Abstract
Plasmid RK2 is unusual in its ability to replicate stably in a wide range of Gram-negative bacteria. The replication origin (oriV) and a plasmid-encoded initiation protein (TrfA; expressed as 33 and 44 kDa forms) are essential for RK2 replication. To examine initiation events in bacteria unrelated to Escherichia coli, the genes encoding the replicative helicase, DnaB, of Pseudomonas putida and Pseudomonas aeruginosa were isolated and used to construct protein expression vectors. The purified proteins were tested for activity along with E.coli DnaB at RK2 oriV. Each helicase could be recruited and activated at the RK2 origin in the presence of the host-specific DnaA protein and the TrfA protein. Escherichia coli or P.putida DnaB was active with either TrfA-33 or TrfA-44, while P.aeruginosa DnaB required TrfA-44 for activation. Moreover, unlike the E.coli DnaB helicase, both Pseudomonas helicases could be delivered and activated at oriV in the absence of an ATPase accessory protein. Thus, a DnaC-like accessory ATPase is not universally required for loading the essential replicative helicase at a replication origin.
Keywords: DnaB helicase/initiation of replication/plasmid RK2/replication initiation protein/replication origin
Introduction
The DnaB protein, a member of the hexameric DNA helicase family, is the major replicative DNA helicase in Escherichia coli (LeBowitz and McMacken, 1986). It interacts with a number of proteins, including the bacterial replication proteins DnaA (Marszalek and Kaguni, 1994), DnaC (Wickner and Hurwitz, 1975) and DnaG (Tougu et al., 1994; Lu et al., 1996), and the tau subunit of DNA polymerase (Kim et al., 1996), as well as the plasmid-encoded replication initiation proteins RepA of pSC101 (Datta et al., 1999) and π of R6K (Ratnakar et al., 1996). The E.coli DnaB helicase is essential for replication initiation from the chromosomal origin of replication (oriC) and is present in vivo as a protein complex with six monomers of the DnaC ATPase protein and six ATP molecules (Wickner and Hurwitz, 1975; Lanka and Schuster, 1983). DNA replication initiation at oriC begins with binding of multiple molecules of the bacterial DnaA initiator protein to five 9 bp repeats (DnaA boxes). This binding promotes destabilization of nearby AT-rich sequences, resulting in unwinding of the DNA double helix and the formation of an open complex (Bramhill and Kornberg, 1988; Krause et al., 1997). Entry of the DnaB–DnaC helicase complex to the unwound oriC is required for establishing bidirectional fork movement. This crucial step is not fully understood. It is known that DnaA protein specifically binds to the DnaB protein and recruits the DnaB–DnaC complex to oriC (Marszalek and Kaguni, 1994). During loading, DnaC protein is released with concomitant hydrolysis of ATP (Funnell et al., 1987; Allen and Kornberg, 1991). It has been shown that a cryptic single-stranded DNA (ssDNA) binding activity of DnaC protein is involved in DnaB helicase loading (Learn et al., 1997). Once loaded onto the melted region, DnaB helicase translocates rapidly in the 5′ to 3′ direction on the ssDNA, separating the strands of duplex DNA in its path, in a process that requires ATP hydrolysis (LeBowitz and McMacken, 1986; Yu et al., 1996).
Several plasmids have also been shown to utilize a bacterial host helicase for unwinding plasmid DNA during replication. In the case of the broad host range plasmid RK2, initiation of DNA replication from the plasmid origin, oriV, in E.coli requires binding of both the host DnaA protein and the plasmid-encoded TrfA protein (Konieczny and Helinski, 1997a). DnaA protein binds to four DnaA boxes present at oriV (Konieczny et al., 1997). This cluster of DnaA boxes serves as the site for the initial formation of the DnaA–DnaB–DnaC complex (Pacek et al., 2001). Two forms of the TrfA protein, of 44 and 33 kDa, generated by two in-frame translational start sites are encoded by the plasmid (Kornacki et al., 1984; Shingler and Thomas, 1984). Either form of the TrfA protein binds to five repeats of a highly conserved 17 base sequence (iterons) located between the DnaA boxes and an AT-rich region at oriV (Perri et al., 1991). This binding results in the formation of an open complex at oriV that is enhanced further by the binding of DnaA protein (Konieczny et al., 1997). The DnaA protein alone cannot form an open complex at oriV. As with replication initiation at oriC (Marszalek and Kaguni, 1994), the E.coli DnaB helicase in complex with DnaC is recruited to the plasmid replication origin as a result of a specific DnaA–DnaB interaction (Konieczny and Helinski, 1997a). However, activation of helicase unwinding activity at oriV requires bound TrfA protein (Konieczny and Helinski, 1997a).
A considerable amount is known about the structure and function of the E.coli DnaB helicase (Egelman, 1998). However, little is known about primary helicases in other bacteria. With the recent proliferation of genome sequencing, many dnaB homologs have been discovered and it appears that DnaB-like helicases are common in bacteria, phages and chloroplasts (Leipe et al., 2000). However, to date, only one dnaB gene has been cloned from a bacterium other than E.coli and the corresponding DnaB protein purified and characterized (Kaplan and Steitz, 1999). In this work, we describe the isolation of dnaB genes from two Gram-negative bacteria in which RK2 replicates and is stably maintained: Pseudomonas aeruginosa and Pseudomonas putida. The Pseudomonas dnaB genes were isolated, His6-tagged versions were constructed and the tagged proteins were purified and examined for in vitro activity at the replication origin of RK2. Whereas loading and activation of the E.coli and P.putida DnaB proteins occur with either the 33 or 44 kDa form of the TrfA protein, loading and activation of the P.aeruginosa DnaB protein were found to be dependent on the TrfA-44 protein. In addition, surprisingly, the DnaB proteins of the two Pseudomonas species could be loaded and activated at the RK2 replication origin in the absence of a DnaC-like accessory ATPase protein.
Results
Isolation of dnaB genes from P.putida and P.aeruginosa
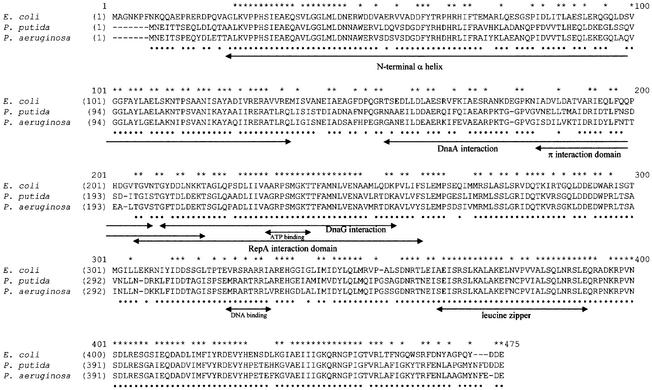
Degenerate primers were used to amplify a conserved portion of the dnaB gene from P.putida chromosomal DNA. For this purpose, the dnaB genes from six Gram-negative bacterial species were aligned and two short well-conserved amino acid sequences were identified (corresponding to positions 235–242 and 407–414 of the E.coli DnaB protein; Figure 1). These conserved sequences were reverse translated and, by making assumptions based on codon frequency in P.putida, the redundancy of the resulting oligonucleotides was reduced. The two designed oligonucleotides were used as PCR primers. One PCR product (pRC30) was found to be 69% identical to the E.coli dnaB gene and, therefore, likely to encode part of the P.putida dnaB gene. The insert of pRC30 was used as a probe against a library of P.putida in the λZap vector to identify a clone, pRC45, which contained the entire dnaB gene, as determined by sequencing.
