Abstract
Telomerase expression represents a good target for cancer gene therapy. The promoters of the core telomerase catalytic [human telomerase reverse transcriptase (hTERT)] and RNA [human telomerase RNA (hTR)] subunits show selective activity in cancer cells but not in normal cells. This property can be harnessed to express therapeutic transgenes in a wide range of cancer cells. Unfortunately, weak hTR and hTERT promoter activities in some cancer cells could limit the target cell range. Therefore, strategies to enhance telomerase-specific gene therapy are of interest. We constructed a Cre/Lox reporter switch coupling telomerase promoter specificity with Cytomegalovirus (CMV) promoter activity, which is generally considered to be constitutively high. In this approach, a telomerase-specific vector expressing Cre recombinase directs excisive recombination on a second vector, removing a transcriptional blockade to CMV-dependent luciferase expression. We tested switch activation in cell lines over a wide range of telomerase promoter activities. However, Cre/Lox–dependent luciferase expression was not enhanced relative to expression using hTR or hTERT promoters directly. Cell-specific differences between telomerase and CMV promoter activities and incomplete sigmoid switch activation were limiting factors. Notably, CMV activity was not always significantly stronger than telomerase promoter activity. Our conclusions provide a general basis for a more rational design of novel recombinase switches in gene therapy.
Keywords: Telomerase, hTR, hTERT, Cre recombinase, gene therapy
Introduction
Telomerase is a ribonucleoprotein reverse transcriptase that is minimally composed of RNA [human telomerase RNA (hTR)] and catalytic [human telomerase reverse transcriptase (hTERT)] subunits, which counteracts cell division–associated attritions of the telomeres of linear chromosomes by synthesizing a new telomere DNA sequences from an internal template sequence in hTR [1–3]. Most normal cells do not express telomerase and are therefore subject to telomere-dependent senescence. However, telomerase activity is essential for immortalization of most cancer cells, and its inhibition results in delayed-onset apoptosis [4–8]. Thus, telomerase represents an exciting target for the development of novel anticancer therapeutics [9–12].
Differential expression of telomerase between normal and cancer cells is attributable to transcriptional regulation of hTR and hTERT. Both transcripts are readily detectable in most cancer cell lines and human malignancies, but are either absent or at very low levels in normal cells and tissues. Cloned hTR and hTERT promoter constructs also show selective activity in cancer cells and have been exploited for transcriptionally targeted gene therapy strategies, with the expectation that therapeutic transgenes can be expressed at high levels, specifically in cancer cells but not in normal cells [9,13–28].
Preclinical results of telomerase-specific gene therapy are encouraging. Therapeutic transgene expression has been demonstrated in multiple human cancer cell lines, whereas normal cells are generally not targeted by the hTR and hTERT promoters [9]. We previously reported that the telomerase promoters drive an efficient and selective expression of the bacterial nitroreductase (NTR) gene in several cancer cell lines and xenografts, but not in normal cells [15,16]. NTR catalyzes the rapid bioactivation of the relatively nontoxic prodrug CB1954, resulting in its conversion to a powerful alkylating agent. Activated CB1954 forms atypical DNA adducts that are poorly repaired, leading to efficient p53-independent apoptosis [29,30].
However, the telomerase promoters exhibit relatively weak activity in some target cells [15,16,24]. This could restrict therapeutic targeting potentials in applications such as enzyme/prodrug therapy, where there is good correlation between high transgene expression levels and significant therapeutic effects. In this respect, we have found the hTERT promoter to be particularly problematic. We have found that several cancer cell lines are inefficiently targeted by the NTR/CB1954 combination using the hTERT promoter. In our model, the stronger hTR promoter results in efficient therapeutic targeting of more cell lines than hTERT. However, the hTR promoter is also inefficient in some cases.
Therefore, strategies to improve the efficacy of telomerase gene therapy in cells with low promoter activity are of interest [21,24]. One approach to improving the expression levels of therapeutic transgenes in tissue-specific gene therapy makes use of the Cre/Lox switch. Phage P1–derived Cre recombinase catalyzes site-specific excision and circularization of stuffer DNA sequences flanked at the 5′ and 3′ ends by a specific 34-bp Cre-binding sequence (the LoxP site). Thus, Cre/Lox technology provides a valuable tool for studies of gene function [31,32].
