Abstract
The expression of 1008 open reading frames (ORFs) from the yeast Saccharomyces cerevisiae has been examined under eight different physiological conditions, using classical northern analysis. These northern data have been compared with publicly available data from a microarray analysis of the diauxic transition in S.cerevisiae. The results demonstrate the importance of comparing biologically equivalent situations and of the standardization of data normalization procedures. We have also used our northern data to identify co-regulated gene clusters and define the putative target sites of transcriptional activators responsible for their control. Clusters containing genes of known function identify target sites of known activators. In contrast, clusters comprised solely of genes of unknown function usually define novel putative target sites. Finally, we have examined possible global controls on gene expression. It was discovered that ORFs that are highly expressed following a nutritional upshift tend to employ favoured codons, whereas those overexpressed in starvation conditions do not. These results are interpreted in terms of a model in which competition between mRNA molecules for translational capacity selects for codons translated by abundant tRNAs.
Keywords: gene expression/genome analysis/mRNA/Saccharomyces cerevisiae/stress responses
Introduction
The availability of the complete genome sequence of the eukaryotic microorganism, Saccharomyces cerevisiae (Goffeau et al., 1996) has allowed researchers to monitor gene transcription on a global (or genome-wide) scale for the first time. The resulting profiles define the complete set of mRNA molecules (the transcriptome; Velculescu et al., 1997) present in the yeast cell under a given set of physiological or developmental conditions (Oliver, 1997). Massively parallel analytical procedures are used in transcriptome analysis that involve the hybridization of labelled mRNA or cDNA molecules to arrays of ‘target’ molecules representing all of the ∼6000 protein-encoding genes defined by the yeast genome (Mewes et al., 1997). These targets may be either oligonucleotides (Wodicka et al., 1997) or PCR products (Lashkari et al., 1997; Hauser et al., 1998) fabricated in either ‘micro’ (on glass slides or chips; Lashkari et al., 1997; Wodicka et al., 1997) or ‘macro’ (on nylon or polypropylene membranes; Hauser et al., 1998) formats. The mRNA or cDNA hybridization probes may be labelled either radioactively (usually with 33P; Hauser et al., 1998) or fluorescently (usually with Cy5 or Cy3; Winzeler et al., 1999). Whatever the experimental protocol employed, all transcriptome analyses using hybridization arrays have in common that they produce massive amounts of data that have to be ‘mined’, using computational techniques, in order to extract meaningful biological information. A number of algorithms have been developed (Eisen et al., 1998; Brown et al., 2000; Kell and King, 2000) to permit the comparison of the transcription patterns of all 6000 protein-encoding genes in different physiological conditions or throughout a time course of development (Cho et al., 1998; Chu, 1998; Spellman et al., 1998) or physiological adaptation (DeRisi et al., 1997). While these algorithms are effective in clustering together genes that show similar patterns of regulation, it is clear that the composition of any particular cluster is enormously sensitive to the thresholds set either for transcript detection or for a significant level of regulation, and thus to the way in which the data have been normalized or otherwise processed.
Because of these concerns about data processing, it is important that we make use of existing biological knowledge in mining hybridization array data. This may be done in two ways, either empirically (e.g. by adjusting threshold levels until genes already known to be co-regulated are clustered together) or, more formally, by using supervised methods of machine learning (Brown et al., 2000; Kell and King, 2000). Whatever approach is used, there is the problem that the prior knowledge has been gained using a different analytical method (most commonly, northern hybridization analysis) and by studying only a few genes at a time. Thus, the clustering algorithms that we wish to employ to mine the array data cannot be applied readily to the data gathered by classical means. What is required in this situation is a large data set, acquired by northern analysis and using a common methodology, to which the clustering and other data mining procedures currently used to analyse hybridization array data may readily be applied. Such a data set could act as a sort of ‘Rosetta Stone’, facilitating the interpretation of data obtained using modern technologies by using knowledge gained from classical experimentation. The acquisition of a large set of gene expression data using classical northern analysis is not a trivial undertaking and is likely to be beyond the manpower resources of any single laboratory. As such, it appeared to be an enterprise suitable for a network approach to research. Therefore, the Transcription Consortium of the EUROFAN network (Oliver 1996) undertook the responsibility of providing such a data set to the S.cerevisiae research community.
We have published a preliminary account of the Consortium’s data (Planta et al., 1999) that dealt with just 250 open reading frames (ORFs) from a single chromosome, S.cerevisiae chromosome XIV (Philippsen et al., 1997). This data set was too small for the types of analyses we now present for the transcription of 1000 ORFs. However, it did demonstrate the efficacy of our ‘control’ genes and showed that defined physiological transients may be more revealing of gene function than the quasi-steady-state conditions of mid-exponential phase batch cultures. Therefore, in this larger study, we have examined gene expression in a total of eight different physiological conditions. These include mid-exponential phase growth on a rich medium containing only non-fermentable carbon sources, the corresponding shift-up to growth on glucose and the subsequent stationary phase. Gene expression in the mid-exponential phase of cultures growing in a glucose-containing minimal medium was examined at two different temperatures (23 and 30°C) and under the following stress conditions: heat shock, hyperosmotic shock and nitrogen starvation. We have used the algorithms written by Eisen et al. (1998) to ‘mine’ these data, and generated clusters of ORFs that show similar patterns of expression under the eight physiological conditions examined. We have paid particular attention to those clusters that contain each of our ‘control’ genes and have searched the upstream regions of the ORFs in these clusters, and those containing only novel genes, for possible transcription factor target sites, using the method of van Helden et al. (1998). Finally, we have compared the clusters containing the ‘control’ genes with the equivalent clusters (generated using the same subset of yeast ORFs and using the same algorithms) from the data of DeRisi et al. (1997). We believe the results to be instructive both generally, in terms of the best way of comparing transcriptome data from different laboratories, and specifically, in terms of the utility of our unique set of northern data for the evaluation of results from hybridization-array analyses.
