Abstract
Sulf-2 is an endosulfatase with activity against glucosamine-6-sulfate modifications within subregions of intact heparin. The enzyme has the potential to modify the sulfation status of extracellular heparan sulfate proteoglycan (HSPG) glycosaminoglycan chains and thereby to regulate interactions with HSPG-binding proteins. In the present investigation, data mining from published studies was employed to establish Sulf-2 mRNA upregulation in human breast cancer. We further found that cultured breast carcinoma cells expressed Sulf-2 mRNA and released enzymatically active proteins into conditioned medium. In two mouse models of mammary carcinoma, Sulf-2 mRNA was upregulated in comparison to its expression in normal mammary gland. Although mRNA was present in normal tissues, Sulf-2 protein was undetectable; it was, however, detected in some premalignant lesions and in tumors. The protein was localized to the epithelial cells of the tumors. In support of the possible mechanistic relevance of Sulf-2 upregulation in tumors, purified recombinant Sulf-2 promoted angiogenesis in the chick chorioallantoic membrane assay.
Keywords: extracellular endosulfatase, Sulf-2, breast cancer, angiogenesis, heparan sulfate
Introduction
Heparan sulfate proteoglycans (HSPGs) exist on cell surfaces and in the extracellular matrix (ECM) [1–3]. Heparan sulfate glycosaminoglycan (GAG) chains consist of repeating disaccharide units of glucuronic/iduronic acid and glucosamine in a linear chain. One or more of these chains are attached covalently to a protein core to form the proteoglycan. HSPGs influence a variety of biologic processes by binding to a multiplicity of molecules. These include signaling molecules such as cytokines, chemokines, growth factors, morphogens, and angiogenic factors [e.g., fibroblast growth factors (FGFs), vascular endothelial growth factor (VEGF), and hepatocyte growth factor (HGF)]. The disaccharide units of HS chains have four different sulfation modification sites, which are substituted on the N-, 3-O, and 6-O positions of glucosamine and the 2-O position of uronic acid residues. The binding interactions of HSPGs require specific patterns of sulfation in subregions of the HS chains (reviewed by Esko and Selleck [4]). Previous work, for example, has revealed that specific sulfation patterns play an essential role in the trimolecular interaction between heparin (a structural analogue of HSPG GAG chains), FGF, and the fibroblast growth factor receptor (FGFR) [3]. Thus, there is a requirement for N- and 2-O-sulfate groups of heparin for binding to FGF-2, and a requirement for 6-O-sulfate for binding to FGFR-1. Sulfation at the 6-O position of glucosamine residues is also required for optimal binding of VEGF, FGF-1, FGF-10, and IL-8 to HS or heparin [5–8]. These observations indicate that modulation of the sulfation status of HSPGs could potentially regulate extracellular signaling.
Classic sulfatases are mainly localized in lysosomes where they hydrolyze sulfate ester bonds from a wide variety of substrates (e.g., GAGs, sulfolipids, and steroid sulfates) [9,10]. QSulf-1 is the first discovered member of a new family of sulfatases that are targeted to the exterior of cells [11,12]. QSulf-1 was identified as a developmentally regulated protein that modulates Wnt signaling, apparently by altering HSPG sulfation. We previously cloned the orthologue of QSulf-1 in mice and humans and found a closely related homologue, called Sulf-2 [13]. The predicted proteins are 870 to 871 amino acids in length, with a sulfatase domain of ∼370 residues at the amino terminus, a basic hydrophilic domain of ∼310 residues, and an additional sulfatase-related domain of ∼110 residues at the C-terminus. The proteins are endoproteolytically processed in the secretory pathway through furin-mediated cleavage. We showed that these Sulfs exhibit both arylsulfatase activity and highly specific endoglucosamine-6-sulfatase activity against heparin with a neutral pH optimum [13]. Subsequently, endosulfatase activity against heparan sulfate was demonstrated [11,14]. Recent studies have reported that the expression of HSulf-1 can downregulate the signaling responses of cells to several growth factors [FGF-2, HGF, and heparin-binding epidermal growth factor (HB-EGF)] [15–18].
Using publicly available serial analysis of gene expression (SAGE) data, we found that the expression of Sulf-2 mRNA is significantly higher in human breast carcinomas, brain tumors, and colon carcinomas than in their normal counterpart tissues [19]. The expression level of Sulf-2 is eight-fold higher in breast carcinomas than in normal tissues. In the present investigation, we further investigate the association of Sulf-2 with breast carcinomas.
Materials and Methods
Preparation of Anti–Sulf-2 Polyclonal Antibodies
Polyclonal antibodies were produced by ProSci (Poway, CA). Rabbits were immunized with KLH conjugates of three peptides derived from the predicted sequence of HSulf-2 (H2.1: CFLSHHRLKGRFQRDRR-COOH; H2.3: GGSRALSNLVPKYYGQGSEAC-COOH; and H2.5: CKWPEMKRPSSKSLGQLWE-COOH, where C denotes the cysteine residues added for coupling). The peptides chosen are highly homologous to the corresponding regions in MSulf-2 (15/16, 20/21, and 18/18 identity, respectively) but lack homology to the corresponding regions in either HSulf-1 or MSulf-1. Antibodies were purified by sequential passage of the rabbit sera through affinity columns, each consisting of one of the peptides coupled to SulfoLink Coupling Gel (Pierce, Rockford, IL), which was used according to the manufacturer's instructions. Antisera and purified antibody titers were determined by ELISA using specific peptides, such as the coated antigen.
Human Breast Carcinoma Cell Lines
MCF-7, BT-20, BT-549, T47-D, MDA-MB-435, MDA-MB-453, MDA-MB-231, and SKBr3 cell lines were obtained from Mark Sternlicht (Department of Anatomy, University of California-San Francisco, San Francisco, CA). They were all shown to be independent cell lines through DNA fingerprinting analysis. All cells were grown under standard conditions in RPMI 1640 or DMEM with 10% fetal bovine serum (Invitrogen, Carlsbad, CA).