Fig. 1. Sequence alignment of DnaB proteins from different bacteria. Alignment of the E.coli DnaB protein with the replicative helicases of P.putida and P.aeruginosa was performed using ClustalX. Regions of the E.coli DnaB protein that have been identified or implicated in interactions with other proteins are indicated by arrows: N-terminal α-helix domain (Miles et al., 1997); DNA-binding and leucine zipper (Biswas et al., 1994; Biswas and Biswas, 1999); DnaA interaction (Seitz et al., 2000); DnaG interaction (Lu et al., 1996); and plasmid-encoded Rep protein interaction (R6K π and pSC101 RepA) (Ratnakar et al., 1996; Datta et al., 1999). An asterisk above the alignment indicates residues that are identical in all three proteins; a dot below the alignment indicates residues that are identical between the two Pseudomonas species only. On the basis of the predicted amino acid sequences, the molecular weight of each of the two Pseudomonas DnaB proteins is 59.6 kDa.
The P.putida dnaB sequence was then used to search against the then unfinished P.aeruginosa genome database. A putative dnaB gene with 83.1% identity to P.putida dnaB was found. PCR primers based on this sequence were used to amplify the gene from P.aeruginosa chromosomal DNA, and the resulting PCR product was cloned into vector pBAD24 and the insert DNA sequenced.
The two Pseudomonas genes are quite similar to each other (83% identity at the nucleic acid level) and are more similar to the E.coli dnaB gene (∼60% identity) than to homologous genes from Gram-positive bacteria (52–54% identity; Table I). At the amino acid level, the products of the Pseudomonas genes still retain 92% similarity with each other and around 75% with E.coli DnaB, while the similarity with the homologous proteins from Gram-positive bacteria decreases to 53–62% (Table I). The similarity among the DnaB proteins is not spread evenly across the protein (Figure 1). The domain least conserved among Gram-negative bacteria is a region implicated in interaction with the DnaA protein (Seitz et al., 2000) (Table I).
Table I. Similarity among several dnaB genes.
| E.coli | P.putida | P.aeruginosa | B.subtilis | S.coelicolor | |
|---|---|---|---|---|---|
| E.coli | 100/100/100a | ||||
| P.putida | 61.8/73.7/58.3 | 100/100/100 | |||
| P.aeruginosa | 63.8/76.4/65.0 | 83.1/92.0/79.3 | 100/100/100 | ||
| B.subtilis | 53.5/60.2/50.0 | 53.4/61.7/55.2 | 53.5/63.4/58.6 | 100/100/100 | |
| S.coelicolor | 50.2/55.6/53.3 | 52.2/53.2/51.7 | 54.3/54.1/50.0 | 48.4/58.3/56.1 | 100/100/100 |
aData are presented in the following format: the first number is the percentage identity at the nucleic acid level for the full gene. The second number is the percentage similarity at the amino acid level for the full protein. The third number is the percentage similarity only among the putative DnaA interaction domains of these proteins. The DnaA interaction domain of E.coli has been shown to be between residues 154 and 210 (Seitz et al., 2000), which corresponds to positions 147–201 in P.putida, 147–201 in P.aeruginosa, 136–189 in B.subtilis and 182–235 in S.coelicolor.
Expression and in vivo activity of DnaB proteins
Genes encoding C-terminal His6-tagged fusions of the DnaB proteins from E.coli, P.putida and P.aeruginosa were constructed to facilitate purification of these proteins. The activity of these His6-tagged DnaB proteins was tested in vivo using complementation of E.coli temperature-sensitive DnaB mutant strains E279 and CR34/313. The E.coli His6-tagged DnaB protein, as well as E.coli wild-type DnaB protein (on plasmid pRLM6), complemented both temperature-sensitive mutants, allowing growth at elevated temperatures (42°C), demonstrating that the E.coli His6-tagged DnaB protein is active in vivo (data not shown). Plasmids expressing the P.putida or P.aeruginosa His6-tagged DnaB proteins did not complement the E.coli mutants, indicating that the Pseudomonas DnaB proteins cannot substitute for E.coli DnaB protein for replication of the E.coli chromosome. We were not able to test the in vivo function of the Pseudomonas proteins in their natural host since temperature-sensitive DnaB mutants of Pseudomonas are not available. However, we were able to verify the in vitro activity of these proteins in the various assays described below.
Both E.coli and Pseudomonas helicases show similar unwinding activity in vitro
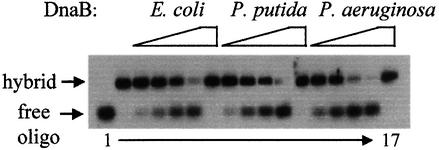
It has been shown that the E.coli DnaB helicase loads on the lagging strand and unwinds DNA with a 5′ to 3′ polarity, and that it can be loaded in vitro on a DNA template provided there is a free single-stranded 3′ tail on the leading strand (LeBowitz and McMacken, 1986). All three His6-tagged DnaB proteins demonstrated ATP-dependent helicase activity, converting a large fraction of the substrate to product during an 8 min reaction period (Figure 2). Substantial differences in enzyme activity were not observed under these conditions or at shorter time periods (data not shown), indicating that the substrate-unwinding activities of the E.coli and Pseudomonas DnaB helicases are similar.
Fig. 2. Strand displacement activity of the E.coli and Pseudomonas sp. helicases. Helicase activity was determined by strand displacement of a radiolabeled synthetic oligonucleotide annealed to phagemid ssDNA. The positions of the hybrid substrate (phagemid ssDNA with annealed oligonucleotide) and the reaction product (free oligonucleotide) are indicated by arrows. Lane 1, hybrid substrate incubated at 95°C for 2 min prior to loading on the gel; lane 2, reaction mix without any source of DnaB; lanes 3–6, the addition of 100, 250, 500 and 1000 ng of E.coli DnaB, respectively; lanes 8–11, 50, 100, 250 and 500 ng of P.putida DnaB, respectively; lanes 13–16, 50, 100, 250 and 500 ng of P.aeruginosa DnaB, respectively. ATP was omitted from the reaction mix containing 1000 ng of E.coli DnaB (lane 7), 500 ng of P.putida DnaB (lane 12) and 500 ng of P.aeruginosa DnaB (lane 17).
Formation of a pre-priming complex by Pseudomonas DnaA and DnaB proteins at the RK2 replication origin does not require DnaC
It is known that E.coli DnaB helicase is delivered to a replication origin in complex with the DnaC accessory protein (Wahle et al., 1989). A DnaA–DnaB–DnaC nucleoprotein complex can be isolated in vitro after incubation of purified proteins with supercoiled oriC plasmid DNA (Marszalek and Kaguni, 1994) or plasmid RK2 oriV DNA (Konieczny and Helinski, 1997a). In view of the broad host range of RK2, the RK2 origin should be functional for recruiting, loading and activation of the Pseudomonas helicases, which allows for a comparison of the delivery mechanism of helicase isolated from different bacterial species.
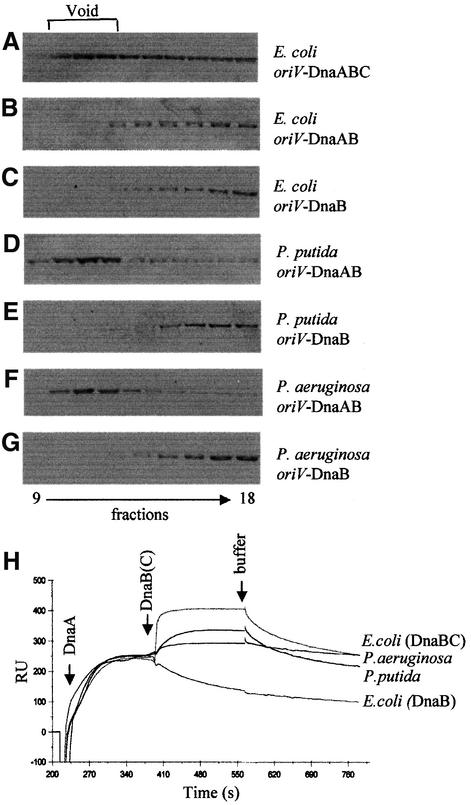
Gel filtration chromatography was used to analyze complexes formed at oriV with the replication initiation proteins from E.coli and the two Pseudomonas species. Similarly to previously published results (Konieczny and Helinski, 1997a), E.coli DnaB helicase formed a complex with the oriV supercoiled DNA template, which is present in the void fractions, only in association with the E.coli accessory ATPase, DnaC, and in the presence of E.coli DnaA (Figure 3A–C). In contrast, the P.putida as well as P.aeruginosa DnaB helicase was present in the void fractions when the incubation mixtures included P.putida or P.aeruginosa DnaA protein, respectively, and no helicase accessory protein (Figure 3D and F). As can be seen in Figure 3E and G, the Pseudomonas helicases do not form a stable complex with supercoiled oriV DNA if the DnaA protein is not present in the incubation mixture.