Harnessed for gene therapy, a therapeutic transgene is separated from a strong constitutive promoter by a LoxP-flanked transcriptional termination signal. A weak tissue-specific promoter of interest drives the expression of Cre from a second vector, resulting in deletion of the stuffer and derepression of transgene expression. Thus, selectivity is neatly coupled with a constitutively high transcription rate. Several groups have applied this strategy in cancer gene therapy approaches [33–37]. Because the hTR and hTERT promoters show weak activity in some cancer cells, we reasoned that the Cre/Lox switch could be adapted to extend the effective target cell range for telomerase-specific gene therapy.
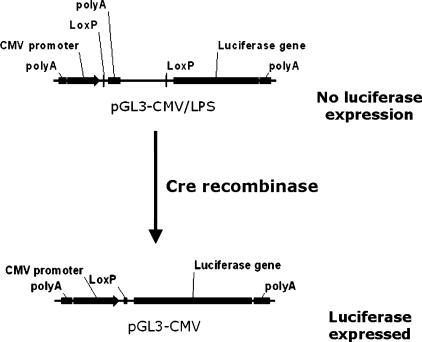
Here we report the development of a telomerase-specific Cre/Lox switch regulating the expression of the luciferase gene. In this system, luciferase expression is controlled by the Cytomegalovirus (CMV) promoter and by a LoxP-flanked stuffer fragment harboring the SV40 late polyadenylation signal upstream of the luciferase gene. Expression of Cre, mediated by hTR, hTERT, or SV40 promoters, excises the stuffer facilitating the luciferase expression (Figure 1). This model allowed us to test quantitatively whether a telomerase-specific Cre/Lox switch could potentially enhance therapeutic transgene expression levels relative to direct expression through the hTR or hTERT promoters.
Figure 1.
Regulation of luciferase expression by the Cre/Lox switch. Transcription of the luciferase gene in vector LPS is repressed by the presence of a LoxP-flanked stuffer fragment harboring the SV40 late polyadenylation signal upstream of the luciferase gene. In the presence of Cre protein expression, in this case directed by the hTR and hTERT promoters, the stuffer and polyadenylation signal are excised, leading to the derepression of CMV-dependent luciferase expression.
We selected a panel of four cancer cell lines with a wide range of hTR and hTERT promoter activities in order to examine switch activation over a range of low to high Cre expression levels. Cre was expressed in a promoter- and cell-specific manner, effectively derepressing luciferase expression over a wide range of concentrations. Unexpectedly, however, the switch did not confer significantly enhanced luciferase expression in any of the cells tested here.
These results were explained by cell-specific differences between telomerase and CMV promoter activities, and by a sigmoid relationship between Cre expression levels and switch activation efficiency. Switch activation saturated at very low Cre doses and increased Cre expression did not confer increased luciferase expression. Because of these attributes of Cre-mediated excision in cells, a very large differential in selective and constitutive promoter activities is required to confidently predict enhancement of gene expression by the Cre/Lox switch. In our model, the CMV promoter was not always significantly stronger than hTR or hTERT. Importantly, because viral promoters have no biomarkers of activity in human tissues, we conclude that Cre/Lox switches using viral promoters cannot be applied in a hypothesis-led fashion as experimental clinical therapeutics. These factors limit the application of the Cre/Lox switch in its current configuration for telomerase-specific gene therapy and are also relevant for successful development of Cre/Lox switches regulated by other promoters. Based on these findings, we highlight possible modifications to significantly improve the performance and predictability of other novel Cre/Lox switches for gene therapy.
Materials and Methods
Cell Lines and Plasmids
In this study, we used the human cancer cell lines 5637 (bladder carcinoma), C33A (cervical carcinoma), A2780 (ovarian adenocarcinoma), and A549 (lung adenocarcinoma). The relative targeting efficiency of these cell lines by hTR- or hTERT-directed enzyme–prodrug therapy has previously been reported [15,16]. All vectors reported in this study are based on the pGL3 reporter vector backbone (Promega, Madison, WI). The hTR- and hTERT-luciferase reporter vectors (pGL3-hTR and pGL3-hTERT) contain 867- and 572-bp fragments of the hTR and hTERT promoters, respectively, which have previously been shown to direct the selective expression of transgenes in tumor cells [15,16]. The control vectors pGL3-SV40 and pGL3-Basic were obtained from Promega. To construct the hTR-, hTERT-, SV40-, and promoterless-Cre expression vectors, the luciferase gene was deleted from the pGL3 series of luciferase reporters by an NcoI/XbaI digest. The ends were made blunt with Klenow fragment, and the vectors were religated. Cre was inserted downstream of each promoter as a HindIII fragment from pTurbo-Cre (a kind gift from Stephen Forrow). The Cre-regulated reporter pGL3-CMV/LPS (vector LPS, “LoxP-Stop”) contains a 588-bp fragment of the CMV immediate-early promoter separated from the luciferase gene by LoxP-flanked stuffer DNA containing the SV40 late polyadenylation signal from pGL3-Basic. The positive control vector pGL3-CMV (vector CMV) was generated by in vitro Cre-mediated deletion of the stuffer DNA using recombinant Cre (no. 69247; Merck Biosciences Ltd., Nottingham, UK).