Results
The EUROFAN Transcription Consortium set itself the target of analysing the expression of some 1000 ORFs of previously unknown function using classical northern analysis. The Consortium decided to examine the expression of these genes under a number of different physiological conditions, both in the quasi-steady state of the mid-exponential phase of batch culture and under a number of defined physiological stress conditions. Within the Consortium, we have taken great care to standardize our protocols of RNA extraction, probe production and labelling, and hybridization. Details of these protocols may be found on the EUROFAN website at MIPS (http://www.mips.biochem.mpg.de/proj/eurofan/eurofan_1/b2/). In addition to standardizing the northern analysis procedures, we have also gone to great lengths to ensure uniformity of cell physiology in the experiments carried out in the five member laboratories of the Consortium. This proved a more exacting task. In achieving this reproducibility, we were helped greatly by the fact that all laboratories used the same yeast strain, distributed from a single source, and common recipes for growth media. The use of ‘control’ genes of known regulatory pattern that all the laboratories monitored using standard hybridization probes enabled the different laboratories to validate the comparability of their experiments with those of their partners in the Consortium. We would suggest that controlling for physiological variability will be one of the greatest problems in generating reproducible transcriptome data that may readily be compared between laboratories that are competitors, rather than collaborators. Thus, we would urge the yeast research community to standardize on strains and media to be used in such analyses.
Transcription in response to physiological transitions
To provide information on the regulation of novel ORFs at the transcription level, all 1008 ORFs were analysed under eight growth conditions (see Materials and methods). These were: glucose derepression (RNA1); glucose upshift (RNA2); stationary phase (RNA3); control at 30°C (RNA4); ammonium starvation (RNA5); hyperosmotic shock (RNA6); control at 23°C (RNA7); and heat shock (RNA8). Several control mRNAs were analysed to test the reproducibility of the physiological stimuli and the consistency of data between research groups. These were ACT1 (encoding actin, and a commonly used standard for mRNA abundance measurements; Moore et al. 1991), CAR1 (encoding arginase, involved in arginine catabolism, a control for nitrogen starvation; Dubois and Messenguy, 1997), HSP12 (encoding a heat-shock protein, a control for stress responses and stationary phase; Praekelt and Meacock, 1990), PCK1 (encoding the gluconeogenic enzyme phosphoenolpyruvate (PEP) carboxykinase, a control for glucose repression; Yin et al., 1996) and RPL25 (encoding ribosomal protein L25, a control for glucose induction and growth rate; Kraakman et al., 1993). The data set for all 1008 ORFs is available at the MIPS website (http://www.mips.biochem.mpg.de/proj/eurofan/eurofan_1/b2/index.html) and also the website http://www.yeastresearch.man.ac.uk/publications/emboj/.
Transcripts were detected under at least one of the conditions examined for 739 of the 1003 previously unknown ORFs (73%) analysed. The absolute level of detected transcripts varied over a 2000-fold range. Of the 739 ORFs with a detectable transcript, 636 ORFs exhibited a detectable transcript under all conditions. One of these 636 ORFs (N3847) has yet to be assigned a name according to the standard yeast ORF nomenclature. Of the 739 ORFs with a detectable transcript, 387 (52%) remain unclassified in the KEGG Database (Ogata et al., 1999).
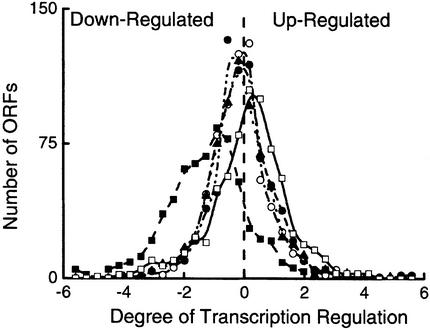
Analysis of transcript levels indicated that, overall, the degree of regulation of ORFs varied according to the physiological transient studied (see Figure 1). These data are summarized in Table I.
Fig. 1. The degree of transcription regulation in S.cerevisiae following various physiological perturbations. The degree of transcription regulation in S.cerevisiae is expressed as a frequency diagram of the number of ORFs plotted against the observed degree of regulation. The degree of regulation was calculated for each ORF for each physiological transient as detailed in Materials and methods. Down-regulated and up-regulated ORFs have negative and positive degrees of regulation, respectively. ORFs were counted in 32 uniform bins. The curves represent ORFs regulated in response to glucose upshift (open squares), heat shock (closed circles), stationary phase (closed squares), hyperosmolarity (open circles) and ammonium starvation (closed triangles).
Table I. Analysis of transcript levels of 1003 ORFs using high-throughput northerns.
| Summary statistics | |
|---|---|
| Total ORFs | 1008 |
| ‘Control’ ORFs (ACT1, HSP12, CAR1, PCK1, RPL25) | 5 |
| ORFs with a detectable transcript under at least one condition | 739 |
| ORFs with a detectable transcript under all conditions | 636 |
| ORFs with a wrong sized transcript | 35 |
| ORFs with no detectable transcript under all conditions | 229 |
| Degree of regulation |
|
|
|---|---|---|
| Transient | No. of ORFs (739) |
|
| Up-regulated | Down-regulated | |
| Glucose up-shift (G) | 413 (56%) | 281 (38%) |
| Heat shock (H) | 319 (43%) | 376 (51%) |
| Stationary phase (S) | 99 (13%) | 546 (74%) |
| Osmoshock (O) | 283 (38%) | 288 (39%) |
| Nitrogen starvation (N) | 284 (38%) | 399 (54%) |
Transcripts corresponding to 229 ORFs (23% of the total ORFs analysed) were undetectable under all conditions investigated. Thirty-five ORFs did not yield a transcript of the expected length. Of the 229 undetectable ORFs, 169 remain unclassified according to the KEGG database. The KEGG classifications of all 1008 ORFs, 264 (229 + 35) ORFs with undetectable transcripts or transcript of the wrong length, and 635 ORFS with a detectable transcript under all conditions are available for download at the website http://www.yeastresearch.man.ac.uk/publications/emboj/. We note that none of the 229 ORFs is classified as a ‘questionable’ gene according to the MIPS database (Mewes et al., 2000).