Fig. 3. An accessory protein is not required for the formation of Pseudomonas pre-priming complexes at the RK2 origin. The formation of E.coli and Pseudomonas helicase complexes at oriV was analyzed by gel exclusion chromatography on Sepharose CL4B. Reactions contained 1200 ng of RK2 supercoiled pTJS42 template and: (A) E.coli DnaA (1300 ng), DnaB (2400 ng) and DnaC (450 ng); (B) E.coli DnaA (1300 ng) and DnaB (2400 ng); (C) E.coli DnaB (2400 ng) only; (D) P.putida DnaA (1300 ng) and DnaB (2400 ng); (E) P.putida DnaB (2400 ng) only; (F) P.aeruginosa DnaA (1300 ng) and DnaB (2400 ng); and (G) P.aeruginosa DnaB (2400 ng) only. Fractions from the column were analyzed by SDS–PAGE followed by western blotting. (H) SPR analyses of pre-priming complex formation with E.coli and Pseudomonas proteins using a 64 bp double-stranded linear DNA fragment containing the four DnaA box sequences of RK2 oriV. Pseudomonas putida DnaA (15 ng) and DnaB (60 ng); P.aeruginosa DnaA (15 ng) and DnaB (60 ng); E.coli DnaA (15 ng) and DnaB (60 ng); and E.coli DnaA (15 ng), DnaB (60 ng) and DnaC (10 ng) were injected over the sensor matrix and tested for binding. Arrows indicate the time of protein or buffer injections.
A comparison of the binding activity of the E.coli and Pseudomonas sp. helicases by surface plasmon resonance (SPR) analysis (Figure 3H) produced similar results. A DNA fragment containing the exact sequence of the four DnaA boxes present at oriV was immobilized on a Biosensor chip surface and proteins were allowed to form complexes that were detected as a change of plasmon resonance, which is proportional to a change of mass. After subsequent protein injections, buffer was passed through the sensor channel, which allowed observation of real-time dissociation of the previously formed nucleoprotein complexes. The Pseudomonas DnaB proteins once again formed a DnaA–DnaB complex in the absence of DnaC. No significant protein binding was observed during control experiments using a DNA fragment without a DnaA box consensus sequence (not shown). In contrast, the E.coli helicase associated with the DnaA–DnaA box complex only in the presence of DnaC.
As shown previously, the affinity of DnaA proteins from E.coli, P.putida and P.aeruginosa for the DnaA boxes is not substantially different (Caspi et al., 2000). However, real-time kinetics by SPR showed variances in helicase binding to the pre-formed DnaA–DnaA box nucleoprotein complex (Figure 3H). DnaB from P.putida and DnaB– DnaC from E.coli showed a relatively high binding affinity. While the response signal seen with the addition of the P.aeruginosa DnaB was relatively low, it was reproducible and significant since (as with all three proteins) the response represents both DnaA dissociation from the DNA (Caspi et al., 2000) and helicase association. The dissociation observed after formation of the helicase complexes and buffer injection differs substantially when comparing the proteins from the three organisms. Eschericha coli DnaA–DnaB–DnaC and P.putida DnaA–DnaB demonstrated a similar dissociation pattern, while the P.aeruginosa DnaA–DnaB complex appeared to be more stable under the conditions employed.
Pseudomonas DnaB helicases do not form a stable complex with the E.coli DnaC accessory protein
We have shown previously that the DnaA proteins of P.putida and P.aeruginosa will not interact with E.coli DnaB–DnaC to form a pre-priming complex at the RK2 origin (Caspi et al., 2000). The availability of DnaB proteins from the two Pseudomonas species allowed us to determine if there was similar specificity in DnaB complex formation with E.coli DnaC. While it is known that the E.coli DnaB hexamer is tightly complexed with E.coli DnaC to form a hetero-hexamer (Wahle et al., 1989; San Martin et al., 1995), specific protein domains responsible for the DnaB–DnaC interaction have not been mapped precisely.
After incubation of E.coli DnaC with either of the two Pseudomonas DnaB proteins followed by chromatography on Bio-Gel 0.5 M agarose, we were not able to detect a DnaB–DnaC complex in the void fractions (Figure 4B and C). In contrast, a stable complex of E.coli DnaB with E.coli DnaC was found in the void fractions (Figure 4A). Incubation of DnaC alone did not result in the presence of the protein in the void volume (Figure 4D). This demonstrates that neither the P.putida nor the P.aeruginosa helicase forms a stable complex with E.coli DnaC, indicating species specificity in the interaction.
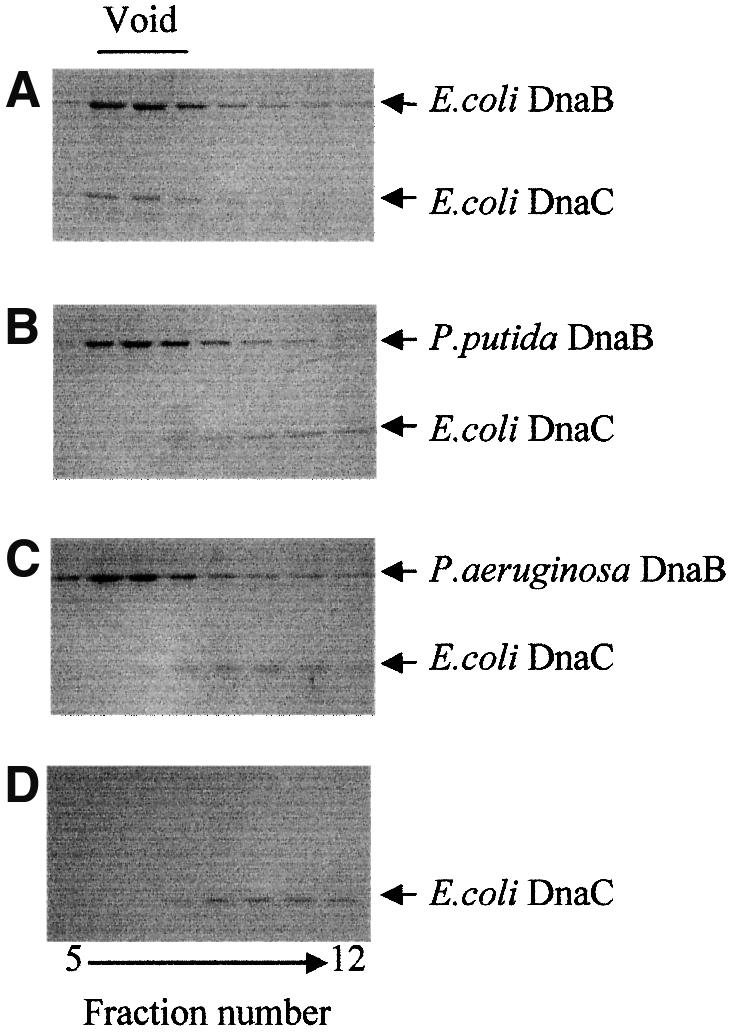
Fig. 4. Escherichia coli DnaC does not form a stable complex with the Pseudomonas DnaB helicases. Fractions eluting from a Bio-Gel 0.5 M column were analyzed for the presence of DnaC and DnaB proteins by SDS–PAGE followed by Coomassie Blue staining. Incubation mixtures contained: (A) E.coli DnaB (2400 ng) and DnaC (450 ng); (B) P.putida DnaB (2400 ng) and E.coli DnaC (450 ng); (C) P.aeruginosa DnaB (2400 ng) and E.coli DnaC (450 ng); and (D) E.coli DnaC (450 ng) alone.