Transgenic Mouse Strains
FVB/N mice expressing the oncogene Wnt1 or Neu in mammary glands under the control of mouse mammary tumor virus (MMTV) LTR were obtained from Jackson Laboratory (Bar Harbor, ME) [20,21].
Reverse Transcription Polymerase Chain Reaction (RT-PCR) for Detection of HSulf-2 mRNA
Total RNA was extracted from cells using RNA-Bee (Tel-Test, Friendwood, TX). One microgram of total RNA was applied for reverse transcription reaction in 20 μl of buffer with oligo(dT), using Superscript II Reverse Transcriptase (Invitrogen). The cDNA were normalized between tissues so as to contain an equal concentration of β-actin transcripts. Amplified were 314-bp HSulf-2 cDNA products, using the following PCR primers: forward, 5′-GAAAAGAGGCAGATTCACGTCGTTTCCAG-3′; reverse, 5′-ATCTGGTGCTTCTTTTGGGATGCGGGAG-3′. The conditions for denaturation, annealing, and extension of the template cDNA were as follows: 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute, for 35 cycles. PCR products were separated by electrophoresis on 2% agarose gels and visualized with ethidium bromide.
Real-Time Quantitative PCR
Real-time quantitative PCR was conducted by the Biomolecular Resource Center at the University of California, San Francisco. For real-time PCR detection of MSulf-2 mRNA expression, the following oligonucleotide primers and TaqMan probe were used: forward primer, 5′-CTCACGGCTCTTCCCCAAT-3′; reverse primer, 5′-TCTGGGTTGGGTGCATAGTTG-3′; probe, 5′-(FAM)- CGTCCCAGCACATCACACCGAGTT-(BHQ1)-3′. Normal mammary tissues were obtained from three independent FVB/N mice (143, 377, and 377 days old); hyperplastic mammary tissues were obtained from three independent MMTV-Wnt1 mice (98, 121, and 176 days old); and mammary gland tumors were obtained from five independent MMTV-Wnt1 mice (98, 121, 176, 193, and 431 days old) and from four independent MMTV-Neu mice (377, 377, 623, and 630 days old). As tumor formation in these two models is stochastic and as we sought tumors of approximately equal sizes, this necessitated the use of mice of different ages. For the Wnt1 model, the excised tumors had the following minimum and maximum dimensions: 12 × 17, 9 × 10, 18 × 20, and 11 × 11 mm. For the Neu tumors, the dimensions were 12 × 15, 15 × 16, 12 × 12, and 16 × 17 mm. All samples were processed individually. Total RNA was extracted from frozen mouse tissues using RNA-Bee (Tel-Test). Two micrograms of total RNA was used for reverse transcription reaction in 100 μl of buffer with random hexamers, using Superscript II Reverse Transcriptase (Invitrogen). PCR was conducted in triplicate with 20-μl reaction volumes of TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 0.9 mM of each primer, 250 nM probe, and 5 μl of cDNA. PCR was performed using the following parameters: 95°C, 10 minutes, 1 cycle; 95°C, 15 seconds; and 60°C, 2 minutes, 40 cycles. Analysis was carried out using a sequence detection software (SDS 2.1) supplied with TaqMan 7900HT (Applied Biosystems). Expression was quantified based on the –CT for MSulf-2 expression relative to the expression of hypoxanthine phosphoribosyltransferase (HPRT).
Western Blot Analysis
Cultured human breast carcinoma cells were lysed in RIPA buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% NP-40, and 0.1% sodium dodecyl sulfate (SDS)] and protease inhibitors for 2 hours and then centrifuged at 12,000g for 20 minutes at 4°C. Cells at ∼80% confluency in a 185-cm2 flask were rinsed with PBS and OptiMEM (Invitrogen) and further cultured in 25 ml of OptiMEM at 37°C in 5% CO2 for 72 hours. Conditioned medium (CM) from COS-7 cells transiently transfected with MSulf-2 cDNA was prepared as described [13]. The lysate or the CM was separated by electrophoresis on reducing SDS–7.5% or 10% polyacrylamide gels (Bio-Rad, Hercules, CA) and then blotted onto ProBlott membrane (Applied Biosystems). Membranes were blocked with 5% nonfat milk/0.2% Tween 20 in PBS (PBS-T; Sigma, St. Louis, MO) and incubated overnight with Sulf-2 antibody (H2.3 for human cells and H2.1 for COS-7 cells) in 5% nonfat milk/PBS-T. The membranes were washed and incubated with horseradish peroxidase–conjugated goat anti–rabbit IgG (Jackson ImmunoResearch, West Grove, PA) before using SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Arylsulfatase Assay
A 100-fold concentrated CM derived from HSulf-2 transiently transfected CHO or MCF-7 cells was dialyzed into 50 mM HEPES (pH 8.0). Sulf-2 was bound to Sulf-2 antibody (H2.3) complexed to protein A-Sepharose beads (IPA-400HC; RepliGen, Needham, MA) by rotation at 4°C for 2 hours and then washed with 50 mM HEPES (pH 8.0). The resins with bound materials were mixed with 10 mM 4-methylumbelliferyl sulfate (4-MUS) and 10 mM MgCl2 in a total volume of 30 μl. The reaction mixtures were terminated by the addition of 100 μl of 0.5 M Na2CO3/NaHCO3 (pH 10.7) to each 20-μl aliquot of the reaction mixture. The fluorescence of 4-methylumbelliferone was measured on a multiwell plate reader CytoFluorII (PerSeptive Biosystems, Framingham, MA), with an excitation wavelength of 360 nm and an emission wavelength of 460 nm.
Immunoprecipitation
Mammary gland tumors were dissected from MMTV-Neu transgenic mouse (456 days old, 14 mm in diameter). Hyperplastic mammary gland and mammary gland tumors were obtained from MMTV-Wnt1 transgenic mice (98, 112, 114, 140, and 166 days old, with dimensions of 12 × 17, 12 × 12, 14 × 15, 15 × 22, and 10 × 12 mm, respectively). Littermate FVB/N mice were used as a source of normal mammary gland. The tissues were homogenized and lysed in RIPA buffer for 30 minutes and centrifuged at 12,000g for 20 minutes at 4°C. The Sulf-2 antibody (H2.1) or nonimmune rabbit IgG was added to protein A-Sepharose beads for 1 hour. The antibody-complexed beads were added to the supernatant, which contained 2.5 mg of protein, and were incubated at 4°C for 2 hours. The immunoprecipitated MSulf-2 was revealed by Western blot analysis to have Sulf-2 antibody, using the procedure described above.