Transfections and Luciferase Assay
All transfections were performed using Lipofectamine transfection reagent, according to the manufacturer's instructions (Invitrogen, Renfreshire, UK). A 1:1 DNA/Lipofectamine ratio was used, and 250 ng/well reporter plasmid was transfected in 96-well luminometer plates (Fisher Scientific UK, Leicestershire, UK) together with varying amounts of hTR-, hTERT-, SV40-, or promoterless-Cre expression vectors. Thirty nanograms of pSV40-Renilla luciferase expression plasmid (Promega) was also cotransfected in each well for the normalization of hTR promoter activity. Forty-eight hours posttransfection, cells were lysed and luciferase activities were determined using Dual Luciferase Assay reagents (Promega), according to the manufacturer's instructions. Each well was normalized to pSV40-Renilla using the formula: LN = LW(RM / RW), which preserves the magnitude of the firefly luciferase activity (LN = normalized firefly luciferase, LW = firefly luciferase activity of a well, RM = mean Renilla activity of the plate, and RW = Renilla value of the well). It should be noted that the regulatory elements in the pGL3-SV40 and SV40-Renilla vectors are not directly comparable because SV40-Renilla contains a chimeric intron used to boost Renilla expression and has a different arrangement of enhancer elements. All transfections were performed in quadruplicate, and all experiments were repeated a minimum of three times.
Western Blot Analysis
A2780, 5637, C33A, or A549 cells were transiently transfected with 5 μg of hTR-, hTERT-, SV40-, or promoterless-Cre expression plasmids using Lipofectamine reagent (Invitrogen). Forty-eight hours posttransfection, protein extracts were prepared in sodium dodecyl sulfate (SDS) lysis buffer (10% SDS, 500 mM EDTA, and 1 M Tris–HCl). Protein concentrations were estimated at OD595 using the Bio-Rad protein assay (Bio-Rad Laboratories Ltd., Hemel Hempstead, UK). Twenty micrograms of protein equivalents was separated alongside 1 U of recombinant Cre (no. 69247; Merck Biosciences Ltd.) by SDS-PAGE then blotted onto PVDF filter (Millipore, Watford, UK) and blocked overnight at 4°C in PBS–Tween containing 5% nonfat dried milk. Filters were probed for 2 hours with 1:3000 dilutions of primary anti-Cre antibody (no. 69050-3; Merck Biosciences Ltd.) or 1:1000 dilutions of anti–α-tubulin antibody (Sigma, Ayrshire, UK), then with a 1:3000 dilution of horseradish peroxidase (HRP)–conjugated antirabbit secondary antibody. HRP was detected using ECL HRP detection reagents (Amersham Pharmacia, Buckinghamshire, UK).
Results
hTR, hTERT, and SV40 Promoters Direct Cre Expression in Cancer Cell Lines
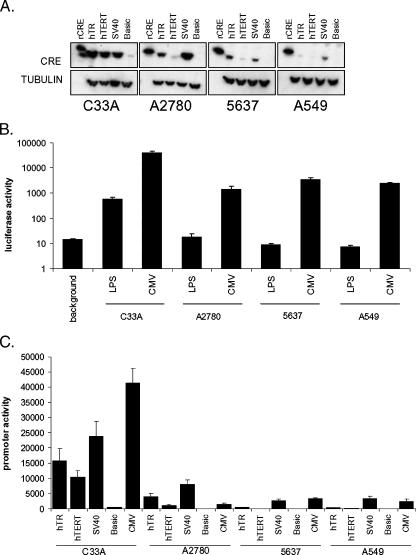
To develop a telomerase-specific Cre/Lox switch system, we first constructed Cre recombinase expression vectors either lacking a promoter (basic) or having Cre controlled by hTR, hTERT, or SV40 promoters. All Cre expression vectors and luciferase expression vectors described here are based on the same vector backbone (pGL3; Promega) that contains an insulating polyadenylation signal upstream of the expression cassette, which is used to reduce background (cryptic) transgene expression. Five micrograms of each vector was transiently transfected in the human cancer cell lines 5637 (bladder carcinoma), C33A (cervical carcinoma), A2780 (ovarian adenocarcinoma), and A549 (lung adenocarcinoma). Western blot analysis was performed to detect the expression of Cre protein. As a control, 1 U of recombinant Cre protein was visualized alongside the extracts of each cell line.