Global controls on transcription
One advantage of such a large set of transcriptional data is that it should be possible to use it to identify global effects on the pattern of gene expression. In our previous paper on a subset of 250 ORFs from the present set of >1000 (Planta et al., 1999), we examined position effects and the relationship between gene expression and codon bias. In analysing this larger set of ORFs, we have revisited these two issues. As in the more limited study, we found no major effect of the position of an ORF on a chromosome arm and its level of expression (details of this investigation may be found at http://www.yeastresearch.man.ac.uk/publications/emboj/). In contrast, the investigation of the relationship between codon bias and expression level did yield results of biological significance.
Codon bias
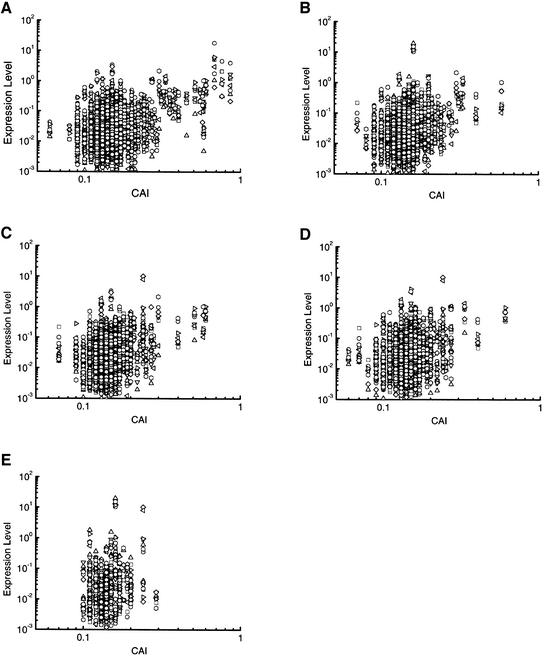
As with the subset of 250 ORFs previously examined (Planta et al., 1999), we found no simple correlation between the level of expression (under any condition) and the codon adaptation index (CAI; Sharp and Cowe, 1991) of the 1008 ORFs examined here. We have refined this analysis to undertake separate assessments of the relationship between CAI and expression level for all ORFs whose expression was regulated under at least one of the physiological conditions that we examined. Figure 2 plots the absolute expression level against CAI for each regulated ORF for each physiological transient. It shows that ORFs whose expression was induced upon glucose up-shift have the highest proportion of members with high CAI values (Figure 2A). In contrast, few or no ORFs whose expression level was induced during the (carbon-limited) stationary phase have a high CAI value (Figure 2E). ORFs whose transcription is elevated in stress conditions, which (in contrast to prolonged starvation) do not cause growth to cease, but merely to slow down, include an intermediate proportion of high CAI members. This applies to hyperosomotic shock and heat shock, two stress conditions that reduce (but do not halt) yeast growth. It also applies to nitrogen starvation; this is because, under our experimental protocol (see Materials and methods), mRNA levels are assessed just 2 h after the transfer to ammonium-free medium—a time when growth has become nitrogen limited but long before it ceases altogether.
Fig. 2. Correlation of transcript expression level with the codon adaptive index (CAI) for up-regulated S.cerevisiae ORFs. The transcript levels for each ORF that was up-regulated following the physiological perturbations: (A) glucose up-shift (413 ORFs); (B) heat shock (319 ORFs); (C) stationary phase (99 ORFs); (D) hyper osmolarity (283 ORFs); and (E) ammonium starvation (284 ORFs) are plotted against the CAI for the ORF. Data points are the transcript levels for ORFs detected in the eight physiological conditions examined, YPGE (squares), glucose up-shift (circles), stationary phase (triangles), YNB-glucose 30°C (inverted triangles), ammonium starvation (wide diamonds), hyperosmolarity (right-facing triangles), YNB-glucose 23°C (left-facing triangles) and heat shock (narrow diamonds).
Cluster analysis
Clustering techniques are used to analyse further the pattern of transcriptome data to detect groups of co-regulated genes or physiological conditions that evoke a similar transcriptional response from the cells. A common computational approach is hierarchical clustering (Eisen et al., 1998). We applied the hierarchical clustering approach to expression profiles for 635 ORFs whose levels were detected in all of the physiological con ditions examined. Cluster analysis was performed using the GeneCluster and TreeView software (Eisen et al., 1998).
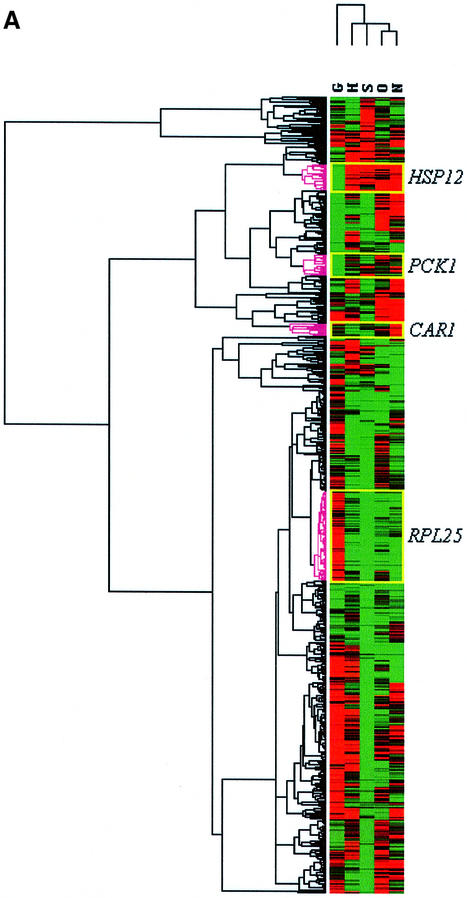
No attempt was made, in this analysis, to determine whether the variation between conditions was statistically significant. ORFs whose transcripts were undetectable in at least one condition were excluded. The result of the cluster analysis for the transcriptome patterns of these 635 ORFs is presented in Figure 3A. Red or green colours denote that the transcription of a given ORF was increased or decreased, respectively, between the two physiological conditions used to define the transient. The intensity of each colour is the relative level of up- or down-regulation. A central feature of Figure 3A is that it may be examined to identify patterns of interest, and it is easy to zoom in on the detailed expression patterns and identify the genes contributing to these patterns. Readers may do this for themselves by visiting the following website: http://www.yeastresearch.man.ac.uk/publications/emboj/. The website provides access to our raw data, and the data files produced by the clustering software. Interestingly, the cluster analysis not only permits the grouping of co-regulated genes, but also clusters physiological transients that evoke a similar pattern of transcription from the yeast cells. Our analysis has grouped the stress transients, hyperosmolarity (O), ammonium starvation (N), stationary phase (S) and heat shock together (H), while glucose up-shift (G) is significantly isolated. The biological significance of these groupings will be considered later (see Discussion).
