Abstract
One of the puzzles in cancer predisposition is that women carrying BRCA-1 mutations preferentially develop tumors in epithelial tissues of the breast and ovary. Moreover, sporadic breast tumors contain lower levels of BRCA-1 in the absence of mutations in the BRCA-1 gene. The problem of tissue specificity requires analysis of factors that are unique to tissues of the breast. For example, the expression of estrogen receptor-α (ERα) is inversely correlated with breast cancer risk, and 90% of BRCA-1 tumors are negative for ERα. Here, we show that estrogen stimulates BRCA-1 promoter activity in transfected cells and the recruitment of ERα and its cofactor p300 to an AP-1 site that binds Jun/Fos transcription factors. The recruitment of ERα/p300 coincides with accumulation in the S-phase of the cell cycle and is antagonized by the antiestrogen tamoxifen. Conversely, we document that overexpression of wild-type p53 prevents the recruitment of ERα to the AP-1 site and represses BRCA-1 promoter activity. Taken together, our findings support a model in which an ERα/AP-1 complex modulates BRCA-1 transcription under conditions of estrogen stimulation. Conversely, the formation of this transcription complex is abrogated in cells overexpressing p53.
Keywords: BRCA-1, ERα, AP-1, p53, sporadic breast cancer
Introduction
The breast and ovarian cancer susceptibility gene BRCA-1 [1,2] encodes for a transcription factor, which contributes to recombination and DNA repair functions [3–5]. Reduced levels of wild-type BRCA-1 protein have been detected in a large percentage of sporadic breast tumors in the absence of mutations in the BRCA-1 gene [6], suggesting that disruption of BRCA-1 expression may be a contributing factor to the onset of mammary carcinogenesis [7]. Exposure to ovarian estrogens has been recognized as one risk factor in breast tumorigenesis based on the evidence that therapies with the estrogen receptor agonist tamoxifen (TMX) reduced the incidence of breast cancer [8]. Effects of estrogen on responsive genes are mediated by two estrogen receptors: estrogen receptor-α (ERα) and estrogen receptor-β (ERβ). In the classic pathway, ERα contacts the DNA at specific estrogen-responsive elements (EREs) comprising target genes and recruits coactivators and cofactors that enhance transcription [9]. Alternatively, ERα may physically interact with p160/p300 proteins bound to an AP-1 (Jun/Fos) complex that contacts DNA [10]. The profile of cofactors and the type of ligand have been shown to influence the transcription activity of ERα [11,12].
The expression of BRCA-1 peaks in the S-phase of the cell cycle [13–15] and is induced by estrogen in breast cancer cell lines [16,17] and estrogen plus progesterone in the mammary gland of ovariectomized mice [18]. Although estrogen depletion reduces BRCA-1 expression [19], the stimulatory effects of estrogen on BRCA-1 expression are believed to be indirect based on the observations that the proximal BRCA-1 promoter lacks consensus EREs that bind ERα and that de novo protein synthesis is required for BRCA-1 upregulation [20]. To clarify the mechanisms of estrogen stimulation of BRCA-1 expression, we investigated whether estrogen regulated BRCA-1 transcription through an alternative pathway involving the recruitment of complexes containing ERα at non-EREs in the BRCA-1 promoter. We report that, in response to estrogen, an ERα/p300 complex is recruited to an AP-1 domain located in the proximal BRCA-1 promoter and activates BRCA-1 transcription, whereas the overexpression of p53 prevents the recruitment of ERα and represses BRCA-1 promoter activity.
Materials and Methods
Cells, Transient Transfections, and Luciferase Assay
HCT116 and HCT116 p53KO cells were a generous gift from B. Vogelstein. MCF-7 and HeLa cells were obtained from the American Type Culture Collection (Rockville, MD). The plasmid pTam67 was originated by cloning the Tam67 cDNA into the EcoRI site of pCR3.1. Transient transfections were performed using the Lipofectamine-Plus procedure according to the manufacturer's instructions (Life Technologies, Inc., Carlsbad, CA). Briefly, 24 hours after cells were plated, each well was cotransfected with the appropriate plasmid and an internal control plasmid pRL-TK (renilla luciferase gene). Cells were incubated with the DNA-liposome complex for 3 hours at 37°C in 5% CO2. Following transfection, cells were maintained in Dulbecco's modified Eagle's medium (DMEM) plus 5% charcoal-stripped fetal bovine serum (FBS) and allowed to recover for 48 hours. Cells were then treated in DMEM containing either control (ethanol vehicle) or 10 nM 17β-estradiol (E2) for the times indicated.
Western Blot Analysis and Flow Cytometry
Western blot analysis for BRCA-1 was performed as described previously [21]. Cell extracts were normalized to protein content and separated by 4% to 12% gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Immunoblotting was carried out with antibodies raised against BRCA-1 (Ab-2; Oncogene Research Products, Cambridge, MA). Normalization of Western blots was confirmed by incubating immunoblots with β-actin antibody-1 (Oncogene Research Products). The immunocomplexes were detected by enhanced chemiluminescence (Amersham Corp., Arlington Heights, IL). Flow cytometry was performed in triplicate as described previously [22]. Briefly, cells were harvested with trypsin and washed in phosphate-buffered saline (PBS). Then cells were treated with RNAse and stained with propidium iodide (70 µM in PBS). Cell cycle distribution profiles were recorded with a FACscan (Becton Dickinson, San Jose, CA), using a CELLQuest program.