Immunohistochemistry
Specimens of mammary carcinoma and hyperplastic tissues from a 176-day-old MMTV-Wnt1 mouse and normal mammary gland tissues from a littermate FVB/N mouse were frozen in OCT embedding medium (Mile, Elkhart, IN). Ten-micrometer frozen sections were cut and fixed in cold acetone for 5 minutes. Sections were blocked with MOM mouse Ig–blocking buffer (Vector, Burlingame, CA) for 1 hour and with MOM diluent containing 5% normal goat serum (staining buffer) for 5 minutes. Sulf-2 antibody (H2.1; 2.5 μg/ml) and anti–pan cytokeratin antibody (10 μg/ml; Biomeda, Foster City, CA) or anti–smooth muscle actin (SMA; 10 μg/ml; Sigma) antibody in staining buffer were incubated with tissues for 1 hour. After washing in PBS, the tissue was incubated with a 1/1000 dilution of biotinylated goat anti–rabbit IgG (Jackson ImmunoResearch) and Cy3 antimouse IgG (Jackson ImmunoResearch) in staining buffer for 45 minutes. After washing in PBS, the tissues were incubated with a 1/1000 dilution of Cy2-SA (Jackson ImmunoResearch) in TBS for 30 minutes. Normal rabbit IgG (Zymed, South San Francisco, CA) or mouse IgG (Zymed) is used as negative control.
Establishment of Sulf-2 Stable Cell Line and Purification of Sulf-2
The sequence encoding the signal peptide was removed from the full-length HSulf-2 cDNA and substituted with a sequence encoding FLAG peptide. The modified cDNA was then ligated into pSecTag2 (N-FLAG-HSulf-2; Invitrogen). An inactive HSulf-2, in which cysteines 88 and 89 were mutated to alanines (designated as HSulf-2ΔCC) [13]), was generated by PCR-based, site-directed mutagenesis using the Quikchange XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Human embryonic kidney 293 cells were grown in DMEM containing 10% fetal calf serum in 10-cm dishes and transfected with 10 μg of plasmid vector (293-N-FLAG-HSulf-2 or HSulf-2ΔCC) and pcDNA 3.1 myc-His(−) using the LipofectAMINE 2000 reagent (Invitrogen) according to the manufacturer's instruction. Stable 293 transfectants were selected with 1 mg/ml G418 (Invitrogen). The CM derived from 293-N-FLAG-HSulf-2 or HSulf-2ΔCC cells was incubated with anti–FLAG/M2 agarose beads (Sigma) by rotation at 4°C overnight and then washed with 50 mM Tris–HCl (pH 7.4) and 150 mM NaCl. A solution of 0.1 mg/ml FLAG peptide (DYKDDDDK) in 50 mM Tris–HCl (pH 7.4) and 500 mM NaCl was used to elute the FLAG-tagged fusion proteins from beads. The eluted material was concentrated and dialyzed to 50 mM HEPES (pH 8.0) using a Centricon-30 microconcentrator (Millipore, Billerica, MA). The purity of wild-type Sulf-2 and HSulf-2ΔCC was 90% and 80%, respectively, as determined by Coomassie blue staining of SDS-PAGE gels. The amount of Sulf-2 was quantified using the BCA assay (Pierce), with BSA as a standard. Comparable amounts of HSulf-2 and HSulf-2ΔCC were determined by performing a capture ELISA using an anti–FLAG M2 antibody (Sigma) as the capture reagent and a mixture of Sulf-2 antibodies (H2.1 and H2.3) for detection.
Chick Chorioallantoic Membrane (CAM) Angiogenesis Assay
Ten-day chicken embryo CAMs were treated with 25, 50, or 100 ng of purified Sulf-2 or 50 ng of Sulf-2ΔCC in 50 μl of 50 mM HEPES (pH 8.0). Three days after treatment, 500 μl of 4% paraformaldehyde was applied to CAMs prior to excision for counting of vessel branch points under a dissection microscope [22]. At least 10 embryos were used per treatment group. Each experiment was performed for a minimum of three times. Data were evaluated in terms of the average number of blood vessel branch points per treatment group ± SEM. VEGF-165 (R&D Systems) was used for as positive control. Statistical analyses were performed using Student's t test.