Fig. 3. Cluster analysis of expression profiles for 635 ORFs in S.cerevisiae expressed following various physiological transients. (A) Cluster analysis of northern data. (B) Cluster analysis of the DeRisi et al. (1997) data subset. (C) Exploded cluster of HSP12 co-regulated ORFs from northern data. ORFs are represented as rows, and physiological conditions as columns in the matrix. Red, black and green elements in the matrix indicate up-regulated, no change and down-regulated ORFs, respectively. Cluster analysis was performed on expression profiles of 635 ORFs across the five physiological transients, glucose upshift (G), heat shock (H), stationary phase (S), hyperosmolarity (O) and ammonium starvation (N). The horizontal and vertical dendrograms indicate the degree of similarity between expression profiles for ORFs and physiological transients, respectively. The yellow boxes show areas of the matrix that correspond to clusters of ORFs closely related to and including the control ORFs, HSP12, CAR1, RPL25 and PCK1.
A major purpose of applying cluster analysis to transcriptional data is to group genes of so far undetermined function with genes of known function in order to provide clues to the function of the novel genes via a process of ‘guilt by association’. Our experiments have concentrated on a set of 1000 ORFs that were of unknown function when the EUROFAN project started; these ORFs were chosen at random from chromosomes sequenced by our European Yeast Genome Sequencing Consortium (see Oliver, 1996). However, since we included five ‘control’ genes, for which not only their function but also their pattern of regulation was well established, it was of interest to explore the clusters into which these ‘control’ genes were grouped.
HSP12 formed a cluster with 20 other ORFs that are down-regulated in response to glucose upshift (Figure 3C), but up-regulated during the stress transients, ammonium starvation, stationary phase, hyperosmolarity and heat shock. Members of this cluster are genes involved in ‘metabolism’ (one), ‘cell growth/division/DNA synthesis’ (one), ‘protein distribution’ (one), ‘cell rescue’ (two) and unclassified (15). The HSP12 cluster consisted of 21 ORFs, of which 18 contained an AGGGG|CCCCT sequence in their upstream region. (as analysed using the RSA Tools package developed by van Helden et al., 1998; http://copan.cifn.unam.mx/Computational_Biology/yeast-tools/; see the website http://www.yeastresearch.man.ac.uk/publications/emboj/ for data on the significance of such occurrences). This pentanucleotide is the known core consensus sequence of the stress response element (STRE), which mediates activation in response to a wide variety of stress conditions (Mager and DeKruijff, 1995), including heat shock and osmotic stress. This observation suggests that the HSP12 cluster obtained from our northern data has a high biological significance.
A cluster of 27 ORFs displayed a similar expression pattern to that of PCK1 (Figure 3A). Four ORFs, including PCK1, were classified in ‘metabolism’, two in ‘energy’ and one each in ‘transcription’, ‘cellular organization’, ‘cell rescue’, ‘intracellular transportation’ and ‘cell growth/cell division/DNA synthesis’. The remainder of the cluster (16) were unclassified. The cluster showed a pattern of down-regulation in the glucose up-shift transient and up-regulation in the stationary phase transient. This pattern of expression is consistent with previously published studies for PCK1 (Yin et al., 1996).
CAR1 is a ‘control’ gene for nitrogen starvation and its transcription should be induced upon ammonium deprivation. A set of 10 ORFs cluster with CAR1 in the N-expression column (red for ammonium starvation condition), indicating that they were significantly induced under that condition (Figure 3). Of the ORFs in this cluster, four are unclassified. Six ORFs (including CAR1) in this cluster fell into the ‘metabolism’, ‘cellular organization’, ‘intracellular transportation’ and ‘cell growth/cell division/DNA synthesis’ classes.
RPL25 encodes the ribosomal protein L25. It was used as a control for glucose-induced genes and for genes that are regulated in response to growth rate changes. This cluster contained 71 ORFs that were classified into 10 different KEGG classes. The top five classes were unclassified (29), ‘transcription’ (10), ‘metabolism’ (eight), ‘protein synthesis’ (seven) and ‘cell growth/cell division/DNA synthesis’ (six). According to Figure 3A, the ORFs that are clustered together with RPL25 show a pattern of being down-regulated when starved for either glucose or ammonium, but up-regulated following a glucose up-shift transient.
The ORFs of the RPL25 cluster have target sites for Abf1p in their upstream regions. This abundant, essential activator was already known to include two other ribosomal protein genes (RPL2A and RPL2B) amongst its targets. However, it is notable that a novel putative target site GACGA|CTGCT occurs in the upstream regions of the ORFs of this cluster at a more significant frequency. None of the target sites found in the upstream regions of the other clusters, including those for which CAR1 and PCK1 are the ‘type’ genes, makes any obvious biological sense (at least, to these authors). However, it is noteworthy that for only one of the clusters comprising entirely genes of unknown function was the target site for any known transcriptional activator found. The upstream regions of a co-regulated cluster of 26 ORFs, including YLR116w, all contained target sites for the Rme1p activator (readers are referred to the website http://www.yeastresearch.man.ac.uk/publications/emboj/ for details and data on the significance of occurrences of target sites for particular transcriptional activators).
Evaluating the ‘Rosetta Stone’: a comparison of northern and microarray data
A comparison of our gene expression data set, obtained by high-throughput northern analysis, with an equivalent data set produced by hybridization array analysis was made for a number of reasons. First, it allows a comparison of the relative sensitivities and dynamic ranges of the two expression analysis systems, so providing a bridge between current genome-wide analyses of the transcriptome and all of the data in the literature that used classical northern techniques to monitor the expression of specific genes. Secondly, it will become increasingly necessary to compare large gene expression data sets both between different experiments and between different laboratories. An exercise that involves two very different technologies may more readily reveal any dangers or pitfalls that may beset such comparisons.