As shown in Figure 2A, all promoters directed the high-level expression of Cre in C33A cells. A weak band was also detected on the transfection of the promoterless vector in these cells, indicating that this vector still harbors a cryptic promoter activity in some cells. A2780 cells also expressed comparatively high levels of Cre from the hTR and SV40 promoters, although the hTERT promoter appeared substantially weaker in this cell line. The 5637 cells expressed even lower levels of Cre from each promoter, with hTERT-directed expression being almost undetectable. The A549 cells showed the lowest expression levels of Cre using all promoters. Thus, the hTR, hTERT, and SV40 promoters direct cell- and promoter-specific Cre expressions. These results correlate well with our previous analyses of therapeutic targeting efficiency using the NTR/CB1954 system in these cell lines [15,16].
Figure 2.
Relative activities of promoter constructs. (A) Expression levels of Cre protein. Cell lines indicated were transiently transfected with 5 μg of expression constructs encoding Cre recombinase under the control of the promoters indicated. Forty-eight hours posttransfection, 20 μg of protein extracts was probed for the expression of Cre and α-tubulin as loading controls. Western blot analyses were repeated three times. (B) Repression of CMV promoter activity by stuffer DNA. Two-hundred fifty nanograms of vector CMV or vector LPS, harboring a polyadenylation signal downstream of the CMV promoter, was transiently transfected in quadruplicate. Forty-eight hours posttransfection, luciferase activities were determined. Results are the pooled means and standard errors of three independent experiments for each cell line. (C) Relative promoter activities. Cell lines were transiently transfected in quadruplicate with 250 ng of hTR-, hTERT-, SV40-, basic (promoterless)–, or CMV-luciferase expression plasmids. Forty-eight hours posttransfection, luciferase activities were determined. Results are the pooled means and standard errors of three independent experiments for each cell line.
Repression of CMV Promoter Activity by LoxP-Flanked Stuffer DNA Containing the SV40 Polyadenylation Signal
We next constructed a luciferase reporter vector in which the CMV promoter is separated from the luciferase gene by a LoxP-flanked stuffer fragment harboring the SV40 late polyadenylation signal (vector LPS, LoxP-Stop; Figure 1). The LoxP sites in this vector were cloned in “reverse” orientation with respect to the CMV promoter, preventing the introduction of false translational initiation codons (which are present in the LoxP forward sequence after stuffer excision) upstream of the luciferase. Incubating this plasmid in vitro with recombinant Cre deleted the stuffer, allowing us to generate clones harboring a positive control plasmid in which the stuffer is completely removed, mimicking the expected product of Cre-mediated recombination in vivo (vector CMV; Figure 1). The activity of this vector is indicative of the maximum possible CMV-dependent expression in each cell line after the cotransfection of the Cre expression vectors.
To determine the extent of luciferase repression by the polyadenylation signal in vector LPS, we compared the luciferase activities of the CMV and LPS vectors (Figure 2B, note the log scale). In C33A and A2780 cells, luciferase activity was repressed by 70- and 80-fold, respectively, whereas in A549 and 5637 cells, the polyadenylation signal repressed luciferase activity by 329- and 388-fold, respectively. Thus, incorporation of the polyadenylation signal upstream of the luciferase gene significantly repressed CMV promoter activity in all cells. For A2780, A549, and 5637 cells, these repressed values were not significantly above the background luminescence detected in untransfected wells. However, in C33A cells, the activity of vector LPS was still substantially above background, suggesting that the CMV promoter is sufficiently strong in these cells for a fraction of transcripts to run through the polyadenylation signal. It should therefore be noted that the CMV promoter/SV40 polyadenylation signal pairing may be leaky in some cells and that this could be a potential source of off-target transgene expression. To optimize the system, it may be necessary to include alternative or additional transcriptional silencing elements.
hTR, hTERT, SV40, and Basic Promoter Activities Relative to CMV
To enhance telomerase-specific transgene expression by this system, the CMV promoter must be more active than hTR or hTERT within target cells. We therefore directly quantified the activity of all promoters in each cell line, using hTR-, hTERT-, SV40-, basic (promoterless)-, and CMV-luciferase reporters. Two hundred fifty nanograms of each reporter was transiently transfected and, 48 hours posttransfection, luciferase activities were determined (Figure 2C). The relative luciferase activities for hTR, hTERT, and SV40 promoters between cell lines correlated well with our Western blot analysis results (compare Figure 2, A and C). It should be noted that above-background luminescence was detected on the transfection of the promoterless luciferase expression vector in both C33A and A2780 cells, although Cre expression was not detected by Western blot analysis on the transfection of the equivalent promoterless Cre expression vector in A2780 cells. Importantly, CMV promoter activity varied significantly between cell lines.