Results
Expression of Sulf-2 mRNA and Protein in Human Breast Cancer
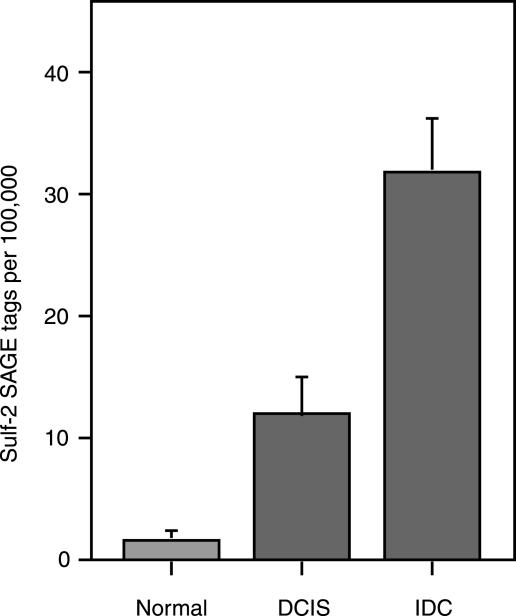
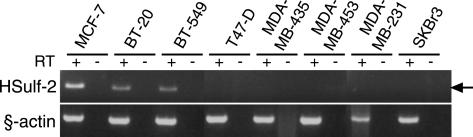
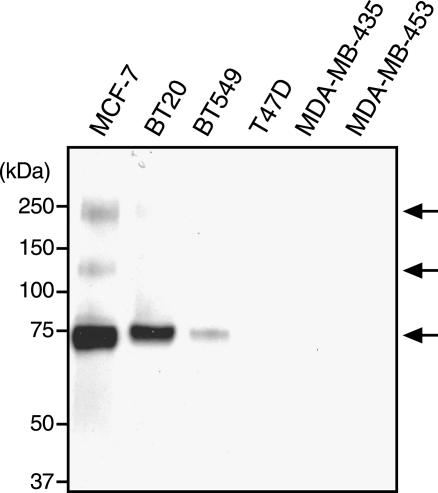
We first confirmed and extended our preliminary observations on Sulf-2 expressions in breast cancer [19]. Data mining from a recent and extensive SAGE analysis [23] showed that there was a seven-fold increase in Sulf-2 SAGE tags between normal breast tissues and ductal carcinoma in situ (DCIS) tissues (P = .018), and a further increase of three-fold between DCIS tissues and invasive ductal carcinoma (IDC) tissues (P = .0001) (Figure 1). Although there was an increase in Sulf-2 expression between DCIS and IDC, one cannot infer a higher expression in carcinoma cells of one tumor versus the other because the proportion of transformed cells in the different tumors is not known. Further data mining from two cDNA microarray profiling studies [24,25] revealed upregulation of Sulf-2 transcripts in 6 of 20 human breast carcinoma specimens relative to normal tissues and significantly higher Sulf-2 expression in estrogen receptor–positive breast tumors compared to receptor-negative ones. We next asked if Sulf-2 was expressed by human breast carcinoma cell lines. We detected Sulf-2 mRNA in three of eight human breast carcinoma cell lines (MCF-7, BT-20, and BT-549), as determined by RT-PCR (Figure 2). We then developed peptide-specific polyclonal antibodies to the predicted protein to examine the expression of Sulf-2 at the protein level (Materials and Methods section). By Western blot analysis with a peptide-specific antibody (Figure 3), we detected Sulf-2 in the CM of cells that expressed Sulf-2 mRNA, but not in the CM of cells lacking mRNA (T47D, MDA-MB-435, and MDA-MB-453). The major protein band had an apparent molecular mass of 75 kDa, with minor species at 240 and 135 kDa. Western blot analysis of a detergent extract of MCF-7 cells revealed a predominant band at 126 kDa (not shown).
Figure 1.
SAGE analysis of Sulf-2 expression in human breast cancer. Data pertinent to Sulf-2 were mined from a published SAGE analysis of human breasts [23]. Sulf-2 tags were enumerated from 4 normal breast tissue libraries, 7 DCIS libraries, and 12 IDC libraries. The total number of SAGE tags in the analysis was 1.75 million. The incidences of Sulf-2 tags in the different tissue libraries were 0.0018% (normal), 0.012% (DCIS), and 0.032% (IDC). The graph shows the mean number of Sulf-2 tags per 100,000 total tags from the libraries for each class of tissue. The error bars indicate SEM. The probabilities provided in the figure and text are based on a t-test comparison in the source study [23].
Figure 2.
Sulf-2 mRNA in human breast carcinoma cell lines. RT-PCR was performed on cDNA that were prepared from eight human breast carcinoma cell lines with primer pairs for Sulf-2 and β-actin primer. A 314-bp HSulf-2 cDNA product (indicated by an arrow) was amplified from MCF-7, BT20, and BT549 cells.
Figure 3.
Sulf-2 in the CM of human breast carcinoma cell lines. The CM of the indicated breast carcinoma cell lines was collected, and Western blot analysis with a Sulf-2 antibody (H2.3) was performed. The apparent molecular mass of the major Sulf-2 species was 72 kDa. Minor species were evident at 135 and 240 kDa.
Recombinant Sulf-1 and Sulf-2 exhibit arylsulfatase activity against the 4-MUS substrate [13]. To determine whether native Sulf-2 from breast carcinoma cells was enzymatically active, we performed an arylsulfatase assay. CM from MCF-7 cells or HSulf-2–transfected CHO cells was collected and incubated with Sulf-2 antibody coupled to protein A-Sepharose. The bead-bound material was assayed for activity on 4-MUS. Both types of CM showed enzyme activity, which was dependent on the volume of CM added (Figure 4). In contrast, no activity was precipitated when a control rabbit IgG was substituted for the specific antibody.
Figure 4.
Arylsulfatase activity of Sulf-2 in MCF-7 CM. Different volumes of CM obtained from MCF-7 cells or HSulf-2–transfected CHO cells were incubated with Sulf-2 antibody (H2.3) or rabbit IgG coupled to protein A beads. Bead-bound material was tested for arylsulfatase activity against a 10-mM 4-MUS substrate. The same results were obtained from three different experiments. (▪) MCF-7 CM + Sulf-2 antibody (H2.3) beads; (□) HSulf-2 CHO CM + Sulf-2 antibody beads; (▴) MCF-7 CM + rabbit IgG beads; (△) HSulf-2 CHO CM + rabbit IgG beads.
Expression of Sulf-2 in the Epithelium of Hyperplastic Tissues and Tumors in Mouse Mammary Carcinoma Models
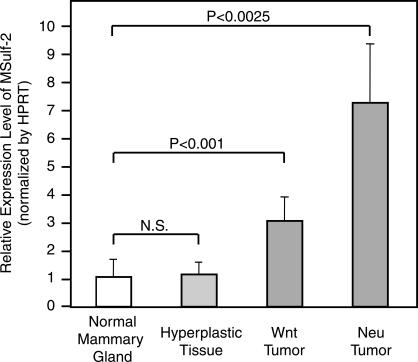
We next investigated if the increase in Sulf-2 expression seen in human tumors and cell lines would generalize to mammary carcinomas in mice. Therefore, we determined whether Sulf-2 was upregulated in two morphologically and mechanistically distinct transgenic models of mammary carcinoma, MMTV-Wnt1 and MMTV-Neu [20,21,26]. We used real-time PCR to measure Sulf-2 mRNA levels in mammary tumors (MMTV-Wnt1 and MMTV-Neu mice), mammary hyperplastic tissues (MMTV-Wnt1 mice), and normal mammary gland tissues (FVB/N mice) and normalized them to the housekeeping gene HPRT. Sulf-2 mRNA was expressed in tumors of MMTV-Wnt1 and MMTV-Neu transgenic mice and was increased by 3.0-fold (P = .00001) and 7.1-fold (P = .001), respectively, compared to that in normal mammary gland tissues (Figure 5). In contrast, Sulf-2 mRNA was present in hyperplastic tissues at the same level as that in normal mammary glands.