Electrophoretic Mobility Shift Assay (EMSA)
Cells were plated in DMEM plus 5% charcoal-stripped FBS. After 24 hours, cells were treated for 24 hours with 10 nM E2 then subsequently harvested. Briefly, cells were trypsinized then washed with ice-cold DPBS. Cells were resuspended in ice-cold 25 mM Hepes buffer containing 1.5 mM EDTA, 1 mM DTT, 0.5 mM PMSF, and 5 µg/ml aprotinin, and placed on ice for 10 minutes. Cells were pelleted and resuspended in 1 ml of ice-cold 25 mM Hepes buffer containing 1.5 mMEDTA, 10% (vol/vol) glycerol, 1 mM DTT, 0.5 mM PMSF, and 5 µg/ml aprotinin. The cell suspension was transferred to a mortar for drilling with a Teflon pestle until more than 90% of the cells in a 2-µl aliquot were unable to exclude trypan blue. After centrifugation, cell pellets were resuspended in 150 µl of ice-cold 25 mM Hepes buffer containing 1.5 mM EDTA, 10% (vol/vol) glycerol, 0.5 M KCl, 1 mM DTT, 0.5 mM PMSF, and 5 µg/ml aprotinin, and placed on ice with intermittent vortexing. Cell debris was removed by centrifugation. Supernatants containing nuclear protein were stored at -70°C. Nuclear protein concentration was determined using the BCA protein assay (Pierce Chemical Company, Rockford, IL). Oligonucleotides used for binding and gel retardation assay were: BRCA-1, 5′-AACCTGAGAGGCGTAAGGCGTT-3′ (sense) and 5′-AACGCCTTACGCCTCTCAGGTT-3′ (antisense); and consensus TRE, 5′-CAAACACATGAGTAATGTGTT-3′ (sense) and 5′-AACACATTACTCATGTGTTTG-3′ from the human collagenase promoter. The complementary oligonucleotides were annealed then phosphorylated at the 5′-end with [γ-32P]ATP and T4 polynucleotide kinase. Unincorporated nucleotides were removed using the TE-10 spin columns (Clontech, Mountain View, CA). Binding assays were performed by incubating 5 µg of nuclear protein in the binding buffer then incubated with the labeled oligonucleotides for 20 minutes. For supershift assays, antibodies (Affinity Bioreagents, Boulder, CO) were incubated with 1-µg nuclear extracts for 2 hours prior to addition of labeled oligonucleotides. For cold competition, a 100-fold excess of the respective unlabeled oligonucleotides was added to the binding reaction 10 minutes prior to addition of the labeled oligonucleotides. Samples were electrophoresed through a 5% nondenaturing polyacrylamide gel at 200 V for 90 minutes. Finally, the gel was dried and exposed to a phosphor screen, and digital phosphorimages were retrieved using the Storm system (Molecular Dynamics, Piscataway, NJ).
Chromatin Immunoprecipitation (ChIP) Assay
Cells were collected after fixation of protein and DNA through the addition of formaldehyde for a final concentration of 1% to the cell culture medium and incubated at 25°C for 10 minutes. Prior to collection, cells were washed twice in cold DPBS. The cells were resuspended in a lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, and protease inhibitor cocktail). After sonication, the dilution of chromatin preparations was either reserved as input (no antibody) material or utilized for immunoprecipitation with the desired antibody. After immunoprecipitation, DNA was recovered and subjected to polymerase chain reaction (PCR) analysis using the following primers: BRCA-1, forward: ATCGGTACCAAGTGATGC TCTGGGGTACTG, reverse: ACTAGATCTACCTCATGACCAGCCGACGTT (237 bp), flanking the AP1-binding site. As a positive control for estrogen treatment and ChIP assay for ERα, we tested for the recruitment of ERα at the ERE region of the estrogen-inducible pS2 gene using the primers: forward: TATGAATCACTTCTGGAGTGA; reverse: GAGCGTTAGATAACATTTGCC (289 bp). As negative controls, we tested for the recruitment of ERα at exon 7 of the BRCA-1 gene using forward: 5′-ATGCAAACAGCTATAATTTTG-3′; reverse, 5′-CAAGGAAGGATTTTCGGGTTC-3′ (140 bp) and through coincubation with IgG.
Results
Estrogen Induces BRCA-1 Promoter Activity
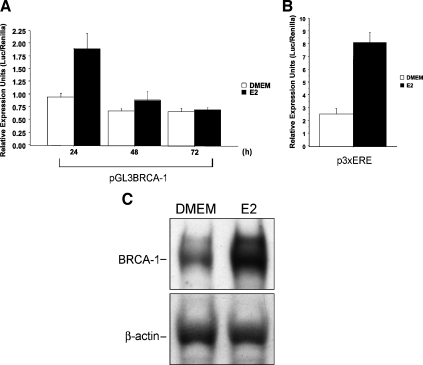
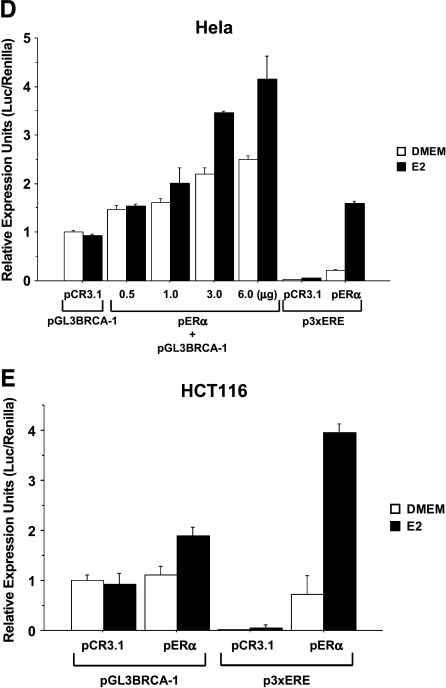
In transient transfection assays with ERα-positive MCF-7 breast cancer cells, we found that the treatment with E2 for 24 hours, but not in earlier time points (data not shown), stimulated by a factor of 2.0 the transcriptional activity of a 1.69-kb BRCA-1 promoter-reporter construct (pGL3BRCA-1) [21] containing both transcription start sites for exons 1A and 1B (Figure 1A). The E2 treatment stimulated luciferase reporter activity from a positive control promoter-reporter construct (p3XERE) containing an array of three consensus EREs (Figure 1B) and the accumulation of BRCA-1 protein (Figure 1C). The requirement for ERα in the E2-dependent regulation of BRCA-1 transcription was confirmed in cervical HeLa and colon HCT116 cancer cells transiently transfected with pGL3BRCA-1 plus a vector encoding for ERα (pERα). For these experiments, we adopted transfection conditions similar to those of previous studies, which examined the role of BRCA-1 on ERα signaling [23]. In detail, we transfected ∼70% confluent 24-well dishes with 0.5 µg of pERα. Our results indicated that in ERα-negative HeLa (Figure 1D) and HCT116(Figure 1E) cells transfected with either pGL3BRCA-1 or an empty vector (pCR3.1), E2 treatment was not sufficient to induce BRCA-1 promoter activity. However, BRCA-1 transcription became responsive to E2 following cotransfection with various amounts of pERα as documented by stimulation of luciferase activity from pGL3BRCA-1. Similarly, transcription from the positive control p3XERE reporter construct was induced by E2 on cotransfection with pERα. The cotransfection of ERα into HeLa and HCT116 cells produced a fold induction in BRCA-1 transcription, similar to that observed in transfected MCF-7 cells expressing endogenous ERα and treated with E2.