In addition to testing species specificity, another rationale for carrying out this analysis was to test for the possibility of contamination of the purified Pseudomonas DnaB preparations with small amounts of bound E.coli DnaC as all three His6-tagged DnaB proteins were purified after being overexpressed in E.coli strain JP313. Con tamination with E.coli DnaC could account for the ability of the Pseudomonas DnaB preparations to form a pre-priming complex at RK2 oriV in the absence of added purified DnaC (Figure 3). Thus, in addition to our inability to immunodetect E.coli DnaC protein in the purified Pseudomonas DnaB preparations (data not shown), the failure of the Pseudomonas DnaB proteins to form a stable complex with E.coli DnaC protein further argues against this possibility.
Loading and unwinding activity of a Pseudomonas helicase is not dependent on an accessory protein
Escherichia coli DnaB helicase activity in vitro on a supercoiled oriV DNA template in the presence of TrfA, E.coli DnaA, DnaC, gyrase, SSB and HU proteins, and ATP results in the formation of a substantially unwound form of supercoiled DNA, designated FI* (Konieczny and Helinski, 1997a). FI* formation is a result of origin opening, helicase loading at the open region and extensive unwinding of the DNA template, and can be detected by agarose gel electrophoresis (Baker et al., 1986). As shown in Figure 5A, the formation of the FI* product by P.putida DnaB in the presence of the P.putida DnaA protein and the 33 kDa TrfA protein (Figure 5A, lane 3) occurred in the absence of a DnaC-like accessory protein. The unwinding activity exhibited by P.putida DnaB requires the presence of TrfA, P.putida DnaA and E.coli gyrase (Figure 5B). The P.aeruginosa DnaB protein, however, in the presence of the P.aeruginosa DnaA protein and the 33 kDa TrfA protein failed to unwind supercoiled oriV DNA in the presence (data not shown) or absence of E.coli DnaC (Figure 5A, lane 4).
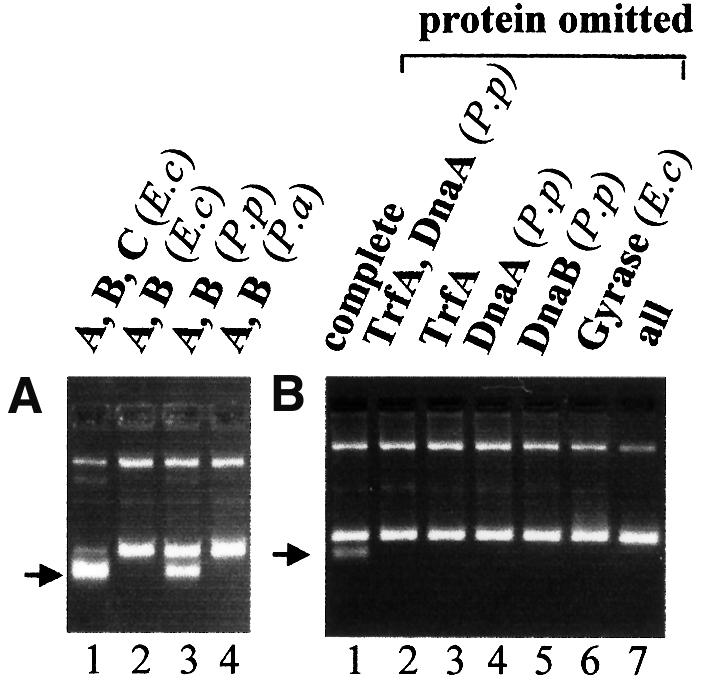
Fig. 5. Requirements for loading and activation of E.coli, P.putida and P.aeruginosa DnaB helicases at the RK2 replication origin. Requirements for helicase-dependent DNA unwinding of supercoiled plasmid DNA containing the RK2 minimal replication origin were tested using the FI* assay. The position of FI*, the extensively unwound covalently closed circular DNA generated by helicase activity, is marked by the arrow. Bands migrating more slowly than FI* (from top to bottom) are: FIII, open circular DNA; FII, linear DNA (faint); and FI, covalently closed circular DNA. All reactions included the supercoiled form of plasmid pTJS42, E.coli DNA gyrase, E.coli SSB, E.coli HU and creatine kinase, except (B), where the component(s) indicated was omitted. (A) The reaction mixture contained the 33 kDa form of TrfA and: lane 1, E.coli DnaA, DnaB and DnaC; lane 2, E.coli DnaB and DnaC; lane 3, P.putida DnaA and DnaB; and lane 4, P.aeruginosa DnaA and DnaB. (B) The reactions contained TrfA-33, P.putida DnaA and DnaB, and E.coli DNA gyrase plus other components of the standard reaction mixture unless an omission is indicated.
Pseudomonas aeruginosa helicase loading and activation differs in its requirement for the plasmid-encoded replication protein TrfA
The experiments described in the previous sections used the 33 kDa form of the TrfA protein. It has been shown in vivo that while in E.coli and P.putida there is little difference in activity between the two forms of the TrfA protein (44 and 33 kDa), in P.aeruginosa there is strong preference for the longer version of the TrfA protein (Shingler and Thomas, 1984; Durland and Helinski, 1987; Fang and Helinski, 1991). In view of these in vivo studies and the failure of TrfA-33 to activate the P.aeruginosa helicase (Figure 5A), the 44 kDa form of TrfA was tested using the FI* assay. As shown in Figure 6, the E.coli DnaA–DnaB–DnaC- and the P.putida DnaA– DnaB-dependent reactions resulting in FI* formation occurred with either TrfA-33 or TrfA-44. In contrast, P.aeruginosa DnaB is loaded and/or activated at the melted oriV region only when TrfA-44 is present in the reaction mixture (Figure 6B, lanes 6–9). No activity was observed in experiments using TrfA-33 over a wide range of concentrations of the P.aeruginosa helicase (data not shown).