Figure 5.
Sulf-2 mRNA in MMTV-Wnt1 and MMTV-Neu transgenic mouse mammary carcinoma tissues. Real-time PCR was performed on cDNA prepared from normal mouse mammary glands (10 independent samples), hyperplastic tissues (9), and tumors from MMTV-Wnt1 (12) and MMTV-Neu (6) transgenic mice. Sulf-2 mRNA expression was normalized relative to HPRT expression. Error bars indicate SEM. Statistical significance was computer with Student's t test.
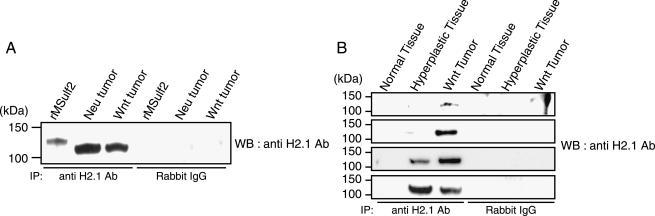
We then analyzed MMTV-Wnt1 and MMTV-Neu tumors for immunoreactive Sulf-2. Immunoprecipitation of detergent lysates followed by Western blot analysis revealed the presence of a 115-kDa protein in both types of tumors (Figure 6A). Recombinant MSulf-2 from COS-7 cells yielded a band at 128 kDa. The bands were absent when nonimmune IgG was substituted for the specific antibody. N-glycanase treatment converted the 115- and 128-kDa bands to a single species at 100 kDa (not shown), demonstrating that the difference in molecular weight between the native and the recombinant protein was due to differential N-glycosylation. Applying the same immunoprecipitation methodology, we compared Sulf-2 expression in mammary tissues from normal FVB/N mice and MMTV-Wnt1 transgenic mice (hyperplastic tissues and tumors). Lysates were prepared from four independent sets of tissues and were normalized for overall protein concentration. The Sulf-2 band was detected in the tumors in all four cases and in hyperplastic tissues in two of four cases. In contrast, Sulf-2 was not present in the extracts of mammary gland from the four normal animals (Figure 6B). Dilution of the tumor lysates up to eight-fold still yielded the specific band (not shown), thus establishing that the expression level of Sulf-2 in mammary tumors was at least eight-fold higher on a per-milligram-of-protein basis than that in normal mammary gland tissues (not shown). The finding that Sulf-2 was detectable in hyperplastic tissues, but not in normal tissues, despite similar levels of mRNA may reflect an increased level of translation or stability of the mRNA in hyperplastic tissues.
Figure 6.
Sulf-2 expression in normal mouse tissues and mammary gland tumors. Detergent lysates of the indicated tissues were subjected to immunoprecipitation and Western blot analysis with a Sulf-2 (H2.1) antibody or normal rabbit IgG, as described in Materials and Methods section. (A) Analysis of expression in MMTV-Neu tumor, MMTV-Wnt1 tumor, and cell extracts of COS-7 cells transfected with MSulf-2. (B) Four sets of normal mammary gland from FVB/N mouse, and hyperplastic and tumor tissues from a littermate MMTV-Wnt1 transgenic mouse were subjected to immunoprecipitation/Western blot analysis, as described above. Sulf-2 was detected in all of the tumors, in two of four of the hyperplastic tissues, and in none of the normal tissue extracts.
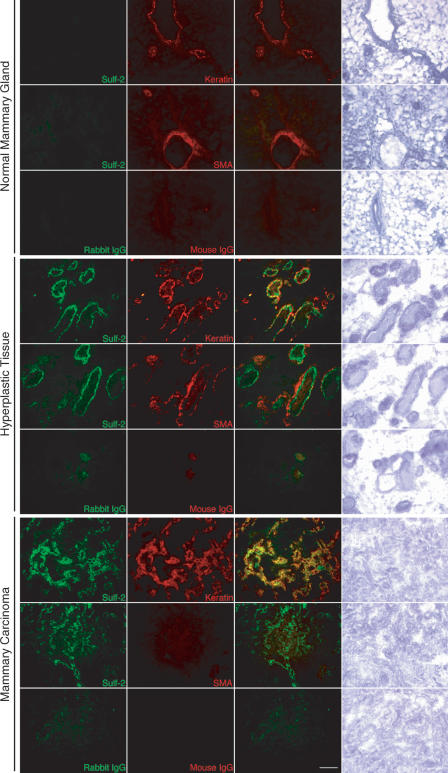
By immunohistochemistry, Sulf-2 staining was evident in hyperplastic tissues and tumors of MMTV-Wnt1 mammary glands, but was undetectable in normal mammary glands from control FVB/N mice (Figure 7). To determine the localization of Sulf-2, we carried out two-color immunofluorescence. We examined the distribution of Sulf-2 (green) relative to cytokeratin (using a pan cytokeratin antibody, in red) as a marker for epithelial cells, or SMA (red) as a marker for myoepithelial cells. In hyperplastic tissues, Sulf-2 was present in luminal epithelial cells but not in myoepithelial cells (Figure 7). In the tumors, Sulf-2 localized to epithelial-derived carcinoma cells. Sulf-2 was also present in mammary tumors of MMTV-Neu mice (not shown), as determined by immunostaining.
Figure 7.
Sulf-2 in the epithelia of hyperplastic mammary tissues and mammary tumors. Sections of mammary tumors and hyperplastic tissues from MMTV-Wnt1 mice and normal mammary glands from FVB/N mice were processed for dual-color immunocytochemistry and bright field histology. The left column shows staining with a Sulf-2 antibody (H2.1; green signal). The second column shows staining with anti–pan cytokeratin (keratin; epithelial cell marker) or anti–SMA (myoepithelial cell marker) antibodies (red signal). The third column shows the overlay of the two fluorescent images. The fourth column shows a bright field image with hematoxylin staining. Scale bar = 100 μm.