Figure 1.
Estrogen induces BRCA-1 promoter activity in transiently transfected MCF-7 cells. (A) MCF-7 cells were precultured for 4 days in phenol red-free DMEM containing 5% charcoal dextran-stripped FBS (Hyclone Laboratories, Logan, UT). Then, a 1.69-kb fragment of the BRCA-1 5′ flanking region driving the expression of a luciferase cassette (pGL3BRCA-1) was transiently transfected, using Lipofectamine Plus (Life Technologies, Inc.), into MCF-7 cells, which were cultured in DMEM or DMEM plus 10 nM E2 (Sigma, St. Louis, MO) for various periods of time. (B) The treatment with E2 induces promoter activity from a positive control vector (p3XERE) transfected into MCF-7 cells. (C) Western blot analysis with antibodies for BRCA-1 (Ab-2) and β-actin (Ab-1) (Oncogene Research Products) documents that BRCA-1 protein levels are induced in MCF-7 cells cultured for 24 hours in DMEM plus 10 nM E2. (D) HeLa and (E) HCT116 cancer cells were cotransfected with pGL3BRCA-1 and either a plasmid encoding for ERα (pERα) or an empty vector (pCR3.1). Transfected cells were cultured in DMEM or DMEM plus 10 nM E2 for 24 hours. Control plates were transfected with p3XERE. Bars represent mean luciferase units corrected for the internal control renilla ±SE from two independent experiments performed in quadruplicate.
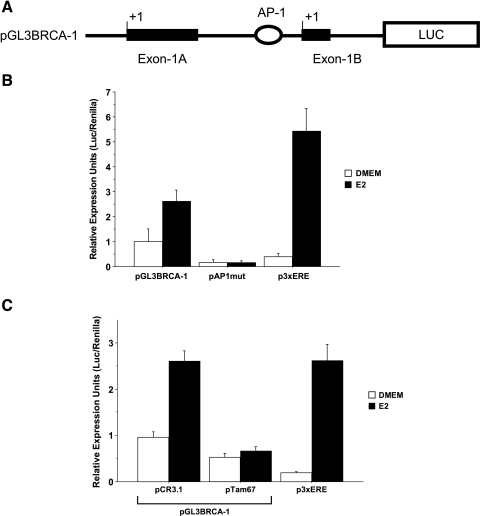
In search of non-ERE sites located in the proximal BRCA-1 promoter that may recruit transcription complexes containing ERα, we mapped a sequence (5′-CTGAG-3′) with significant homology to consensus sequences for AP-1 transcription factors [24] at positions -27/-31 upstream of the transcription start site on exon 1B (Figure 2A). Site-directed mutagenesis of the candidate AP-1-like (CTGAG to CACTA) site abrogated basal and E2-dependent activity of the BRCA-1 promoter, as evidenced by the significant reduction in luciferase activity observed in MCF-7 cells transfected with a mutated BRCA-1 promoter-luciferase reporter construct (pAP1mut) (Figure 2B). Moreover, cotransfection with an expression vector encoding for a dominant-negative variant (pTam67) of c-Jun abrogated basal and E2-induced BRCA-1 promoter activity (Figure 2C), confirming the requirement for AP-1 in basal and estrogen-dependent regulation of BRCA-1 transcription.
Figure 2.
An AP-1-like site contributes to estrogen-induced BRCA-1 promoter activity. (A) Position of the candidate AP-1 element in the BRCA-1 promoter. (B) MCF-7 cells were precultured for 4 days in DMEM with 5% FBS and then transiently transfected with pGL3BRCA-1 or pGL3BRCA-1 mutated at the AP-1 site (pAP1mut). p3XERE is a positive control for treatment with 10 nM E2. (C) Transient transfection of MCF-7 cells with a dominant-negative c-Jun plasmid (pTam67) represses basal and estrogen-regulated BRCA-1 transcription. (B and C) Data represent mean luciferase units corrected for the internal control renilla ±SE from two independent experiments performed in quadruplicate.
Estrogen Stimulates the Recruitment of ERα to an AP-1 Site in the BRCA-1 Promoter Region
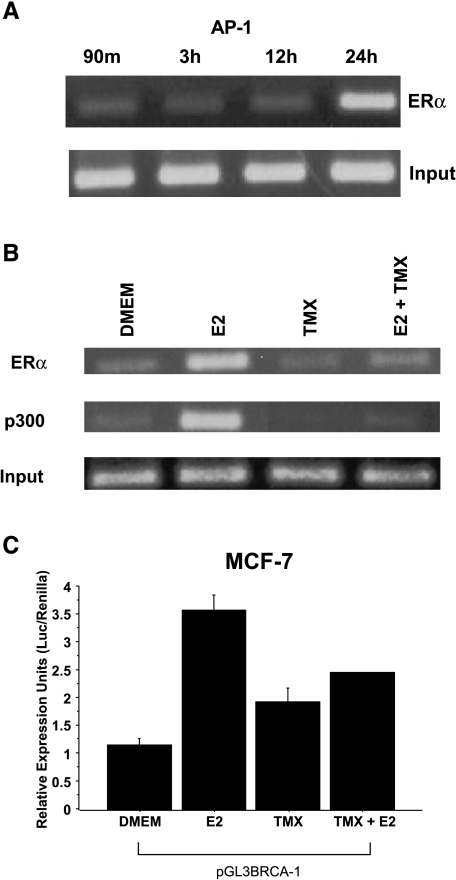
We used ChIP assays to investigate whether or not estrogen stimulated the recruitment of ERα to the BRCA-1 promoter region containing the AP-1 domain. Based on the information that BRCA-1 levels peak in S-phase [25], we enriched the fraction of MCF-7 cells positioned in the G1 phase of the cell cycle (Table 1). Time course experiments showed that E2 stimulated the recruitment of ERα to a BRCA-1 promoter fragment comprising the AP-1-like motif (Figure 3A). This was accompanied by recruitment of p300 (Figure 3B) and accumulation of cells in S-phase (71.5%) (Table 1). Conversely, the E2-induced accumulation in S-phase and the recruitment of ERα and p300 were antagonized by cotreatment with TMX (Figure 3C). Even though the treatment with TMX was stimulated by ∼50% BRCA-1 promoter activity, in combination with E2, it antagonized the activation of BRCA-1 transcription. These agonist/antagonist effects of TMX were similar to those exerted by this compound on the expression of other estrogen-responsive genes [10].