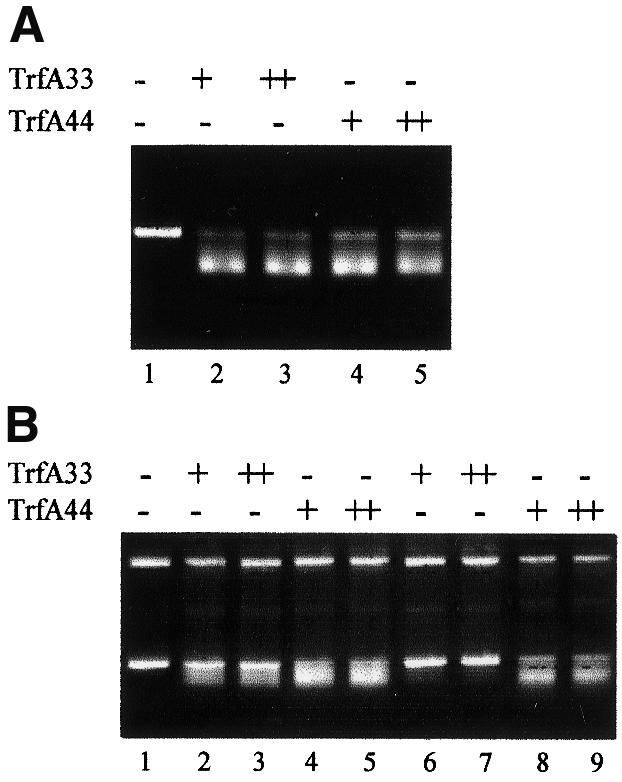
Fig. 6. TrfA requirements for loading and activation of DnaB helicases at the RK2 replication origin. The FI* assay was used to test the requirements for the two forms of the TrfA protein for loading and activation of the E.coli, P.putida and P.aeruginosa DnaA and DnaB proteins at the RK2 replication origin on plasmid pKD19L1. TrfA-33 or TrfA-44 was added at 100 (+) or 250 ng (++) to reactions as noted. (A) Escherichia coli DnaA and DnaB proteins with no TrfA protein (lane 1), TrfA-33 (lanes 2 and 3) or TrfA-44 (lanes 4 and 5). (B) Lanes 1–5, P.putida DnaA and DnaB proteins with no TrfA (lane 1) or increasing amounts of TrfA-33 (lanes 2 and 3) or TrfA-44 protein (lanes 4 and 5). Lanes 6–9, P.aeruginosa DnaA and DnaB proteins and increasing amounts of TrfA-33 (lanes 6 and 7) or TrfA-44 (lanes 8 and 9).
To determine whether the requirement for TrfA-44 for FI* formation by P.aeruginosa DnaB is due to direct involvement of TrfA protein in helicase loading and/or activation, or due to a difference in open complex formation by TrfA-33 and TrfA-44 and the P.aeruginosa DnaA protein, strand opening activity with the two forms of the TrfA protein and the DnaA proteins from E.coli, P.putida and P.aeruginosa was determined. As shown in Figure 7, no difference in opening activity by the two forms of the TrfA protein was observed with any of the three DnaA proteins used. This result indicates that the inactivity of TrfA-33 in FI* formation is not due to the inability of this protein to form an open complex in the presence of the P.aeruginosa DnaA protein, but is due to its inactivity in the loading and activation of the P.aeruginosa DnaB protein.

Fig. 7. Formation of an open complex at the RK2 replication origin by both forms of the TrfA protein and the E.coli or Pseudomonas DnaA protein. DnaA protein (250 ng) from E.coli, P.putida or P.aeruginosa was incubated as indicated with 300 ng of supercoiled plasmid pKD19L1 in the presence of E.coli HU protein (5 ng) and TrfA-33 (500 ng) (lanes 2–5) or TrfA-44 (500 ng) (lanes 6–9). Bands indicate polymerase stops due to KMnO4 modifications and cleavage of pyrimidines in the ssDNA of the open region. Lane 1, TrfA and DnaA were not added to the reaction mix; lanes 2 and 6, DnaA protein was not added to the reaction mix.
Discussion
Different requirements for helicase recruitment and activation
The broad host range plasmid RK2 has the ability to transfer into and be stably maintained in >30 Gram-negative bacterial species (Thomas and Helinski, 1989). It previously was not known whether the mechanism of RK2 replication initiation was similar in different bacterial hosts. This work demonstrates that while the overall sequence of replication events is the same, the replication initiation mechanism differs in each of the three bacteria examined. In the case of the Pseudomonas DnaB helicases, the recruitment of each helicase to the RK2 replication origin and the activation of the helicase at that origin occur in the absence of an accessory protein. Furthermore, whereas either of the two forms of the RK2 initiation protein, TrfA, is functional with the DnaA and DnaB initiation proteins of E.coli or P.putida, the 44 kDa form alone is active with these proteins from P.aeruginosa.
The TrfA initiation protein is not required for the initial recruitment of the DnaB helicase. This is in contrast to what was found for the narrow host range plasmids R6K (Lu et al., 1996) and pSC101 (Datta et al., 1999) where the replication initiators π and RepA, respectively, are required for helicase recruitment. It has been known that the 44 kDa TrfA protein is required for stable replication of RK2 in P.aeruginosa, while either the 44 or the 33 kDa forms of the TrfA protein are functional in E.coli and P.putida (Shingler and Thomas, 1984; Durland and Helinski, 1987; Fang and Helinski, 1991). In this study, we show that the formation of an open complex at RK2 oriV in the presence of any of the three DnaA proteins occurs with either of the two forms of the TrfA protein (Figure 7). However, the formation of a pre-priming complex at the RK2 origin, as indicated by FI* formation (Figures 5 and 6), requires the 44 kDa form of the TrfA protein in the case of the DnaA and DnaB proteins of P.aeruginosa. This result indicates that the requirement specifically for TrfA-44 in P.aeruginosa occurs at the level of helicase loading and/or activation, possibly involving direct protein–protein interactions. Thus, the RK2 replicon, at least in vitro, functions without a DnaC-like accessory ATPase when Pseudomonas DnaA and DnaB proteins are utilized. Furthermore, the RK2 replicon shows a differential requirement for the two forms of the TrfA protein depending on which of the two Pseudomonas species is the source of the DnaA and DnaB proteins.
Homologous and analogous proteins of DnaB and DnaC
Unlike dnaB homologs, homologous dnaC genes are not found readily in other organisms. Some enterobacteria such as Salmonella typhimurium and Klebsiella pneumoniae possess genes similar to E.coli dnaC (77 and 67.3% identity, respectively; data not shown). Similar genes, however, are missing from many other bacteria whose genomes have been sequenced in their entirety, raising the question of whether or not DnaC is a universal replication protein. It should be noted, however, that there are many examples in both prokaryotes and eukaryotes of proteins that perform analogous functions at replication forks without sharing much sequence homology (Baker and Bell, 1998; Leipe et al., 1999).
The analysis of DNA replication of the E.coli chromosome, bacterial plasmids, viruses and eukaryotic chromosomes reveals a number of analogous events and similarities in replisome architecture. It has been demonstrated for many systems that initiation proteins, including the bacterial DnaA protein, phage λO protein, SV40 T antigen and the eukaryotic origin recognition complex (ORC), assemble large complexes at their origins that serve as platforms for all subsequent initiation events (Baker and Bell, 1998). Binding of a replication protein(s) to the replication origin facilitates origin opening at an AT-rich region to provide the entry site for a helicase, which unwinds DNA and provides an extended open region for the polymerization reaction. In E.coli, the DnaC and λP accessory ATPases, which share no sequence similarity, are involved in DnaB helicase recruitment to the E.coli origin and the replication origin of bacteriophage λ, respectively. The gp59 protein plays a similar role as a helicase loading factor for T4 gp41 helicase (Kreuzer and Morrical, 1994) as does the DnaI protein in Bacillus subtilis (Imai et al., 2000). Cdc6 has been proposed to recruit the MCM complex in Xenopus and Saccharomyces (Baker and Bell, 1998). The amino acid sequences of these proteins are not similar.
Our results demonstrate that even though the E.coli and Pseudomonas helicases show 60% identity, the E.coli DnaC protein does not form a stable complex with the helicases of P.putida or P.aeruginosa (Figure 4). It is still not known with certainty which regions of the E.coli DnaB protein interact with DnaC. The results of this study suggest that it is a region with relatively low homology to the Pseudomonas DnaB proteins. It has been observed that overexpression of DnaC can overcome the temperature-sensitive phenotype of the mutant DnaB252 in vivo (Sclafani and Wechsler, 1981). This mutation was traced to position 299, changing it from glycine to aspartic acid (Saluja and Godson, 1995). It is, therefore, possible that amino acid residue 299 is part of the DnaB–DnaC inter action domain of the E.coli DnaB protein. Interestingly, residue 299 and the region surrounding it are highly divergent in different DnaB proteins. This suggests that there would be little sequence conservation of putative DnaC proteins in different bacteria.