DeRisi et al. (1997) investigated the ‘diauxic shift’ that occurs when a batch culture of S.cerevisiae exhausts the supply of fermentable sugar in the growth medium and turns to the consumption of ethanol by aerobic respiration. In their experiments, the Stanford group allowed an initial 9 h of growth and then cells were harvested at seven successive two hourly intervals. Microarrays of PCR products of all 6000 yeast ORFs permitted a comprehensive analysis of mRNA abundance. We have examined a wider range of physiological conditions than in the paper from the Stanford group. However, there are points of equivalence between the two data sets; for instance, the seventh sample taken in the DeRisi et al. (1997) experiment represents a similar condition to that of the glucose-limited stationary phase (RNA3) in our own experiments. The DeRisi et al. (1997) data were extracted from the Stanford database (http://cmgm.stanford/edu/pbrown/explore/).
To further our analysis, we were interested in determining whether those ORFs with undetectable transcripts in our northern hybridization data were also undetectable in the DeRisi et al. (1997) data set. To select ORFs for which the presence of a transcript was questionable in the DeRisi data, a threshold criterion of mean expression level (signal – background) ≤1.960 SDs (equivalent to a 95% confidence limit) was applied. Of the 6153 ORFs reported by DeRisi et al., 1760 (29%) fell below this threshold. However, only 52 (23%) of these were common with the 229 ORFs for which a transcript was undetected in our data. The mean expression level in the DeRisi data for our undetected 229 ORFs was 1983 ± 2187 (SD) counts. The mean expression level for all ORFS was 3133 ± 3389 (SD) counts. This, and the observation that only 52 ORFs were common in both ‘undetectable’ transcript data sets, suggests that the 229 ORFs undetected in our study are a random selection of genes that, overall, are expressed at low levels. It is noteworthy that the fraction of ORFs that we consider undetectable from the data of DeRisi (29%) is greater than that obtained with our northern analysis (23%). This suggests that northern analysis is marginally more sensitive than microarray analysis at detecting low-level transcripts. Clearly, this comparison is dependent upon the stringency of the criterion used for selecting ORFs for which the presence of a transcript is questionable. Furthermore, currently, we are unable to exclude the possibility that the observed differences between our data and the DeRisi data are due to unknown differences in the physiological conditions examined and/or yeast strain used.
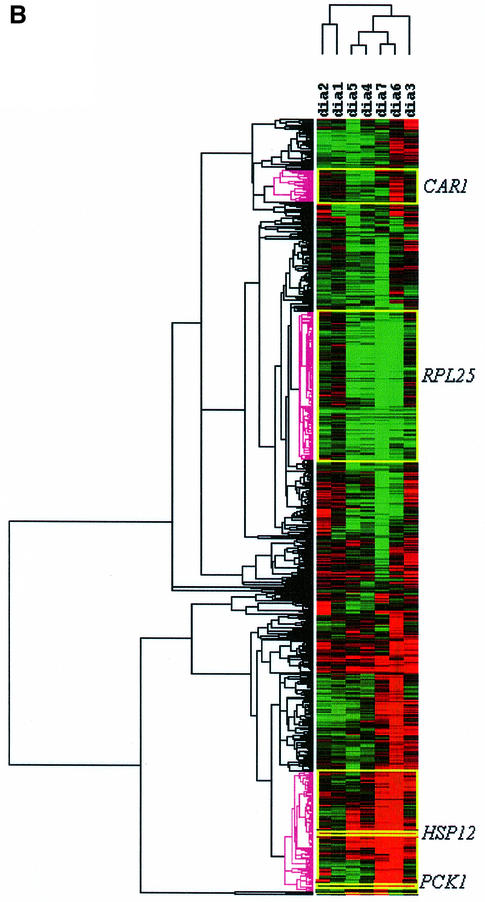
To compare northern hybridization data with microarray data further, we examined the clusters of co-regula ted ORFs produced following hierarchical clustering of our data and the DeRisi et al. (1997) data. In an initial analysis of the output from the clustering software, we examined the clusters that contained our ‘control’ genes (PCK1, HSP12, RPL25 and CAR1) and compared them with the clusters containing the same genes from the DeRisi et al. (1997) data. The results of the cluster analysis for the expression profiles of 635 ORFs from the data of DeRisi et al. (1997) are presented in Figure 3B.
Praekelt and Meacock (1990) reported that HSP12 gene expression is induced by heat shock and on entry into stationary phase. A similar response was observed in both our data set and that of DeRisi et al. (1997). However, analysis of the DeRisi data yielded a much larger cluster of 98 ORFs, which exhibited co-regulation with HSP12. We note that the Pearson correlation coefficients for our HSP12 cluster and the DeRisi HSP12 cluster were 0.84 and 0.86, respectively. The DeRisi HSP12 cluster contained 66 ORFs (67%) with the STRE activator sequence. In contrast, a significantly higher fraction of our HSP12 cluster contained the STRE activator sequence (18 of 21; 86%). Comparison of the two clusters of ORFs identified 14 in common. The ORFs with the STRE activator sequence uniquely identified in our cluster were RER2, YJR008w, YLR057w and AGP2. As our experimental conditions were more tailored to the analysis of stress conditions, it is highly likely that the different physiological conditions used in preparing the two data sets contributed to the differences observed in the clusters obtained.