Table 1.
Estrogen Stimulates G1-Phase to S-Phase Transition of MCF-7 Cells.
| Treatment | G1 | S | G2/M |
| 4 days in DMEM | 65.0 | 34.5 | 0.5 |
| After 24 hr in DMEM | 40.5 | 59.0 | 0.5 |
| E2 | 28.0 | 71.5 | 0.5 |
| TMX | 54.6 | 42.0 | 3.4 |
| TMX + E2 | 35.2 | 61.2 | 3.6 |
MCF-7 cells were precultured for 4 days in phenol red-free DMEM containing 5% charcoal dextran-stripped FBS. Then cells were cultured for 24 hours in basal DMEM or DMEM supplemented with 10 nM E2, 1 µM TMX, or their combination. Cell were stained with propidium iodide and used for flow cytometry. Cell cycle distribution profiles were recorded with a FACscan (Becton Dickinson) using a CELLQuest program.
Figure 3.
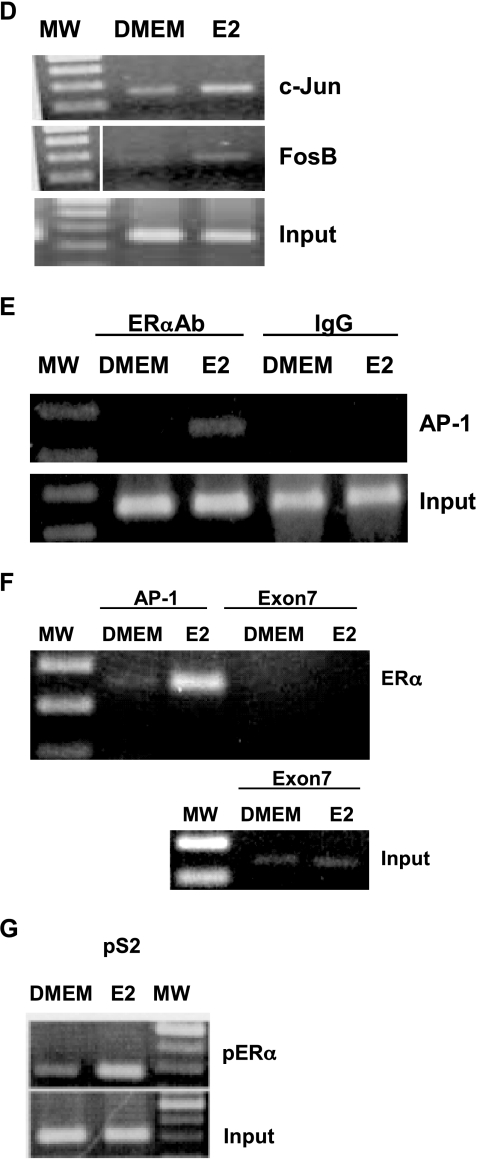
Treatment of MCF-7 cells with estrogen stimulates BRCA-1 promoter occupancy by ERα and p300 to an AP-1 site. (A) MCF-7 cells were precultured for 4 days in DMEM with 5% FBS and then treated with 10 nM E2 for various periods of time. Cells were processed for ChIP assay using an antibody against ERα (Neomarkers, Fremont, CA). Inputs are control bands generated by PCR from cross-linked chromatin. (B) At 24 hours, estrogen stimulates the recruitment of ERα and p300 (antibody from Affinity Bioreagents), whereas 1 µM TMX (Sigma) antagonizes E2-dependent recruitment of ERα and p300. (C) E2-induced BRCA-1 transcription in transiently transfected MCF-7 cells is antagonized by cotreatment with TMX. (D) E2 stimulates the recruitment of c-Jun and FosB. (E) Coincubation with IgG followed by PCR amplification does not produce a band comprising the AP-1 segment. (F) The recruitment of ERα to a region of exon 7 in the BRCA-1 gene (negative control) is not stimulated by E2, which stimulates (G) the recruitment of ERα to an ERE in the pS2 gene (positive control). The size of the amplicon was 237 bp for BRCA-1 (-98 to +139 bp from +1 on exon 1B), 289 bp for the pS2 ERE, and 140 bp for exon 7 of the BRCA-1 gene. (H) EMSA for ERα at the BRCA-1 promoter. MCF-7 cells were precultured for 4 days in DMEM with 5% FBS and then treated for 24 hours with E2. Nuclear extracts were coincubated with a 32P-labeled BRCA-1 oligonucleotide (-40/-19 bp) plus various amounts of mouse IgG or an antibody for ERα. The ERα antibody supershifted a complex (band A) in a dose-dependent manner, thus confirming the presence of ERα at this region; FP, free probe.
The E2 treatment stimulated the recruitment of c-Jun and FosB (Figure 3D), confirming that AP-1 contributed to the formation of a transcription complex at this region. Control experiments indicated that the coincubation of cross-linked chromatin with preimmune IgG did not generate a corresponding BRCA-1 amplification product (Figure 3E). Neither did E2 stimulate the recruitment of ERα to the coding region of exon 7 in the BRCA-1 gene (Figure 3F). However, the treatment with E2 triggered the recruitment of ERα to an ERE in the pS2 gene (Figure 3G), thus confirming the efficacy of the E2 treatment and the experimental conditions for the ChIP assay.