Helicase recruitment: is there a need for an accessory ATPase?
It is known that the E.coli DnaB helicase has an intrinsic ATP-dependent ssDNA-binding activity; however, this activity alone is not sufficient for helicase loading at the replication origin of the E.coli chromosome. Additionally, E.coli DnaC protein is essential for E.coli helicase recruitment at several plasmid origins, including RK2 (Konieczny and Helinski, 1997a), R6K (Lu et al., 1998) and pSC101 (Datta et al., 1999). Furthermore, during bacteriophage λ DNA replication, the bacteriophage protein λP binds the E.coli DnaB helicase and, via interaction with the λ replication initiation protein λO, delivers the helicase to the λ origin (Dodson et al., 1985). Thus, in these cases, the E.coli DnaB helicase requires an accessory ATPase for delivery to the open initiation site.
Since E.coli has been used traditionally as a model organism, it has been assumed that an accessory protein is a universal requirement in all prokaryotes, even though there is no supporting evidence from other organisms. A similar requirement has been proposed for replication initiation in eukaryotic systems (Baker and Bell, 1998). Our results, however, question a universal requirement for helicase recruitment. These studies demonstrate that although the E.coli and Pseudomonas helicases share >60% homology, the recruitment and loading mechanisms at the origin of plasmid RK2 are different. In contrast to E.coli DnaB, the P.putida and P.aeruginosa DnaB proteins form a DnaA–DnaB complex on double-stranded DNA in the absence of a DnaC homolog (Figure 3). Our studies demonstrated that not only are DnaA–DnaB complexes formed at oriV with the Pseudomonas proteins, but also the Pseudomonas helicases are active in DNA unwinding, which indicates loading at ssDNA in the absence of DnaC. Whether this novel, accessory protein-independent mechanism for Pseudomonas primary replicative helicase recruitment and loading is true for the chromosomal origins of the two Pseudomonas strains is currently being investigated.
Materials and methods
Bacterial strains and growth
Escherichia coli strain XL1-Blue (Stratagene) was used throughout as the host for gene cloning. The E.coli DnaB temperature-sensitive mutants E279 (dnaB279) and CR34/313 (dnaB313) (obtained from Dr R.McMacken) were used to test functionality of the His6-tagged DnaB proteins in vivo. Other E.coli strains used are as noted. All bacteria were grown on LB medium. Antibiotics were 250 µg/ml penicillin or 100 µg/ml ampicillin.
Isolation and cloning of the P.putida and P.aeruginosa dnaB genes
A conserved region of the dnaB gene was amplified from chromosomal DNA of P.putida mt2 (Nieto et al., 1990) by PCR using degenerate oligonucleotide primers dnaB5′ (5′-CCCCCGGGATGGGIAARACSACSTTYGCIATG-3′), dnaB3′ (5′-CTTCTAGAARRTCIGCRTCYTGYTCRATIS-3′) and Taq 2000 polymerase (Stratagene). The PCR amplification resulted in two products, which were cloned separately into pBluescript II SK– (Stratagene) using SmaI and XbaI, and then sequenced. The larger product (545 bp in plasmid pRC30) was homologous to E.coli dnaB.
The insert of pRC30 was used as a probe in a Southern hybridization against a genomic library of P.putida mt2, prepared in λZap (Stratagene). Positive plaques were isolated and excised according to the manufacturer’s instructions, and analyzed by restriction enzyme cleavage, Southern hybridization and, lastly, DNA sequencing at both ends of the insert. One construct, pRC45, had an insert with sequences corresponding to the N- and C-terminal ends of P.putida DnaB, consistent with the presence of a complete gene. This insert was fully sequenced in both directions.
For the purpose of purification of the DnaB protein, the P.putida dnaB gene in plasmid pRC45 was amplified by PCR, using the primers PPdnaBHis5′ (5′-AACGAGATCACCACCTCCGAA-3′) and PPdnaB3′ (5′-GCTCTAGATCAGTGGTGGTGGTGGTGGTGTTCATCATCATCAAAGTTGTA-3′) prior to cloning into the pBAD24 expression vector (Guzman et al., 1995) as described below.
The P.aeruginosa dnaB gene was identified by BLAST searches using the sequence of the P.putida dnaB gene against an unfinished P.aeruginosa genome database. PCR primers were designed based on the putative reading frame, and the gene was amplified from chromosomal DNA of P aeruginosa PA0116 (Isaac and Holloway, 1968) using the primers PAdnaBHis5′ (5′-AACGAGATCACCAGCCCCGAGCAATA-3′) and PAdnaBHis3′ (5′-GCTCTAGATCAGTGGTGGTGGTGGTGGTGTTCGTCCTCGAAGTTGTACAT-3′).
The E.coli dnaB gene, cloned in plasmid pRLM6 (kindly provided by Dr R.McMacken), was amplified by PCR from this plasmid using the primers ECdnaBHis5′ (5′-GCAGGAAATAAACCCTTCAAC-3′) and ECdnaBHis3′ (5′-GCTCTAGATCAGTGGTGGTGGTGGTGGTGTTCGTCGTCGTACTGCGGCCC-3′). In all three cases, P.putida, P.aeruginosa and E.coli, the 5′ primers started at the fourth nucleotide of the gene, and the 3′ primers added codons for six histidine residues immediately upstream of the stop codon, which was followed by an XbaI restriction site. All PCRs for cloning purposes were performed using high fidelity Pfu DNA polymerase (Stratagene).
The PCR products of the dnaB genes of E.coli, P.putida and P.aeruginosa were cleaved with the restriction endonuclease XbaI and cloned into the vector pBAD24, which was prepared as described (Guzman et al., 1995), resulting in plasmids pRC50, pRC52 and pRC58, respectively. The inserts in these plasmids were verified by sequencing to ensure that no errors occurred during PCR.
The dnaB sequence data have been submitted to the GenBank database under accession Nos AF229444 for the P.aeruginosa gene and AF229443 for the P.putida gene.
Purification of His6-tagged DnaB proteins
C-terminal His6-tagged DnaB proteins from E.coli, P.putida and P.aeruginosa were overexpressed in E.coli JP313 (Economou et al., 1995). One liter cultures were grown with selection at 37°C to an OD of 0.6 (at 600 nm) and l-arabinose was added to a final concentration of 0.2% to induce expression. Cells were grown for an additional 4 h before harvesting. The cell pellet was resuspended in 3 ml of lysis buffer (50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 10 mM imidazole) and lysed by sonication. DnaB-His6 protein was purified on Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions. Following elution with 0.5 ml of 50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 250 mM imidazole and 2 mM ATP, the protein was dialyzed against 2 l of 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 2 mM ATP and 20% glycerol, and stored at –70°C.
Other proteins
C-terminal His6-tagged DnaA proteins from E.coli, P.putida and P.aeruginosa were purified as described (Caspi et al., 2000). The monomeric mutant form of the 33 kDa TrfA protein, His6-TrfA(G254D/S267L), was purified as described (Blasina et al., 1996). This protein, used in previous RK2 biochemical analyses, initiates replication in vitro with kinetics similar to those of the largely dimeric wild-type protein (Konieczny and Helinski, 1997b), but, unlike the wild-type protein, is present largely in the form of a monomer, which is the active form of TrfA for binding to the iterons at the origin (Toukdarian et al., 1996). The 44 kDa version of this protein, His6-TrfA44(M98L/G254D/S267L), was purified after l-arabinose induction of expression from plasmid pGC1, which contains a gene encoding the His6 fusion protein carrying the M98L/G254D/S267L mutations under the control of the pBAD promoter. The N-terminal His6-tagged G254D/S267L copy-up mutant forms of TrfA-33 and TrfA-44 were used throughout this study.