Cluster analysis of the DeRisi et al. (1997) data set produced clusters of 19, 60 and 26 ORFs that were co-regulated with PCK1, RPL25 and CAR1, respectively. The two RPL25 clusters showed that 16 ORFs were found in both clusters, five of which remain unclassified. In contrast to the HSP12 and RPL25 clusters, PCK1 and CAR1 showed only two and three ORFs in common. It was not unexpected that the two CAR1 clusters had little in common. Our study looked specifically at ammonium starvation, and also ammonium limitation (the consequence of a glucose up-shift), while it can be inferred that the cells in the Stanford group’s batch culture (DeRisi et al., 1997) were always in conditions of nitrogen excess. However, the lack of congruence between the two PCK1 clusters was unexpected. This might be explained readily in terms of the way in which the data from the two laboratories were normalized. DeRisi et al. (1997) used two-colour fluorescence ratios to quantify the data from their microarrays. The reference sample used was their time zero point and, therefore, all of their data are referenced to the lag phase condition. In this condition, the glucose concentration in the growth medium was at its maximum. In contrast, the data for our cluster analysis were calculated using a mid-exponential phase sample (see Materials and methods) as the reference. We expected the glucose concentration to be reduced in the medium in mid-exponential phase. PCK1 encodes the gluconeogenic enzyme PEP carboxykinase, and its expression is subject to glucose repression (Yin et al., 1996). Hence, we might expect expression of PCK1 and its co-regulated genes to be induced in our reference condition. Thus, this difference may explain the lack of congruence in the two PCK1 clusters. This illustrates the importance of the effect of data normalization procedures on the interpretation of transcription data.
Discussion
This northern analysis of the expression levels of >1000 yeast genes under eight different physiological conditions is probably the largest set of transcription data that has ever been collected using this classical technique. Moreover, it is unlikely to be surpassed since, although a large number of ORFs have been analysed, it is still well short of the complete yeast transcriptome of some 6000 mRNA molecules that may be studied readily using either macro- or microarray technology (Lashkari et al., 1997; Wodicka et al., 1997; Hauser et al., 1998). This data set, therefore, serves two purposes. It is a valuable resource, in its own right, that may be mined for new biological information concerning the regulation of gene expression in S.cerevisiae. Secondly, since it is a large enough data set to allow analysis using the clustering algorithms commonly used to mine hybridization array data, it provides a means of relating transcriptome data to all of the yeast gene expression data, obtained by northern analysis, that exist in the literature.
To demonstrate the utility of our data in the evaluation of transcriptome studies, we have compared the gene clusters produced, using the Eisen algorithm (Eisen et al., 1998), from 635 ORFs showing variable levels of expression in our eight physiological conditions, with the clusters produced from the same set of 635 ORFs using the microarray data of DeRisi et al. (1997). If the clusters containing our control genes are examined, it is found that there is reasonable agreement between the DeRisi et al. (1997) data and our own for both the RPL25 and HSP12 clusters. However, for the clusters containing CAR1 and PCK1, there is only one ORF (besides the control gene itself) in common between the two clusters. In the case of CAR1 and genes co-regulated with it, this is likely to reflect the fact that only the northern experiments provided data relevant to their functions. Our experiments included both ammonium starvation (RNA5) and glucose up-shift (RNA2); the latter condition is likely to result in ammonium limitation, as is confirmed by the fact that the Eisen algorithm clusters the RNA5 and RNA2 conditions together. In contrast, the diauxic shift examined by DeRisi et al. (1997) involves a transition to a slower growth rate, and it is likely that nitrogen is in excess throughout their batch growth experiment. Thus, their experiment would have produced no data relevant to nitrogen regulation of gene expression. This emphasizes the need to compare biologically equivalent situations when mining transcriptome data from a number of different experiments, and also to ensure the experimental design is appropriate to the biological questions being posed.
The gene clusters generated from our northern data on gene expression have been used to identify putative regulatory sequences in the promoter regions upstream of the ORFs. For those clusters that included one of our ‘control’ genes, we discovered target sites for known transcriptional activators in their upstream regions (although the cognate activator did not always appear appropriate to the pattern of gene regulation observed). In contrast, for the clusters comprised entirely of ORFs of unknown function, only in one case did we identify the target site of a known transcriptional activator. This leads to the rather sobering conclusion that not only do these novel ORFs have so far undiscovered functions, but that they are also organized into regulatory networks that have yet to be revealed by experimental science. Thus, there may be entire pathways in yeast that so far have evaded experimental analysis.
We would like to make some general observations about cluster analysis and the comparison of clusters derived from different experiments and/or different laboratories. The first is that the clusters become more refined, and are more likely to contain ORFs that have similar transcription factor target sites in their upstream regions, the more conditions that are relevant to a particular physiological response are analysed. Thus the northern data, which included four different stress conditions, produced an HSP12 cluster that contained a significantly higher fraction of ORFs with an STRE element in their upstream regions. Secondly, while different numbers of ORFs included in a cluster analysis will obviously produce different clusters, we would note that the number of ORFs analysed is a function of the threshold of detectability employed. We have always excluded ORFs whose northern signals are so low as to be considered undetectable from our quantitative analyses. In contrast, it is common practice in analysing array data by the two-colour ratio technique to include all data points in the ratios calculations. We have found that very different cluster patterns are produced according to whether data below the threshold of detection (in either northern or array data) are included in the analysis. We intend to offer a more detailed assessment of this problem in a future publication.
It is to be hoped that large-scale analyses of gene expression in yeast will tell us something about the global controls that operate on transcription in this model eukaryotic organism. While it is more likely that such controls will be revealed by the comprehensive analyses provided by the hybridization arrays, our northern data set was sufficiently large to make it worthwhile examining some possible global phenomena.
Our analysis of the effect of chromosomal position on the maximum level of gene expression failed to reveal any obvious effect on the maximum level of expression of the 1008 ORFs studied.
Our studies of the correlation between the CAI of the ORFs studied and their expression levels revealed three classes of effect. Those ORFs whose expression is induced upon a growth rate up-shift include the highest proportion of members with high CAI values, while no such members are found in the class of ORFs that are induced during stationary phase. The class of ORFs whose expression is induced by stress conditions that reduce, but do not halt, growth includes an intermediate proportion of members with a high CAI. These data are compatible with a model in which genes require a favourable codon bias if their expression needs to be significantly increased under conditions where there is competition between mRNAs for a limiting translational capacity. This is the case for glucose up-shift, where an increase in ribosome biogenesis is required to support the increase in growth rate. According to this model, those genes induced under conditions where there is excess translational capacity (such as starvation) do not require a favourable codon bias. Previous work in one of our laboratories (Dickson and Brown, 1998) has indicated that there is excess translational capacity in stationary phase yeast cells.