Proangiogenic Activity of Sulf-2
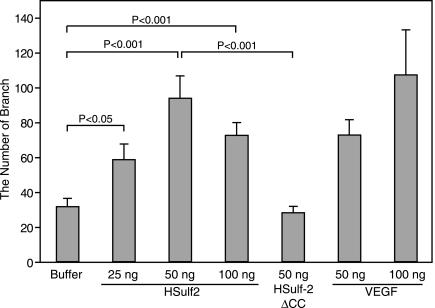
We have shown elsewhere that recombinant Sulf-2 can modulate the interaction of certain growth factors and chemokines (e.g., FGF-1, VEGF165, and SDF-1) with heparin [27]. Thus, pretreatment of heparin with Sulf-2 prevents the binding of each of these factors, and Sulf-2 treatment of the preformed heparin growth factor complex of each type mobilizes the growth factor into solution. HSPGs in the ECM are known to sequester growth and angiogenic factors from their signal-transducing cellular receptors (reviewed by Bernfield et al. [28] and Vlodavsky et al. [29]). The release of HSPG-bound factors (e.g., VEGF) through the activity of an extracellular enzyme, such as a metalloproteinase or a heparanase, provides a mechanism for regulating tumor angiogenesis [30–32]. In view of the activity of Sulf-2 in reversing the binding of VEGF and other growth factors to heparin, we hypothesized that Sulf-2 could potentially promote angiogenesis when exogenously provided to a tissue site with angiogenic potential. We took advantage of the CAM assay, which is widely used to test the angiogenic activity of exogenous factors [33]. CAMs of 10-day-old chick embryos were subjected to topical application of a test substance, and angiogenesis was quantified by counting the number of blood vessel branching points 72 hours later. CAMs exposed to recombinant HSulf-2 formed new extensive branches in a dose-dependent manner (Figure 8). Fifty nanograms of HSulf-2 showed the maximum activity. The activity of Sulf-2 on a per-unit-of-protein-mass basis was comparable to that of VEGF-165, which served as a positive control. HSulf-2ΔCC, which was mutated at two critical cysteines and was inactive as both an arylsulfatase and a heparin endosulfatase [13], was inactive in the CAM assay when tested at 50 ng.
Figure 8.
Angiogenic activity of Sulf-2 in the CAM assay. Ten-day-old chicken embryo CAMs were treated with purified Sulf-2, Sulf-2ΔCC, or VEGF in 50 mM HEPES (pH 8.0). Three days after the treatment, the number of vessel branch points was counted. Data were pooled from three independent experiments. Means and SEM are shown. Statistical comparisons were performed with Student's t test.
Discussion
Three broad categories of function are attributed to HSPGs: 1) assembly and integrity of basement membranes; 2) sequestration and storage of signaling molecules (growth factors, morphogens, angiogenic factors, and so forth) from their site of action; and 3) coreceptors for cell surface tyrosine kinases [28]. Mutations in enzymes involved in the biosynthesis of GAG chains have established that HSPGs play essential roles in development [4,34]. The sulfation state of the HS GAG chains strongly influences the protein ligands with which they interact. An enormous diversity of HS sequences is possible [3], and considerable variegation has, in fact, been documented in normal adult tissues [35]. Furthermore, HS chains are dynamically regulated during development and tumorigenesis [36,37], consistent with regulatory changes in the binding of signaling molecules. Considerable interest therefore exists in the mechanisms that generate a diversity of GAG chains. One possible mechanism is through regulation of the expression of specific sulfotransferases [38–40]. Another potential mechanism would be the removal of specific sulfation modifications through the action of extracellular sulfatases. Prior to the discovery of the Sulfs, the only known GAG sulfatases were exosulfatases, which remove sulfates from the nonreducing termini of chains during catabolic breakdown in lysosomes [10]. The newly discovered Sulfs fulfill requirements for enzymes that could dynamically edit the sulfation status of HSPG chains and thereby modulate ligand interactions.
QSulf-1 was the first member of this new class of sulfatases to be discovered. Sonic hedgehog induces its expression during somite formation in the quail embryo [12]. It is also expressed in the notochord, floor plate, and ventral neural tube. QSulf-1 is required for the activation of MyoD. It was initially suggested that QSulf-1 might be involved in Wnt signaling [12]. In fact, recombinant QSulf-1 promotes Wnt signaling in in vitro assays [11,12]. The mechanism apparently involves the action of QSulf-1 in remodeling the glucosamine-6-O sulfation status of HS chains, decreasing Wnt binding to the chains and thereby allowing ligand access to its receptors on the target cells. QSulf-1 is thought to act on a cell autonomous basis because the recombinant protein is restricted to the cell surface of the transfected cell and is not secreted into the medium; moreover, HS chains are modified only on the cell that expresses the enzyme, not on neighboring cells [11,12]. A second example of signal enhancement through QSulf-1 has been reported by Viviano et al. [14]. In this system, transfection of QSulf-1 promotes responses of target cells to BMP through release of an HS-bound inhibitor of BMP (noggin) from the cell surface. Contrasting with these signal-promoting activities, Sulf-1 has been shown to be a negative regulator of signaling for other factors. Thus, transfection of Sulf-1 into various cell types reduces signaling by FGF-2, HB-EGF, or HGF [15–18]. The effects on FGF-2 signaling were predictable because the HS coreceptor for FGFR-1 (the tyrosine kinase receptor for FGF-2) required 6-O sulfation [5].
Sulf-1 mRNA is expressed in a number of tissues during development [12,41]. It is upregulated in breast and brain tumors, as determined by SAGE analysis [19]. In ovarian tumors, however, downregulation is common in primary ovarian cancer specimens relative to normal ovarian epithelial samples [16]. Enforced expression of Sulf-1 in ovarian cancer lines results in diminished signaling in response to FGF-2 and HB-EGF. The picture in hepatocellular carcinoma is mixed, with decreased levels of Sulf-1 mRNA relative to normal tissues in one third of the samples and increased levels in the remaining two thirds [18].