Escherichia coli DnaC protein was purified as previously described (Konieczny and Helinski, 1997a). Commercially available proteins were HU, SSB and DNA gyrase from Enzyco, Inc., creatine kinase and bovine serum albumin (BSA) (fraction V) from Sigma, Inc., and DNA restriction and modification enzymes from various commercial sources.
Helicase strand displacement assay
Single-strand pBluescript II SK– phagemid DNA was prepared according to the manufacturer’s instructions. A synthetic 60mer oligonucleotide (5′-ACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGTTAATACGTCACGACGGTATTGG-3′) was labeled at the 3′ end with [α-32P]ATP by terminal transferase. The first 37 bases of this oligonucleotide are complementary to bases 2479–2515 of the phagemid, while the last 23 bases form a free 3′ tail upon hybridization. Phagemid DNA and labeled oligonucleotide were combined at a molar ratio of 20:1. The mixture was heated to 95°C for 2 min, placed at 65°C for 15 min, followed by 1 h at 42°C, and then allowed to cool slowly to room temperature. Free oligonucleotide was removed from the hybrid substrate using a Sephacryl-400 spin column (Promega). Increasing amounts of helicase were added to a reaction mix containing 40 mM HEPES–KOH pH 7.6, 150 mM NaCl, 11 mM magnesium acetate, 50 µg/µl BSA, 3.4 mM ATP and 0.15 pmol of substrate in a total volume of 15 µl. Reactions were incubated at 30°C for 8 min and then terminated by the addition of 5 µl of stop solution (1% SDS, 50 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol FF and 40% glycerol). Samples were loaded onto a 1% neutral agarose gel and electrophoresed (LeBowitz and McMacken, 1986).
Helicase unwinding assay
Helicase unwinding assays (FI* formation) were performed as previously described (Konieczny and Helinski, 1997a) using 300 ng of either plasmid pTJS42 (Schmidhauser et al., 1983) or pKD19L1 (Doran et al., 1998) as the DNA template. Unless noted otherwise, proteins were added at: DnaA (320 ng), DnaB (600 ng), DnaC (120 ng), 33 kDa TrfA (G254D/S267L) (500 ng), 44 kDa TrfA (G254D/S267L) (500 ng), HU (5 ng), gyrase (120 ng) and SSB (230 ng).
Gel exclusion chromatography
Column gel filtration was used to isolate helicase pre-priming complexes as described previously (Konieczny and Helinski, 1997a). After incubation, the reactions containing the specified proteins and plasmid pTJS42 were run through a Sepharose CL-4B column. Fractions were collected and analyzed by SDS–PAGE, followed by a protein transfer and immunoblotting with rabbit antiserum specific against E.coli DnaB protein. Bound rabbit antibody was detected by a colorimetric reaction with a horseradish peroxidase-conjugated goat anti-rabbit IgG.
Analysis of the interaction of E.coli DnaC with the three helicases was performed as described in Konieczny and Marszalek (1995). Reaction mixtures containing the specified proteins were incubated at 30°C for 10 min, applied to a Bio-Gel 0.5 M column, followed by analysis of the fractions by SDS–PAGE and Coomassie Blue staining.
Surface plasmon resonance (SPR)
Standard SPR analyses were performed as previously described (Caspi et al., 2000) using a double-stranded 64 bp linear fragment containing the four DnaA boxes from oriV immobilized on a streptavidin matrix-coated Sensor Chip S.A. (BiaCore). SPR analysis was performed in a BiaCore 3000 instrument by injecting 10 µl of protein solutions in binding buffer (40 mM HEPES–KOH pH 8.0, 25 mM Tris–HCl pH 7.4, 80 µg/ml BSA, 4% sucrose, 4 mM dithiothreitol, 11 mM magnesium acetate and 2 mM ATP) for 2 min at room temperature. Protein injections were followed by SPR buffer (10 mM HEPES–KOH pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20). The results are presented as the sensogram obtained after subtraction of the background response signal (obtained in control experiments). For control of non-specific protein–DNA interaction, complementary oligonucleotides with scrambled DnaA box sequences were synthesized, annealed and analyzed. No significant response signal was detected in the control experiments.
Strand-opening assay
The strand-opening assay based on permanganate (KMnO4) footprinting followed by primer extension using PCR and electrophoresis of the products on a denaturing acrylamide gel was performed as described previously (Doran et al., 1998).
Computer analysis
DNA and protein sequence analysis and alignments were performed with the programs VectorNTI (Informax) and ClustalX (Thompson et al., 1997). Sequence searches were performed over the Internet using BLAST and gapped-BLAST (Altschul et al., 1997).
Acknowledgments
Acknowledgements
We thank Drs E.Biswas and S.Biswas for advice on helicase assay template preparation. We are grateful to Drs L.Burns and S.Taylor for their advice and assistance with the SPR analyses. This work was supported by National Institutes of Health Research Grant AI-07194 to D.R.H., a Polish State Committee for Scientific Research Grant 6P04A01115 to I.K. and a US–Polish Maria Sklodowska-Curie Fund II Grant 98-349.
References
- Allen G.C. and Kornberg,A. (1991) Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J. Biol. Chem., 266, 22096–22101. [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Scheaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T.A. and Bell,S.P. (1998) Polymerases and the replisome: machines within machines. Cell, 92, 295–305. [DOI] [PubMed] [Google Scholar]
- Baker T.A., Sekimizu,K., Funnell,B.E. and Kornberg,A. (1986) Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell, 45, 53–64. [DOI] [PubMed] [Google Scholar]
- Biswas E.E. and Biswas,S.B. (1999) Mechanism of DNA binding by the DnaB helicase of Escherichia coli: analysis of the roles of domain γ in DNA binding. Biochemistry, 38, 10929–39. [DOI] [PubMed] [Google Scholar]
- Biswas S.B., Chen,P.H. and Biswas,E.E. (1994) Structure and function of Escherichia coli DnaB protein: role of the N-terminal domain in helicase activity. Biochemistry, 33, 11307–14. [DOI] [PubMed] [Google Scholar]
- Blasina A., Kittell,B.L., Toukdarian,A.E. and Helinski,D.R. (1996) Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc. Natl Acad. Sci. USA, 93, 3559–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D. and Kornberg,A. (1988) Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E.coli chromosome. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- Caspi R., Helinski,D.R., Pacek,M. and Konieczny,I. (2000) Interactions of DnaA proteins from distantly related bacteria with the replication origin of the broad host range plasmid RK2. J. Biol. Chem., 275, 18454–18461. [DOI] [PubMed] [Google Scholar]
- Datta H.J., Khatri,G.S. and Bastia,D. (1999) Mechanism of recruitment of DnaB helicase to the replication origin of the plasmid pSC101. Proc. Natl Acad. Sci. USA, 96, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Roberts,J., McMacken,R. and Echols,H. (1985) Specialized nucleoprotein structures at the origin of replication of bacteriophage λ: complexes with λO protein and with λO, λP and Escherichia coli DnaB proteins. Proc. Natl Acad. Sci. USA, 82, 4678–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran K.S., Konieczny,I. and Helinski,D.R. (1998) Replication origin of the broad host range plasmid RK2: positioning of various motifs is critical for initiation of replication. J. Biol. Chem., 273, 8447–8453. [DOI] [PubMed] [Google Scholar]
- Durland R.H. and Helinski,D.R. (1987) The sequence encoding the 43-kilodalton trfA protein is required for efficient replication or maintenance of minimal RK2 replicons in Pseudomonas aeruginosa. Plasmid, 18, 164–9. [DOI] [PubMed] [Google Scholar]
- Economou A., Pogliano,J.A., Beckwith,J., Olover,D.B. and Wickner,W. (1995) SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell, 83, 1171–1181. [DOI] [PubMed] [Google Scholar]