In all, our high-throughput northern analysis of yeast gene expression has generated a number of hypotheses that make specific predictions that are amenable to further experimental tests. The data have also revealed a number of important factors that must be taken into account when comparing transcriptome data between different laboratories, the most important of which are data normalization regimes and physiological equivalence. In the latter context, it cannot be emphasized too strongly that the comprehensive nature of transcriptome analysis demands greater care in the design of experiments and much more stringent controls than have been necessary when studying only a small subset of an organism’s genes.
Materials and methods
Growth conditions
All analyses were performed on S.cerevisiae strain FY73 (MATα, his3Δ200, ura3-52; Winston et al., 1995). Analyses were performed under eight ‘transient’ conditions. Cells were grown in YPGE (1% yeast extract, 1% bacto-peptone, 2% glycerol, 1% ethanol) at 30°C until the OD600 reached 0.8, whereupon one-third of the cells were harvested for analysis (RNA1; glucose derepression). The remaining cells were washed in sterile distilled water, resuspended in an equal volume of YPD (1% yeast extract, 1% bacto-peptone, 2% glucose) and grown for 60 min at 30°C. Half of these cells were harvested for analysis (RNA2; glucose up-shift), whereas the remaining cells were harvested after a further 24 h of growth (RNA3; stationary phase). Separate cultures were set up in 0.67% yeast nitrogen base without amino acids, 4% glucose, 20 µg/ml uracil, 20 µg/ml histidine, and cells were grown at 30°C until the OD600 reached 0.8. One-third of these cells were harvested for analysis at this point (RNA4; 30°C control). The second third of these cells were washed in sterile distilled water, resuspended in 2 vols of 0.67% yeast nitrogen base without amino acids or ammonium sulfate, 4% glucose, 20 µg/ml uracil, 20 µg/ml histidine, and starved for 2 h at 30°C (RNA5; nitrogen starvation). NaCl was added (0.7 M final concentration) to the final third of cells, which were harvested after 60 min at 30°C (RNA6; osmo stress). Finally, separate cultures were grown at 23°C in 0.67% yeast nitrogen base without amino acids, 4% glucose, 20 µg/ml uracil, 20 µg/ml histidine, to an OD600 of 0.8, whereupon half of the cells were taken for analysis (RNA7; 23°C control). The other half was incubated at 36°C for 30 min before harvesting (RNA8; heat shock).
Northern analysis
RNA was extracted from yeast cells as described previously (Lindquist, 1981; Brown, 1995). Northern analyses and quantification of transcript levels were also performed as described previously (Planta et al., 1999). Transcript levels were measured relative to total RNA by loading approximately equal amounts of RNA on northern gels. Details of the oligonucleotide primers used to generate each PCR probe are available at the MIPS WWW site: (http://www.mips.biochem.mpg.de/proj/eurofan/eurofan_1/b2/). This site also contains the complete set of raw data generated from this study. Hybridization signals were quantified by direct two-dimensional phosphorimaging of northern membranes using the Bio-Rad GS-525 Molecular Imager® System or PhosphorImager™ 425 from Molecular Dynamics.
Data analysis
Two types of data were calculated for each transcript using the signals obtained by phosphorimaging: absolute and relative mRNA levels. The absolute mRNA level was calculated based on the maximum level observed for the target transcript under the conditions analysed. Although, it is known that yeast ACT1 mRNA levels change during growth and starvation (Delbruck and Ernst, 1993), ACT1 is still a commonly used reference for mRNA abundance measurements, and was, similarly, our choice in this work. The absolute mRNA level for each ORF was calculated using the equation given in Planta et al. (1999). Relative mRNA levels were obtained by comparing the signals for each target transcript under the various growth conditions analysed. These were calculated relative to the maximum signal obtained for this transcript; the relevant method for this may be found in Planta et al. (1999).
To determine the degree of regulation of each ORF between the different conditions analysed, we processed the data as follows. The expression level of each ORF was calculated by multiplying the absolute and relative mRNA levels. This gave the expression level as total mRNA hybridized to the membrane for each ORF in a particular condition. We note that this expression level value was relative to the reference, ACT1, in exponential phase. The data set from one condition was divided by the data set from a reference condition, and converted to logarithm (base 2). We term this value the degree of regulation. Positive and negative values refer to up- and down-regulated ORFs with respect to the reference condition. ORFs whose transcripts were undetectable in at least one condition were excluded. As a result, only 635 ORFs were considered in our analysis of the degree of regulation.
The degree-of-regulation data across five transients produced a matrix from which a gene expression similarity metric (the Pearson correlation coefficient) was calculated using the implementation reported by Eisen et al. (1998). Subsequently, we analysed the similarity metrics to determine clusters of ORFs with similar expression profiles. The cluster analysis was performed using the software tools written by Eisen (Eisen et al., 1998; http://rana.stanford.edu/clustering/), GeneCluster and TreeView. All data used for clustering (including calculated values of similarity) are available for download at our website, http://www.yeastresearch.man.ac.uk/publications/emboj/.
The functions of the ORFs were categorized by using the 16 functional classes defined by the KEGG (Ogata et al., 1999) and the MIPS Yeast Genome (Mewes et al., 2000) databases. Using the Eisen et al. (1998) software, it was possible to cluster genes according to similarities in their expression profiles. We examined upstream regulatory sequences on clusters of ORFs that exhibited similar expression profiles to the ‘control’ ORFs, HSP12, CAR1, PCK1 and RPL25. We hypothesized that such ORFs would have similar transcription factor-binding sites, of 5–25 bp, within several hundred base pairs upstream of the respective initiator ATGs (Mellor 1993). Identification of possible upstream gene regulatory sites in groups of co-regulated genes was performed using the RSA Tools (http://www.ucmb.ulb.ac.be/bioinformatics/rsa-tools/). We considered the region located between the transcription start and immediate upstream coding sequence as putative regulatory sequences. We chose upstream regions from –800 to –1 bp, based on the knowledge that 99% of the known upstream elements exist between these limits (van Helden et al., 1998). We scanned oligonucleotide sequences of between 4 and 8 bp. The occurrences of each oligonucleotide, on both DNA strands, were summed.