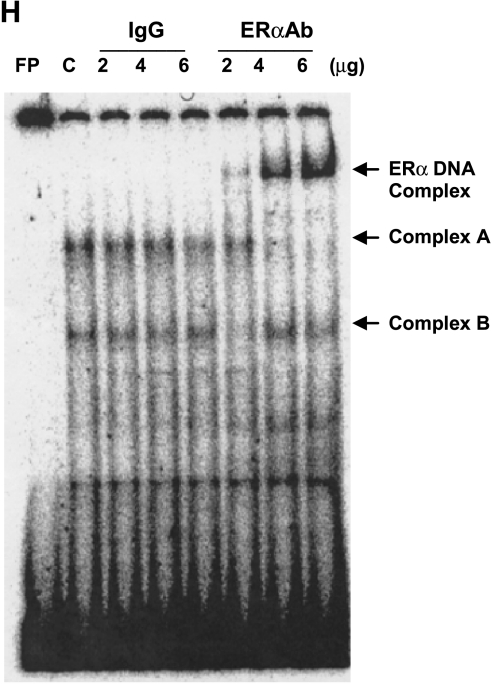
To obtain additional evidence that the BRCA-1 promoter region containing the AP-1 site was targeted for binding by ERα, we incubated nuclear extracts obtained from E2-treated MCF-7 cells with a BRCA-1 oligonucleotide spanning 22 bp (-40/-19) upstream of the exon 1B transcription start site. The incubation of nuclear extracts with the BRCA-1 oligonucleotide produced two distinct complexes (bands A and B). Band A was supershifted in a dose-dependent fashion following coincubation of the BRCA-1 oligonucleotide with increasing amounts of an ERα antibody (ERαAb) (Figure 3H), suggesting that this complex contained ERα. These results mapped the binding region for ERα to the BRCA-1 promoter segment comprised between -40 and -19 bp, which included the AP-1-like domain. Conversely, the coincubation of nuclear extracts with control preimmune IgG did not produce a supershifted band. The formation of complex A was competed by coincubation with excess cold oligonucleotide containing the consensus AP-1 sequence 5′-TGACTCA-3′ from the human collagenase promoter (data not shown), confirming that the BRCA-1 oligonucleotide was a target for AP-1. Taken together, these results suggested that induction of BRCA-1 transcription by estrogen required the concomitant expression of ERα and its occupancy along with p300 at an AP-1 site in the proximal BRCA-1 promoter.
Overexpression of p53 Prevents the Recruitment of ERα to AP-1 and Represses BRCA-1 Transcription
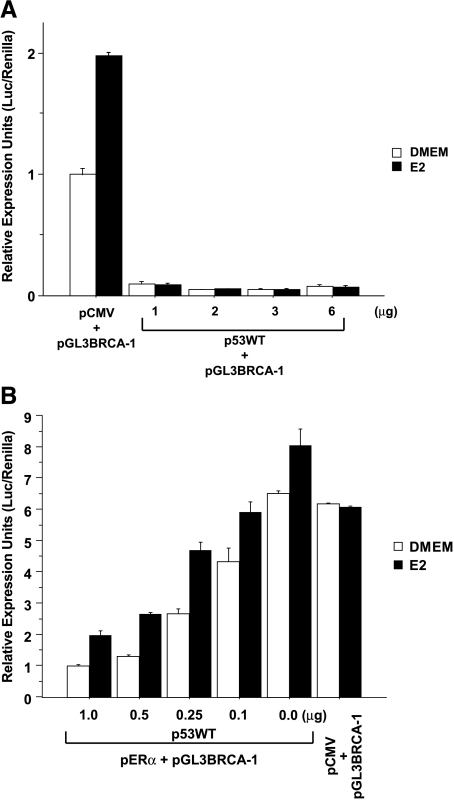
In previous studies, p53 has been shown to repress BRCA-1 expression [21,26], through yet unknown mechanisms. Therefore, we examined whether or not p53 interfered with E2 stimulation of BRCA-1 transcription. In transient transfection experiments with MCF-7 cells, we found that basal and E2-induced BRCA-1 promoter activities were repressed following cotransfection with various amounts of a vector encoding for wild-type p53 (p53WT) (Figure 4A). In parallel transfection experiments with HCT116 cells lacking p53 (HCTKO, p53-/-), the treatment with E2 did not influence BRCA-1 transcription in cells cotransfected with pGL3BRCA-1 plus the empty vector pCMV (Figure 4B). The cotransfection with pGL3BRCA-1 plus pERα did not alter basal promoter activity, but resulted in a modest increase (∼20%) in BRCA-1 transcription following E2 treatment. Conversely, the cotransfection of HCTKO cells with various amounts of p53WT repressed BRCA-1 promoter activity in a dose-dependent fashion. However, the relative induction by E2 was greater in cells transfected with higher amounts (0.25–1.0 µg) of p53WT, suggesting that E2 regulation of BRCA-1 transcription was influenced by the relative expression levels of p53 and ERα.
Figure 4.
Overexpression of wild-type p53 represses basal and estrogen-induced BRCA-1 transcription. (A) MCF-7 and (B) HCTKO (p53-/-) cells were precultured for 4 days in DMEM with 5% FBS and then transiently transfected with pGL3BRCA-1 or pGL3BRCA-1 plus various amounts of a plasmid encoding for wild-type p53 (p53WT) (gift from Bert Vogelstein, John Hopkins University, Baltimore, MD) under the control of the cytomegalovirus (CMV) promoter or an empty plasmid (pCMV). Where indicated, the HCTKO cells were cotransfected with 3.0 µg of pERα. Transfected cells were cultured in DMEM or DMEM supplemented with 10 nM E2 for 24 hours. Bars represent mean luciferase units corrected for the internal control renilla ±SE from two independent experiments performed in quadruplicate.
Based on the information that p53 physically interacts with ERα [27], we asked whether or not overexpression of p53 modulated the recruitment of ERα to the AP-1 site. ChIP experiments with MCF-7 cells indicated that E2-induced recruitment of ERα to this site was repressed following overexpression of exogenous p53WT (Figure 5, A and B). These findings suggested that p53 represses E2-induced BRCA-1 transcription by preventing the recruitment of ERα to the BRCA-1 promoter. Western blot analysis confirmed that in MCF-7 cells overexpressing p53, the induction of BRCA-1 protein by E2 was greatly reduced compared with MCF-7 cells transfected with the empty plasmid pCMV. These data suggested that the interaction of ERα at the AP-1 was critical for the E2-dependent increase in BRCA-1 protein observed in Western blots.
Figure 5.