In the present study, we have focused our attention on Sulf-2. At the protein level, Sulf-2 is 63% to 65% identical to Sulf-1 in mice and humans [13]. A survey of GenBank sequences reveals the existence of Sulf-2 homologues in rat (XP230861), quail (AAV37455), zebrafish (AAR04058), and Drosophila (EAL28608). Previously, we showed that recombinant Sulf-2 exhibits arylsulfatase and heparin endosulfatase activities, which are indistinguishable from those of Sulf-1 [13]. Here we report the upregulation of Sulf-2 transcripts in human breast cancer tissues and in two mouse models of mammary carcinoma. Moreover, we establish Sulf-2 expression at the protein level in mammary hyperplastic and carcinoma tissues from mammary glands of MMTV-Wnt1 transgenic mice. Immunohistochemical analysis showed that expression was present in epithelial cells. Interestingly, Sulf-2 could not be detected either by Western blot analysis or by tissue staining in normal adult mammary glands, although transcripts for the gene were detectable.
Examination of human breast carcinoma cell lines led to the identification of Sulf-2 at both the mRNA and protein levels in three of eight cell lines. Importantly, the protein and corresponding sulfatase activity were readily detected in CM from the positive cell lines. The protein was released into CM in a processed form with a subunit molecular mass of 75 kDa. This compares to the 126-kDa protein observed in cell lysates, which corresponds to precursor polypeptide [13]. Our detection of active Sulf-2 in CM contrasts with several reports for recombinant Sulf-1, in which the protein and its enzymatic activity are confined to the cells that synthesize it [11,12,16,41].
The secretion of active Sulf-2 by carcinoma cells raises the possibility that the enzyme is used by the cells to remodel the ECM in their environment, thereby affecting the carcinoma cells themselves or the neighboring host cells. A strong parallelism can be drawn with mammalian heparanase, an enzyme that is preferentially expressed in metastatic cell lines and human tumor tissues [42,43]. Overexpression of heparanase cDNA in nonmetastatic tumor cells leads to increased Matrigel invasion as well as augmented metastasis and tumor angiogenesis in vivo [31]. Moreover, converse effects are observed in the gene silencing of endogenous heparanase in tumor cells: reduced invasiveness in vitro and less vascularized tumors in vivo [30]. A hypothesis for the increased invasiveness is the disassembly of physical barriers through the cleavage of heparan sulfate chains by the enzyme [31]. The effects on angiogenesis have been attributed to the ability of the enzyme to mobilize heparan sulfate–bound angiogenic factors from sequestration in the ECM [44]. In contrast to heparanase, Sulf-2 is an endosulfatase that selectively acts on glucosamine-6-sulfate units within highly sulfated subregions of heparin and heparan sulfate [11,13,14]. As noted above, glucosamine-6-sulfate modification is required for the high-affinity binding of heparin chains to a number of protein ligands, including VEGF. A prior report demonstrated that QSulf-1 treatment of heparin or an HSPG prevents the binding of a Wnt ligand [11]. We have shown that Sulf-2 reverses the interaction of heparin with VEGF and other growth factors [27]. Therefore, proangiogenic effects of Sulf-2, which are demonstrated in the present study, are plausibly explained by the release of VEGF or another angiogenic factor from HSPG sequestration, thereby making it available to act on blood vessels. Our results thus rationalize one potential mechanism by which cancer cells could utilize Sulf-2 to favor tumor growth, namely, by promoting tumor angiogenesis. Therefore, the upregulation of Sulf-2 expression in breast tumors (rather than Sulf-2 being merely a correlate of tumor progression) could be mechanistically critical for the growth of tumors. Because a myriad of extracellular events are influenced by heparan sulfate interactions with growth factors, morphogens, cell adhesion molecules, and so forth [1], we can envisage other mechanisms by which cancer cells could exploit the activity of Sulf-2. In this light, it is notable that the Sulf-2 locus was recently identified in an insertional mutagenesis screen for candidate cancer-causing genes in malignant brain tumors [45]. Further study of Sulf-2 role in tumor progression is clearly warranted.
Abbreviations
- CAM
chick chorioallantoic membrane
- CM
conditioned medium
- DCIS
ductal carcinoma in situ
- ECM
extracellular matrix
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- GAG
glycosaminoglycan
- HGF
hepatocyte growth factor
- HB-EGF
heparin-binding epidermal growth factor
- HSPG
heparan sulfate proteoglycan
- SAGE
serial analysis of gene expression
- HPRT
hypoxanthine phosphoribosyltransferase
- IDC
invasive ductal carcinoma
- MMTV
mouse mammary tumor virus
- 4-MUS
4-methylumbelliferyl sulfate
- PBS-T
0.2% Tween 20 in PBS
- SMA
smooth muscle actin
- VEGF
vascular endothelial growth factor
Footnotes
This work was supported by a grant from the Mizutani Foundation (S.D.R.); a UCSF Comprehensive Cancer Center Intramural Award from the Alexander and Margaret Stewart Trust (S.D.R.); a grant from the National Cancer Institute (CA72006, to Z.W.); and fellowships from the US Department of Defense Breast Cancer Program (BC021285, to D.L.), the California Breast Cancer Research Program (M.E.), and the Japan Society for the Promotion of Science (M.M.-T. and K.U.).
