JF., Jr Continuing climb in rates of esophageal adenocarcinoma: an update. JAMA. 1993;270(11):13–20. [PubMed] [Google Scholar]
- 10.Farhadi A, Fields J, Banan A, Keshavarzian A. Reactive oxygen species: are they involved in the pathogenesis of GERD, Barrett's esophagus, and the latter's progression toward esophageal cancer? Am J Gastroenterol. 2002;97(1):22–26. doi: 10.1111/j.1572-0241.2002.05444.x. [DOI] [PubMed] [Google Scholar]
- 11.Lee JS, Oh TY, Ahn BO, Cho H, Kim WB, Kim YB, Surh YJ, Kim HJ, Hahm KB. Involvement of oxidative stress in experimentally induced reflux esophagitis and Barrett's esophagus: clue for the chemoprevention of esophageal carcinoma by antioxidants. Mutat Res. 2001;480–481:189–200. doi: 10.1016/s0027-5107(01)00199-3. [DOI] [PubMed] [Google Scholar]
- 12.Dreher D, Junod AF. Role of oxygen free radicals in cancer development. Eur J Cancer. 1996;32A(1):30–38. doi: 10.1016/0959-8049(95)00531-5. [DOI] [PubMed] [Google Scholar]
- 13.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 14.Malins DC, Polissar NL, Gunselman SJ. Progression of human breast cancers to the metastatic state is linked to hydroxyl radical-induced DNA damage. Proc Natl Acad Sci USA. 1996;93(6):2557–2563. doi: 10.1073/pnas.93.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerutti PA. Prooxidant states and tumor promotion. Science. 1985;227(4685):375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 16.Sies H. Oxidative stress: oxidants and antioxidants. Exp Physiol. 1997;82(2):291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 17.Drge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 18.Takebe G, Yarimizu J, Saito Y, Hayashi T, Nakamura H, Yodoi J, Nagasawa S, Takahashi K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem. 2002;277(43):41254–41258. doi: 10.1074/jbc.M202773200. [DOI] [PubMed] [Google Scholar]
- 19.Mörk H, Lex B, Scheurlen M, Dreher I, Schutze N, Kohrie J, Jakob F. Expression pattern of gastrointestinal selenoproteins—targets for selenium supplementation. Nutr Cancer. 1998;32(2):64–70. doi: 10.1080/01635589809514720. [DOI] [PubMed] [Google Scholar]
- 20.Tham DM, Whitin JC, Kim KK, Zhu SX, Cohen HJ. Expression of extracellular glutathione peroxidase in human and mouse gastrointestinal tract. Am J Physiol. 1998;275(6 (Part 1)):G1463–G1471. doi: 10.1152/ajpgi.1998.275.6.G1463. [DOI] [PubMed] [Google Scholar]
- 21.Bjornstedt M, Xue J, Huang W, Akesson B, Holmgren A. The thioredoxin and glutaredoxin systems are efficient electron donors to human plasma glutathione peroxidase. J Biol Chem. 1994;269(47):29382–29384. [PubMed] [Google Scholar]
- 22.Yamamoto Y, Takahashi K. Glutathione peroxidase isolated from plasma reduces phospholipid hydroperoxides. Arch Biochem Biophys. 1993;305(2):541–545. doi: 10.1006/abbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 23.Esworthy RS, Chu FF, Paxton RJ, Akman S, Doroshow JH. Characterization and partial amino acid sequence of human plasma glutathione peroxidase. Arch Biochem Biophys. 1991;286(2):330–336. doi: 10.1016/0003-9861(91)90048-n. [DOI] [PubMed] [Google Scholar]
- 24.Wetscher GJ, Hinder RA, Klingler P, Gadenstatter M, Perdikis G, Hinder PR. Reflux esophagitis in humans is a free radical event. Dis Esophagus. 1997;10(1):29–32. doi: 10.1093/dote/10.1.29. (discussion, 33) [DOI] [PubMed] [Google Scholar]
- 25.Olyaee M, Sontag S, Salman W, Schnell T, Mobarhan S, Eiznhamer D, Keshavarzian A. Mucosal reactive oxygen species production in oesophagitis and Barrett's oesophagus. Gut. 1995;37(2):168–173. doi: 10.1136/gut.37.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Ding YW, Yang G, Bondoc F, Lee MJ, Yang CS. Oxidative damage in an esophageal adenocarcinoma model with rats. Carcinogenesis. 2000;21(2):257–263. doi: 10.1093/carcin/21.2.257. [DOI] [PubMed] [Google Scholar]
- 27.Wetscher GJ, Hinder RA, Bagchi D, Hinder PR, Bagchi M, Perdikis G, McGinn T. Reflux esophagitis in humans is mediated by oxygen-derived free radicals. Am J Surg. 1995;170(6):552–556. doi: 10.1016/s0002-9610(99)80014-2. (discussion, 556–557) [DOI] [PubMed] [Google Scholar]
- 28.Goldstein SR, Yang GY, Curtis SK, Reuhl KR, Liu BC, Mirvish SS, Newmark HL, Yang CS. Development of esophageal metaplasia and adenocarcinoma in a rat surgical model without the use of a carcinogen. Carcinogenesis. 1997;18(11):2265–2270. doi: 10.1093/carcin/18.11.2265. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21(2):163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 30.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 31.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60(18):5021–5026. [PubMed] [Google Scholar]
- 32.Wong DJ, Barrett MT, Stoger R, Emond MJ, Reid BJ. p16INK4a promoter is hypermethylated at a high frequency in esophageal adenocarcinomas. Cancer Res. 1997;57(13):2619–2622. [PubMed] [Google Scholar]
- 33.Klump B, Hsieh CJ, Holzmann K, Gregor M, Porschen R. Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett's esophagus. Gastroenterology. 1998;115(6):1381–1386. doi: 10.1016/s0016-5085(98)70016-2. [DOI] [PubMed] [Google Scholar]
- 34.El-Rifai W, Moskaluk CA, Abdrabbo M, Harper JC, Yoshida C, Riggins G, Frierson HF, Jr, Powell SM. Gastric cancers overexpress S100A calcium binding proteins. Cancer Res. 2002;62:6823–6826. [PubMed] [Google Scholar]
- 35.Schneider-Stock R, Boltze C, Lasota J, Miettinen M, Peters B, Pross M, Roessner A, Gunther T. High prognostic value of p16INK4 alterations in gastrointestinal stromal tumors. J Clin Oncol. 2003;21(9):1688–1697. doi: 10.1200/JCO.2003.08.101. [DOI] [PubMed] [Google Scholar]
- 36.McCord JM. The biology and pathology of oxygen radicals. Ann Intern Med. 1978;89(1):122–127. doi: 10.7326/0003-4819-89-1-122. [DOI] [PubMed] [Google Scholar]
- 37.van Lieshout EM, Tiemessen DM, Witteman BJ, Jansen JB, Peters WH. Low glutathione and glutathione S-transferase levels in Barrett's esophagus as compared to normal esophageal epithelium. Jpn J Cancer Res. 1999;90(1):81–85. doi: 10.1111/j.1349-7006.1999.tb00669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brigelius-Flohe R. Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med. 1999;27(9–10):951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi K, Avissar N, Whitin J, Cohen H. Purification and characterization of human plasma glutathione peroxidase: a seleno-glycoprotein distinct from the known cellular enzyme. Arch Biochem Biophys. 1987;256(2):677–686. doi: 10.1016/0003-9861(87)90624-2. [DOI] [PubMed] [Google Scholar]