Overexpression of p53 in MCF-7 cells prevents estrogen-induced accumulation of ERα at the AP-1 site and BRCA-1 protein. (A) Bands are PCR products following ChIP assay for ERα. Inputs are control bands generated by PCR from cross-linked chromatin. (B) Western blot analysis with antibodies for p53 and BRCA-1 documents that p53 protein levels are overexpressed in MCF-7 cells transfected with p53WT, whereas E2-induced BRCA-1 protein is repressed. (A and B) MCF-7 cells were cultured for 24 hours in DMEM or DMEM plus 10 nM E2. (C) Proposed model that integrates E2 regulation of BRCA-1 expression with G1-phase to S-phase transition. Our data suggest that E2 stimulates the recruitment of an ERα/p300 transcription complex to an AP-1 site in the BRCA-1 promoter. In turn, increased BRCA-1 levels may activate G1/S-phase checkpoints to allow time for DNA repair [40] and to block ERα transcriptional activity [23]. However, accumulation of p53 interferes with the recruitment of ERα, leading to transcriptional repression of BRCA-1.
Discussion
The objective of the present study was to investigate whether or not E2 activated BRCA-1 promoter activity and the potential involvement of ERα. Previous studies have documented that E2 stimulates BRCA-1 expression [16,17]. However, the lack of EREs in the promoter of BRCA-1 has led to the suggestion that E2 activation of BRCA-1 expression is indirect [20]. In this study, we used a 1.7-kb BRCA-1 promoter fragment containing both transcriptional start sites of exons 1A and 1B [21] to investigate the E2 regulation of the BRCA-1 promoter. The results of transfection studies indicated that E2 stimulated BRCA-1 promoter-reporter activity in breast MCF-7 cells expressing endogenous ERα, and in colon (HCT116) and cervical (HeLa) cells cotransfected with a plasmid encoding for ERα. Our transfection conditions were similar to those used in previous studies, which examined the 876 Estrogen Regulation of BRCA-1 Transcription Jeffy et al. interplay between BRCA-1 and ERα [23]. In the current study, the efficacy of transfection conditions and E2 treatment was confirmed by induction in transfected cells of transcription activity from a control expression construct containing an array of three consensus EREs (p3XERE), as well as accumulation of BRCA-1 protein in MCF-7 cells. Conversely, cotreatment with the antiestrogen TMX abrogated E2-induced BRCA-1 promoter activity in MCF-7 cells, confirming the involvement of ERα in the regulation of BRCA-1 transcription.
The action of ERα at an ERE (5′-GGTCAnnnTGACC-3′) is well understood and involves the direct binding of ER homodimer to DNA and the recruitment of coactivators including CBP/p300 and p160 [9,28]. Alternatively, ERα has been shown to activate the transcription of several E2-inducible genes including IGF-1 [29], collagenase [30], and cyclin D [31] at AP-1 sequences that bind members of the Jun and Fos families. Inspection of the proximal BRCA-1 promoter revealed its presence in close proximity to the exon 1B transcription start site of an element (CTGAG) homologous to a sequence that binds AP-1 factors [24]. Several observations presented in this report support the notion that this site is important for basal and E2-induced regulation of BRCA-1 transcription. First, mutation of the candidate AP-1 site, as well as overexpression of a dominant-negative variant of c-Jun (pTam67), led to repression of BRCA-1 promoter activity in transfected MCF-7 cells. These results were in agreement with those of previous investigations documenting that mutation of sites binding Jun/Fos proteins and expression of Tam67 reduced basal and E2-inducible AP-1 transactivation [32–34]. Second, the results of ChIP assays provided direct evidence that the BRCA-1 promoter region flanking the AP-1 site was targeted by c-Jun and FosB in the treatment of MCF-7 cells with E2. Compared to other members of the Jun and Fos families, c-Jun and FosB are considered strong transactivators [35], and their recruitment to the BRCA-1 gene highlights the role of AP-1 in E2-dependent activation of BRCA-1 transcription. Third, treatment with E2 induced the recruitment of ERα and its cofactor p300 to the BRCA-1 promoter region comprising the AP-1, whereas the antiestrogen TMX antagonized these effects. Results of EMSAs mapped the recruitment of ERα to a 21-bp promoter region (-40 and -19 bp) spanning the AP-1 site. These cumulative data are consistent with a model that attributes to ERα and its coactivator p300 a key role in the activation of BRCA-1 transcription through an AP-1. The physiological relevance of these findings is that ERα/AP-1 interactions may lead to activation of BRCA-1 promoter activity, which is paralleled by an increase in BRCA-1 protein. Unlike gel mobility studies with segments of the BRCA-1 promoter, the ChIP assay offered the opportunity to examine the recruitment of these transcription factors to the BRCA-1 promoter in the context of the native chromatin.
The dynamics of cofactor recruitment to the AP-1 site is likely complex and involves an orderly recruitment of transcription factors and alterations in chromatin state [9]. For example, the participation of p300, which possesses intrinsic histone acetyl transferase (HAT) activity [36], in the formation of the transcription complex recruited at the AP-1 may be a key event that may lead to chromatin remodeling and activation of BRCA-1 transcription. This notion is supported by the current observations that cotreatment with TMX antagonized the recruitment of p300 to the AP-1 site and antagonized the stimulatory effects of E2 on BRCA-1 promoter activity. In this study, although we did not assess the presence of p160s, members of this family of transcription factors have been shown to physically interact with ERα at AP-1 sites [28]. One possibility is that Jun/Fos heterodimers bound to the BRCA-1 DNA may recruit p300-p160s, whereas ERα may be recruited to the complex through contacts with the coactivator p160 [37]. Furthermore, it is plausible that ERα may form bridges with accessible transcription factors recruited at adjacent sites in the BRCA-1 promoter. For example, we identified just upstream of the AP-1 site a consensus sequence (5′-GGGCGG-3′) for the transcription factor Sp1, which has been shown to interact with ERα [38]. This potential interaction may stabilize the ERα/p300 transcription complex formed at the AP-1. Finally, another factor that may influence the degree and temporal activation of the BRCA-1 gene by E2 is the relative abundance and profile of AP-1 proteins recruited to the BRCA-1 promoter. Regardless of the precise complement of cofactors and coactivators recruited at the AP-1 site, our studies showed that ERα and p300 play an important role in E2-dependent activation of the BRCA-1 promoter. The recruitment of an ERα/p300 complex to an AP-1 rather than an ERE site may integrate the role of AP-1 in the regulation of cell proliferation [37] with that of BRCA-1 in cell growth response. The fact that the kinetics and magnitude of BRCA-1 induction by E2 are different from those of inducible genes containing EREs [20] may be due to the type of transcriptional response that is required for BRCA-1. Elevation of BRCA-1 expression as cells enter S-phase may offer a control mechanism that activates DNA repair and cell cycle checkpoints before DNA replication occurs [14].