- Egelman E.H. (1998) Bacterial helicases. J. Struct. Biol., 124, 123–128. [DOI] [PubMed] [Google Scholar]
- Fang F.C. and Helinski,D.R. (1991) Broad-host-range properties of plasmid RK2: importance of overlapping genes encoding the plasmid replication initiation protein TrfA. J. Bacteriol., 173, 5861–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B.E., Baker,T.A. and Kornberg,A. (1987) In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem., 262, 10327–10334. [PubMed] [Google Scholar]
- Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Ogasawara,N., Ishigo-Oka,D., Kadoya,R., Daito,T. and Moriya,S. (2000) Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids. Mol. Microbiol., 36, 1037–48. [DOI] [PubMed] [Google Scholar]
- Isaac J.H. and Holloway,B.W. (1968) Control of pyrimidine biosynthesis in Pseudomonas aeruginosa. J. Bacteriol., 96, 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.L. and Steitz,T.A. (1999) DnaB from Thermus aquaticus unwinds forked duplex DNA with an asymmetric tail length dependence. J. Biol. Chem., 274, 6889–97. [DOI] [PubMed] [Google Scholar]
- Kim S.S., Dallmann,H.G., McHenry,C.S. and Marians,K.J. (1996) Coupling of a replicative polymerase and helicase—a tau–DnaB interaction mediates rapid replication fork movement. Cell, 84, 643–650. [DOI] [PubMed] [Google Scholar]
- Konieczny I. and Helinski,D.R. (1997a) Helicase delivery and activation by DnaA and TrfA proteins during the initiation of replication of the broad host range plasmid RK2. J. Biol. Chem., 272, 33312–33318. [DOI] [PubMed] [Google Scholar]
- Konieczny I. and Helinski,D.R. (1997b) The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc. Natl Acad. Sci. USA, 94, 14378–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny I. and Marszalek,J. (1995) The requirement for molecular chaperones in λ DNA replication is reduced by the mutation π in λP gene, which weakens the interaction between λP protein and DnaB helicase. J. Biol. Chem., 270, 9792–9799. [DOI] [PubMed] [Google Scholar]
- Konieczny I., Doran,K.S., Helinski,D.R. and Blasina,A. (1997) Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J. Biol. Chem., 272, 20173–20178. [DOI] [PubMed] [Google Scholar]
- Kornacki J.A., West,A.H. and Firshein,W. (1984) Proteins encoded by the trans-acting replication and maintenance regions of broad host range plasmid RK2. Plasmid, 11, 48–57. [DOI] [PubMed] [Google Scholar]
- Krause M., Reuckert,B., Lurz,R. and Messer,W. (1997) Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol., 274, 365–380. [DOI] [PubMed] [Google Scholar]
- Kreuzer K.N. and Morrical,S.W. (1994) Initiation of DNA replication. In Karam,J.D. (ed.), Molecular Biology of Bacteriophage T4. ASM Press, Washington, DC, pp. 28–42.
- Lanka E. and Schuster,H. (1983) The DnaC protein of Escherichia coli. Purification, physical properties and interaction with DnaB protein. Nucleic Acids Res., 11, 987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learn B.A., Um,S.-J., Huang,L. and McMacken,R. (1997) Cryptic single-stranded-DNA binding activities of the phage λP and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E.coli DnaB helicase onto DNA. Proc. Natl Acad. Sci. USA, 94, 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBowitz J.H. and McMacken,R. (1986) The Escherichia coli DnaB replication protein is a DNA helicase. J. Biol. Chem., 261, 4738–4748. [PubMed] [Google Scholar]
- Leipe D.D., Aravind,L. and Koonin,E.V. (1999) Did DNA replication evolve twice independently? Nucleic Acids Res., 27, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D.D., Aravind,L., Grishin,N.V. and Koonin,E.V. (2000) The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res., 10, 5–16. [PubMed] [Google Scholar]
- Lu Y.B., Ratnakar,P.V., Mohanty,B.K. and Bastia,D. (1996) Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc. Natl Acad. Sci. USA, 93, 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.-B., Datta,H.J. and Bastia,D. (1998) Mechanistic studies of initiator–initiator interaction and replication initiation. EMBO J., 17, 5192–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek J. and Kaguni,J.M. (1994) DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem., 269, 4883–4890. [PubMed] [Google Scholar]
- Miles C.S., Weigelt,J., Stamford,N.P.J., Dammerova,N., Otting,G. and Dixon,N.E. (1997) Precise limits of the N-terminal domain of DnaB helicase determined by NMR spectroscopy. Biochem. Biophys. Res. Commun., 231, 126–130. [DOI] [PubMed] [Google Scholar]
- Nieto C., Fernandez-Tresguerres,E., Sanchez,N., Vicente,M. and Diaz,R. (1990) Cloning vectors, derived from a naturally occurring plasmid of Pseudomonas savastanoi, specifically tailored for genetic manipulations in Pseudomonas. Gene, 87, 145–149. [DOI] [PubMed] [Google Scholar]
- Pacek M., Konopa,G. and Konieczny,I. (2001) DnaA-box sequences as the site for helicase delivery during plasmid RK2 replication initiation in Escherichia coli. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Perri S., Helinski,D.R. and Toukdarian,A. (1991) Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J. Biol. Chem., 266, 12536–12543. [PubMed] [Google Scholar]
- Ratnakar P.V.A.L., Mohanty,B.K., Lobert,M. and Bastia,D. (1996) The replication initiator protein π of the plasmid R6K specifically interacts with the host-encoded helicase DnaB. Proc. Natl Acad. Sci. USA, 93, 5522–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluja D. and Godson,G.N. (1995) Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutants dnaB8, dnaB252, dnaB70, dnaB43 and dnaB454. J. Bacteriol., 177, 1104–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin M.C., Stamford,N.P.J., Dammerova,N., Dixon,N.E. and Carazo,J.M. (1995) A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J. Struct. Biol., 114, 167–176. [DOI] [PubMed] [Google Scholar]
- Schmidhauser T.J., Filutowicz,M. and Helinski,D.R. (1983) Replication of derivatives of the broad host range plasmid RK2 in two distantly related bacteria. Plasmid, 9, 325–330. [DOI] [PubMed] [Google Scholar]
- Sclafani R.A. and Wechsler,J.A. (1981) Deoxyribonucleic acid initiation mutation dnaB252 is suppressed by elevated dnaC+ gene dosage. J. Bacteriol., 146, 418–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H., Weigel,C. and Messer,W. (2000) The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol., 37, 1270–1279. [DOI] [PubMed] [Google Scholar]
- Shingler V. and Thomas,C.M. (1984) Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J. Mol. Biol., 175, 229–249. [DOI] [PubMed] [Google Scholar]
- Thomas C.M. and Helinski,D.R. (1989) Vegetative replication and stable inheritance of IncP plasmids. In Thomas,C.M. (ed.), Promiscuous Plasmids of Gram-negative Bacteria. Academic Press, San Diego, CA, pp. 1–25.
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougu K., Peng,H. and Marians,K.J. (1994) Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J. Biol. Chem., 269, 4675–4682. [PubMed] [Google Scholar]
- Toukdarian A.E., Helinski,D.R. and Perri,S. (1996) The plasmid RK2 initiation protein binds to the origin of replication as a monomer. J. Biol. Chem., 271, 7072–7078. [DOI] [PubMed] [Google Scholar]
- Wahle E., Lasken,R.S. and Kornberg,A. (1989) The dnaB–dnaC replication protein complex of Escherichia coli. I. Formation and properties. J. Biol. Chem., 264, 2463–2468. [PubMed] [Google Scholar]
- Wickner S. and Hurwitz,J. (1975) Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc. Natl Acad. Sci. USA, 72, 921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Jezewska,M.J., Bujalowski,W. and Egelman,E.H. (1996) The hexameric E.coli DnaB helicase can exist in different quarternary states. J. Mol. Biol., 259, 7–14. [DOI] [PubMed] [Google Scholar]