Acknowledgments
Acknowledgements
This paper is dedicated to the memory of Rudi Planta, whose untimely death occurred during the preparation of the manuscript. This work was supported by the CEC within the framework of the EUROFAN Programme. The Spanish group is grateful for additional support from CICYT (BIO96.2022-CE). The Manchester group was also supported by a grant to S.G.O. from the Engineering and Biological Systems Committee of the BBSRC. F.R. thanks the Government of Indonesia for a post-graduate scholarship.
References
- Brown A.J.P. (1995) Preparation of total RNA. In Evans,I. (ed.), Methods in Yeast Molecular Biology. Humana Press, Totowa, NJ, pp. 269–267.
- Brown M.P.S., Grundy,W.N., Lin,D., Cristianini,N., Sugnet,C.W., Furey,T.S., Ares,M. and Haussler,D. (2000) Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl Acad. Sci. USA, 97, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey H. et al. (1995) The nucleotide-sequence of chromosome I from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 92, 3809–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R.J. et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- Chu S. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- Clarke L. (1998) Centromeres: proteins, protein complexes and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev., 8, 212–218. [DOI] [PubMed] [Google Scholar]
- Delbruck S. and Ernst,J.F. (1993) Morphogenesis-independent regulation of actin transcript levels in the pathogenic yeast Candida albicans. Mol. Microbiol., 10, 859–866. [DOI] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Dickson L.M. and Brown,A.J.P. (1998) mRNA translation in yeast during entry into stationary phase. Mol. Gen. Genet., 259, 282–293. [DOI] [PubMed] [Google Scholar]
- Dubois E. and Messenguy,F. (1997) Integration of the multiple controls regulating the expression of the arginase gene CAR1 of Saccharomyces cerevisiae in response to different nitrogen signals: role of Gln3p, ArgRp-Mcm1p and Ume6p. Mol. Gen. Genet., 253, 568–580. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A. et al. (1996) Life with 6000 genes. Science, 274, 546–563. [DOI] [PubMed] [Google Scholar]
- Hauser N.C., Vingron,M., Scheideler,M., Krems,B., Hellmuth,K., Entian,K.D. and Hoheisel,J.D. (1998) Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast, 14, 1209–1221. [DOI] [PubMed] [Google Scholar]
- Kell D.B. and King,R.D. (2000) On the optimization of classes for the assignment of unidentified reading frames in functional genomics programmes: the need for machine learning. Trends Biotechnol., 18, 93–98. [DOI] [PubMed] [Google Scholar]
- Kraakman L.S., Griffioen,G., Zerp,S., Groeneveld,P., Thevelein,J.M., Mager,W.H. and Planta,R.J. (1993) Growth related expression of ribosmoal protein genes in Saccharomyces cerevisiae. Mol. Gen. Genet., 239, 196–204. [DOI] [PubMed] [Google Scholar]
- Lashkari D.A., DeRisi,J.L., McCusker,J.H., Namath,A.F., Gentile,C., Hwang,S.Y., Brown,P.O. and Davis,R.W. (1997) Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl Acad. Sci. USA, 94, 13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. (1981) Regulation of protein synthesis during heat shock. Nature, 293, 311–314. [DOI] [PubMed] [Google Scholar]
- Mager W.H. and DeKruijff,A.J.J. (1995) Stress-induced transcriptional activation. Microbiol. Rev., 59, 506–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J. (1993) Multiple interactions control the expression of yeast genes. In Broda,P.M.A., Oliver,S.G. and Sims,P.F.G. (eds), The Eukaryotic Genome: Organisation and Regulation. Cambridge University Press, Cambridge, UK, pp. 275–320.
- Mewes H.W. et al. (1997) Overview of the yeast genome. Nature, 387, 7–8. [DOI] [PubMed] [Google Scholar]
- Mewes H.W. et al. (2000) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 28, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.A., Sagliocco,F.A., Wood,R.M.C. and Brown,A.J.P. (1991) Yeast glycolytic mRNAs are differentially regulated. Mol. Cell. Biol., 11, 5330–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., Goto,S., Sato,K., Fujibuchi,W., Bono,H. and Kanehisa,M. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res., 27, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. (1996) A network approach to the systematic analysis of yeast gene function. Trends Genet., 12, 241–242. [DOI] [PubMed] [Google Scholar]
- Oliver S.G. (1997) From gene to screen with yeast. Curr. Opin. Genet. Dev., 7, 405–409. [DOI] [PubMed] [Google Scholar]
- Philippsen P. et al. (1997) The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature, 387, 93–98. [PubMed] [Google Scholar]
- Planta R.J. et al. (1999) Transcript analysis of 250 novel yeast genes from chromosome XIV. Yeast, 15, 329–350. [DOI] [PubMed] [Google Scholar]
- Praekelt U.M. and Meacock,P.A. (1990) HSP12, a new small heat shock gene of Saccharomyces cerevisiae—analysis of structure, regulation and function. Mol. Gen. Genet., 223, 97–106. [DOI] [PubMed] [Google Scholar]
- Sharp P.M. and Cowe,E. (1991) Synonymous codon usage in Saccharomyces cerevisiae. Yeast, 7, 657–678. [DOI] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J., Andre,D. and Collado-Vides,J. (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol., 281, 827–842. [DOI] [PubMed] [Google Scholar]
- Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E., Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Characterization of the yeast transcriptome. Cell, 88, 243–251. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard,C. and Ricuperohovasse,S.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Schena,M. and Davis,R.W. (1999) Fluorescence-based expression monitoring using microarrays. Methods Enzymol., 306, 3–18. [DOI] [PubMed] [Google Scholar]
- Wodicka L., Dong,H.L., Mittmann,M., Ho,M.H. and Lockhart,D.J. (1997) Genome-wide expression monitoring in Saccharomyces cerevisiae. Nature Biotechnol., 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- Yin Z.K., Smith,R.J. and Brown,A.J.P. (1996) Multiple signalling pathways trigger the exquisite sensitivity of yeast gluconeogenic mRNAs to glucose. Mol. Microbiol., 20, 751–764. [DOI] [PubMed] [Google Scholar]