Previous studies by other investigators [39,40] and our laboratory [21] documented that BRCA-1 expression levels were downregulated in response to p53 induction and that these effects were due to transcriptional repression by p53. However, no consensus p53-binding sites have been found in the BRCA-1 promoter [39], suggesting that the repressive effects of p53 on BRCA-1 transcription are not mediated through direct binding to target DNA sequences. In this study, we found that overexpression of p53 counteracted the E2-dependent upregulation of BRCA-1 promoter activity. Results of ChIP assays documented that the recruitment of ERα to the AP-1 site in the BRCA-1 promoter was abrogated in MCF-7 cells overexpressing exogenous p53. One potential explanation for these results is that p53 may physically interact with ERα, thus hampering the transcriptional activity of the liganded ERα [27]. The interaction between p53 and ERα occurs at multiple sites on the ERα protein and interferes with the ability of ERα to bind to EREs or other proteins in an ERα-mediated transcription complex [41]. In addition, p53 may interfere with the recruitment of p300 and factors associated with AP-1, while stimulating the recruitment of corepressors and histone deacetylases. For example, the physical interaction of p53 with histone deacetylase-1 and mSin3a has been reported to mediate transcriptional repression of the Map4 gene [42]. Other reports documented that p53 binds to p300 in a region that is required for its intrinsic HAT activity [36]. The formation of p53–p300 complexes has been shown to reduce the amount of p300 available and repress transcription from an AP-1 site, whose activation is p300-dependent [43].
Overall, these findings provided evidence for a direct role of ERα in the regulation of BRCA-1 promoter activity by E2. The increased expression of BRCA-1 may lead to activation of S-phase checkpoints, including p53 and p21 [40,44–46] and DNA damage-responsive genes [47] (Figure 5). This signaling may be of particular significance in cells of the breast, which undergo cyclic proliferative pressure by ovarian estrogens. Without the positive regulation of BRCA-1 expression by ERα, it is conceivable that E2 may stimulate progression through S-phase without proper control, and increase the risk for cancer growth in E2-responsive tissues. Studies have shown that 90% of BRCA-1 tumors are likely to be ER-negative [48] and may explain the lower levels of BRCA-1 observed in sporadic breast tumors [6]. In turn, increased expression of BRCA-1 may inhibit the transcriptional activity of ERα as suggested by earlier studies [23].
Our results also indicated that endogenous p53 levels appeared to have no effects of ERα recruitment on the BRCA-1 promoter. Conversely, overexpression of p53 antagonized the recruitment of ERα to the AP-1 site. These data may explain, at least in part, how accumulation of p53 leads to repression of BRCA-1 transcription [39]. Interestingly, upregulation of BRCA-1 has been shown to stabilize wild-type p53, providing a feedback loop in which these tumor-suppressor genes regulate each other [26]. Overall, the interplay between the positive regulation and the negative regulation by ERα and p53, respectively, on BRCA-1 expression may be part of an integral signaling pathway that is mediated by AP-1 and determines whether the cell undergoes checkpoints/DNA repairs or apoptosis should the damage be irreparable. Disruption of this trafficking may predispose to the onset of breast cancer.
Acknowledgements
We thank B. Vogelstein for plasmid encoding for wild-type p53 (p53WT) and HCTKO cells.
Abbreviations
- AP-1
activator protein-1
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco's modified Eagle's medium
- E2
17β-estradiol
- ERα
estrogen receptor-α
- ERE
estrogen-responsive element
- FCS
fetal calf serum
Footnotes
This study was supported by a National Institutes of Health grant (ES009966, to D.F.R), an Arizona Disease Research Commission grant (8015, to D.F.R.), the Graduate Training Program Grant ES007091 (B.D.J. and J.K.H) and Southwest Environmental Health Sciences Center Grant ES06694 (to D.F.R.).
Brandon D. Jeffy and Jennifer K. Hockings contributed equally to this work.
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Ting NS, Zheng L, Chen PL, Ziv Y, Shiloh Y, Lee EY, Lee WH. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- 4.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 5.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 6.Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen JJ, Scully R, Livingston D, et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- 7.Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9:444–450. doi: 10.1038/ng0495-444. [DOI] [PubMed] [Google Scholar]
- 8.Strassmer-Weippl K, Goss PE. Prevention of breast cancer using SERMs and aromatase inhibitors. J Mammary Gland Biol Neoplasia. 2003;8:5–18. doi: 10.1023/a:1025727103811. [DOI] [PubMed] [Google Scholar]
- 9.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 10.Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- 11.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 12.Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Farmer AA, Chen CF, Jones DC, Chen PL, Lee WH. BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner. Cancer Res. 1996;56:3168–3172. [PubMed] [Google Scholar]
- 14.Vaughn JP, Davis PL, Jarboe MD, Huper G, Evans AC, Wiseman RW, Berchuck A, Iglehart JD, Futreal PA, Marks JR. BRCA1 expression is induced before DNA synthesis in both normal and tumor-derived breast cells. Cell Growth Differ. 1996;7:711–715. [PubMed] [Google Scholar]
- 15.Gudas JM, Li T, Nguyen H, Jensen D, Rauscher FJ, III, Cowan KH. Cell cycle regulation of BRCA1 messenger RNA in human breast epithelial cells. Cell Growth Differ. 1996;7:717–723. [PubMed] [Google Scholar]
- 16.Gudas JM, Nguyen H, Li T, Cowan KH. Hormone-dependent regulation of BRCA1 in human breast cancer cells. Cancer Res. 1995;55:4561–4565. [PubMed] [Google Scholar]