References
- 1.Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 3.Gallagher JT. Heparan sulfate: growth control with a restricted sequence menu. J Clin Invest. 2001;108:357–361. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esko JD, Selleck SB. ORDER OUT OF CHAOS: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 5.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 6.Spillmann D, Witt D, Lindahl U. Defining the interleukin-8–binding domain of heparan sulfate. J Biol Chem. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- 7.Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J Biol Chem. 2001;276:30744–30752. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- 8.Ashikari-Hada S, Habuchi H, Kariya Y, Itoh N, Reddi AH, Kimata K. Characterization of growth factor–binding structures in heparin/heparan sulfate using an octasaccharide library. J Biol Chem. 2004;279:12346–12354. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- 9.Parenti G, Meroni G, Ballabio A. The sulfatase gene family. Curr Opin Genet Dev. 1997;7:386–391. doi: 10.1016/s0959-437x(97)80153-0. [DOI] [PubMed] [Google Scholar]
- 10.Hanson SR, Best MD, Wong CH. Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl. 2004;43:5736–5763. doi: 10.1002/anie.200300632. [DOI] [PubMed] [Google Scholar]
- 11.Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 13.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J Biol Chem. 2004;279:5604–5611. doi: 10.1074/jbc.M310691200. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Ai X, Freeman SD, Pownall ME, Lu Q, Kessler DS, Emerson CP., Jr QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc Natl Acad Sci USA. 2004;101:4833–4838. doi: 10.1073/pnas.0401028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai J, Chien J, Staub JK, Avula R, Greene EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR, Shridhar V. Loss of HSulf-1 upregulates heparin binding growth factor signaling in cancer. J Biol Chem. 2003;21:21. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 17.Lai JP, Chien J, Strome SE, Staub J, Montoya DP, Greene EL, Smith DI, Roberts LR, Shridhar V, Chien JR, et al. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 18.Lai JP, Chien JR, Moser DR, Staub JK, Aderca I, Montoya DP, Matthews TA, Nagorney DM, Cunningham JM, Smith DI, et al. hSulf1 sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology. 2004;126:231–248. doi: 10.1053/j.gastro.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 19.Morimoto-Tomita M, Uchimura K, Rosen SD. Novel extracellular sulfatases: potential roles in cancer. Trends Glycosci Glycotechnol. 2003;15:159–164. [Google Scholar]
- 20.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 21.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudreau NJ, Varner JA. The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis. J Biol Chem. 2004;279:4862–4868. doi: 10.1074/jbc.M305190200. [DOI] [PubMed] [Google Scholar]
- 23.Abba MC, Drake JA, Hawkins KA, Hu Y, Sun H, Notcovich C, Gaddis S, Sahin A, Baggerly K, Aldaz CM. Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression. Breast Cancer Res. 2004;6:R499–R513. doi: 10.1186/bcr899. Epub 2004 July 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–856. doi: 10.1158/0008-5472.can-03-2361. [DOI] [PubMed] [Google Scholar]
- 25.Nagai MA, Da Ros N, Neto MM, de Faria Junior SR, Brentani MM, Hirata R, Jr, Neves EJ. Gene expression profiles in breast tumors regarding the presence or absence of estrogen and progesterone receptors. Int J Cancer. 2004;111:892–899. doi: 10.1002/ijc.20329. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchimura K, Morimoto-Tomita M, Bistrup A, Li J, Lyon M, Gallagher J, Werb Z, Rosen SD. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 29.Vlodavsky I, Bar-Shavit R, Ishai-Michaeli R, Bashkin P, Fuks Z. Extracellular sequestration and release of fibroblast growth factor: a regulatory mechanism? Trends Biochem Sci. 1991;16:268–271. doi: 10.1016/0968-0004(91)90102-2. [DOI] [PubMed] [Google Scholar]
- 30.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 31.Goldshmidt O, Zcharia E, Abramovitch R, Metzger S, Aingorn H, Friedmann Y, Schirrmacher V, Mitrani E, Vlodavsky I. Cell surface expression and secretion of heparanase markedly promote tumor angiogenesis and metastasis. Proc Natl Acad Sci USA. 2002;99:10031–10036. doi: 10.1073/pnas.152070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z, et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol. 2000;2:737–744. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks PC, Clark RAF, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 34.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 35.Dennissen MA, Jenniskens GJ, Pieffers M, Versteeg EM, Petitou M, Veerkamp JH, van Kuppevelt TH. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J Biol Chem. 2002;277:10982–10986. doi: 10.1074/jbc.M104852200. [DOI] [PubMed] [Google Scholar]
- 36.Brickman YG, Ford MD, Gallagher JT, Nurcombe V, Bartlett PF, Turnbull JE. Structural modification of fibroblast growth factor–binding heparan sulfate at a determinative stage of neural development. J Biol Chem. 1998;273:4350–4359. doi: 10.1074/jbc.273.8.4350. [DOI] [PubMed] [Google Scholar]
- 37.Jayson GC, Vives C, Paraskeva C, Schofield K, Coutts J, Fleetwood A, Gallagher JT. Coordinated modulation of the fibroblast growth factor dual receptor mechanism during transformation from human colon adenoma to carcinoma. Int J Cancer. 1999;82:298–304. doi: 10.1002/(sici)1097-0215(19990719)82:2<298::aid-ijc23>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 38.Holmborn K, Ledin J, Smeds E, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate synthesized by mouse embryonic stem cells deficient in NDST1 and NDST2 is 6-O-sulfated but contains no N-sulfate groups. J Biol Chem. 2004;279:42355–42358. doi: 10.1074/jbc.C400373200. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Shworak NW, Sinay P, Schwartz JJ, Zhang L, Fritze LM, Rosenberg RD. Expression of heparan sulfate d-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem. 1999;274:5185–5192. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 40.Nogami K, Suzuki H, Habuchi H, Ishiguro N, Iwata H, Kimata K. Distinctive expression patterns of heparan sulfate O-sulfotransferases and regional differences in heparan sulfate structure in chick limb buds. J Biol Chem. 2004;279:8219–8229. doi: 10.1074/jbc.M307304200. [DOI] [PubMed] [Google Scholar]
- 41.Ohto T, Uchida H, Yamazaki H, Keino-Masu K, Matsui A, Masu M. Identification of a novel nonlysosomal sulphatase expressed in the floor plate, choroid plexus and cartilage. Genes Cells. 2002;7:173–185. doi: 10.1046/j.1356-9597.2001.00502.x. [DOI] [PubMed] [Google Scholar]
- 42.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 43.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis [see comments] Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 44.Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 45.Johansson FK, Brodd J, Eklof C, Ferletta M, Hesselager G, Tiger CF, Uhrbom L, Westermark B. Identification of candidate cancer-causing genes in mouse brain tumors by retroviral tagging. Proc Natl Acad Sci USA. 2004;101:11334–11337. doi: 10.1073/pnas.0402716101. [DOI] [PMC free article] [PubMed] [Google Scholar]