- 17.Romagnolo D, Annab LA, Thompson TE, Risinger JI, Terry LA, Barrett JC, Afshari CA. Estrogen upregulation of BRCA1 expression with no effect on localization. Mol Carcinog. 1998;22:102–109. doi: 10.1002/(sici)1098-2744(199806)22:2<102::aid-mc5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 18.Marquis ST, Rajan JV, Wynshaw-Boris A, Xu J, Yin GY, Abel KJ, Weber BL, Chodosh LA. The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nat Genet. 1995;11:17–26. doi: 10.1038/ng0995-17. [DOI] [PubMed] [Google Scholar]
- 19.Spillman MA, Bowcock AM. BRCA1 and BRCA2 mRNA levels are coordinately elevated in human breast cancer cells in response to estrogen. Oncogene. 1996;13:1639–1645. [PubMed] [Google Scholar]
- 20.Marks JR, Huper G, Vaughn JP, Davis PL, Norris J, McDonnell DP, Wiseman RW, Futreal PA, Iglehart JD. BRCA1 expression is not directly responsive to estrogen. Oncogene. 1997;14:115–121. doi: 10.1038/sj.onc.1200808. [DOI] [PubMed] [Google Scholar]
- 21.Jeffy BD, Chirnomas RB, Chen EJ, Gudas JM, Romagnolo DF. Activation of the aromatic hydrocarbon receptor pathway is not sufficient for transcriptional repression of BRCA-1: requirements for metabolism of benzo[a]pyrene to 7r,8t-dihydroxy-9t,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene. Cancer Res. 2002;62:113–121. [PubMed] [Google Scholar]
- 22.Jeffy BD, Chen EJ, Gudas JM, Romagnolo DF. Disruption of cell cycle kinetics by benzo[a]pyrene: inverse expression patterns of BRCA-1 and p53 in MCF-7 cells arrested in S and G2. Neoplasia. 2000;2:460–470. doi: 10.1038/sj.neo.7900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, et al. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1353–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 24.Weisz A, Rosales R. Identification of an estrogen response element upstream of the human c-fos gene that binds the estrogen receptor and the AP-1 transcription factor. Nucleic Acids Res. 1990;18:5097–5106. doi: 10.1093/nar/18.17.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajan JV, Wang M, Marquis ST, Chodosh LA. Brca2 is coordinately regulated with Brca1 during proliferation and differentiation in mammary epithelial cells. Proc Natl Acad Sci USA. 1996;93:13078–13083. doi: 10.1073/pnas.93.23.13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLachlan TK, Takimoto R, El-Deiry WS. BRCA1 directs a selective p53-dependent transcriptional response towards growth arrest and DNA repair targets. Mol Cell Biol. 2002;22:4280–4292. doi: 10.1128/MCB.22.12.4280-4292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu CL, Driggers P, Barrera-Hernandez G, Nunez SB, Segars JH, Cheng S. The tumor suppressor p53 is a negative regulator of estrogen receptor signaling pathways. Biochem Biophys Res Commun. 1997;239:617–620. doi: 10.1006/bbrc.1997.7522. [DOI] [PubMed] [Google Scholar]
- 28.Webb P, Nguyen P, Shinsako J, Anderson CM, Nguyen MP, McKinerney E, Katzenellenbogen BS, Stallcup M, Kushner PJ. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocriol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 29.Umayahara Y, Kawamori R, Watada H, Imano E, Iwama N, Morishima T, Yamasaki Y, Kajimoto Y, Kamada T. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J Biol Chem. 1994;269:16433–16442. [PubMed] [Google Scholar]
- 30.Uht RM, Anderson CM, Webb P, Kushner PJ. Transcriptional activities of estrogen and glucocorticoid receptors are functionally integrated at the AP-1 response element. Endocrinology. 1997;138:2900–2908. doi: 10.1210/endo.138.7.5244. [DOI] [PubMed] [Google Scholar]
- 31.Geum D, sun W, Paik SK, Lee CC, Kim K. Estrogen-induced cyclin D1 and D3 gene expression during mouse uterine cell proliferation in vivo: differential induction mechanism of cyclin D1 and D3. Mol Reprod Dev. 1997;46:450–458. doi: 10.1002/(SICI)1098-2795(199704)46:4<450::AID-MRD2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 32.Brown PH, Alani R, Preis LH, Szabo E, Birrer MJ. Suppression Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- 33.Cooper SJ, MacGowan J, Ranger-Moore J, Young MR, Colburn NH, Bowden GT. Expression of dominant negative c-jun inhibits ultraviolet B-induced squamous cell carcinoma number and size in an SKH-1 hairless mouse model. Mol Cancer Res. 2003;1:848–854. [PubMed] [Google Scholar]
- 34.Thompson EJ, MacGowan J, Young MR, Colburn N, Bowden GT. A dominant negative c-jun specifically blocks okadaic acid-induced skin tumor promotion. Cancer Res. 2002;62:3044–3047. [PubMed] [Google Scholar]
- 35.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 36.Ogryzko VV, Schiltz RL, Russanova V, Howard B, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 37.Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- 38.Porter W, Saville B, Hoivik D, Safe S. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol Endocrinol. 1997;11:1569–1580. doi: 10.1210/mend.11.11.9916. [DOI] [PubMed] [Google Scholar]
- 39.Arizti P, Fang L, Park I, Yin Y, Solomon E, Ouchi T, Aaronson SA, Lee SW. Tumor suppressor p53 is required to modulate BRCA1 expression. Mol Cell Biol. 2000;20:7450–7459. doi: 10.1128/mcb.20.20.7450-7459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLachlan TK, Dash BC, Dicker DT, El-Deiry WS. Repression of BRCA1 through a feedback loop involving p53. J Biol Chem. 2000;275:31869–31875. doi: 10.1074/jbc.M003338200. [DOI] [PubMed] [Google Scholar]
- 41.Liu G, Schwartz JA, Brooks SC. p53 down-regulates ER-responsive genes by interfering with the binding of ER to ERE. Biochem Biophys Res Commun. 1999;264:359–364. doi: 10.1006/bbrc.1999.1525. [DOI] [PubMed] [Google Scholar]
- 42.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avantaggianti ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signaling pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 44.Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p53-dependent gene expression. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, Weber BL, El-Deiry WS. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
- 46.Somasundaram K, Zhang H, Zeng YX, Houvras Y, Peng Y, Zhang H, Wu GS, Licht JD, Weber BL, El-Deiry WS. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 47.Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, Haber DA. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 48.Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]