The CMV promoter was frequently stronger than hTR or hTERT, although the differential was small in some cases. In C33A cells, for example, the CMV promoter was only 2.6- and 4-fold stronger than the hTR and hTERT promoters, respectively. Interestingly, in A2780 cells, CMV promoter activity was lower than hTR activity. Thus, in A2780 cells, use of the CMV promoter to drive therapeutic transgene expression in the context of this system will be less efficient than using hTR alone. The CMV promoter was also only 1.3-fold more active than the hTERT promoter in these cells. Specific implications of this finding are considered in the Discussion section. In contrast, in the cells with weaker hTR and hTERT activities, CMV activity was 8.7- and 15.7-fold stronger than hTR and hTERT in A549 cells, respectively, and was 7.2- and 44-fold stronger than hTR and hTERT in 5637 cells, respectively. It was therefore expected that the telomerase-specific Cre/Lox recombination should derepress the stronger CMV promoter activity and thus significantly enhance luciferase expression in these latter cells, relative to use of the hTR or hTERT promoters alone.
Telomerase-Specific Cre Expression Activates the Recombinase Switch
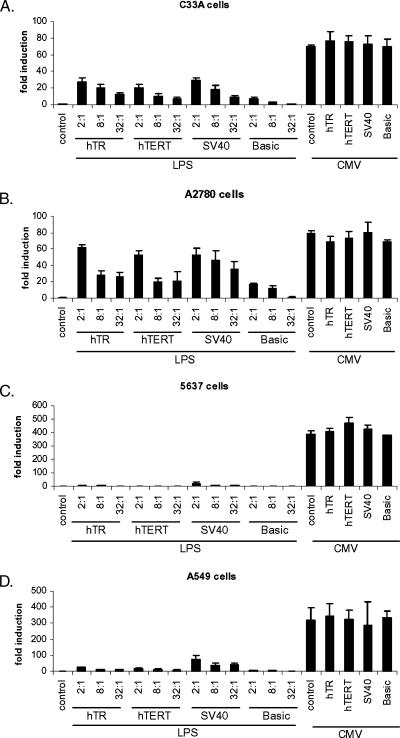
To test the performance of the switch, we cotransfected the LPS reporter with a titration of each Cre expression vector and measured the fold induction of luciferase activity (2:1, 8:1, and 32:1 vector LPS/Cre expression vector ratios). The CMV reporter with and without Cre expression was also included to estimate the activation efficiency of the switch. As shown in Figure 3, A–D, cotransfection of the hTR-, hTERT-, or SV40-Cre vectors induced the activity of the LPS reporter in a promoter-, cell-, and dose-dependent manner, indicating that the switch was functional. As expected, Cre transfection had no significant effect on control CMV promoter activity.
Figure 3.
Activation of the Cre/Lox switch. Two-hundred fifty nanograms of vector LPS or vector CMV was transiently transfected in quadruplicate in each cell line alone or in combination with the indicated hTR-, hTERT-, SV40-, or basic (promoterless)–Cre expression vectors at 2:1, 8:1, or 32:1 ratios (LPS or CMV: Cre expression vector), as indicated. For vector CMV, only the 2:1 ratio is shown (rightmost four bars). Results are pooled means and standard errors of three independent experiments for each cell line. (A) C33A cells; (B) A2780 cells; (C) 5637 cells; (D) A549 cells.
Cre expression led to a strong induction of luciferase activity in C33A cells. At the highest ratio tested in this experiment, the fold inductions ranged from 20- to 29-fold. However, these values were significantly lower than the maximum 70-fold difference between LPS and CMV activities. Interestingly, fold activation profiles were relatively similar between promoters and across the concentration range despite differences in the activities of the hTR, hTERT, and SV40 promoters. We also noted that the high concentration of basic-Cre led to a sevenfold induction of activity. In A2780 cells, a strong induction was observed for all promoters (range, 52- to 62-fold). Again, these values were lower than the maximal 80-fold difference between CMV and LPS promoter activities. Despite more pronounced differences in hTR and hTERT promoter activities than in C33A cells, however, both the hTR- and hTERT-Cre expression vectors resulted in similar switch activations across the concentration range. In these cells, SV40-Cre induced luciferase expression more than hTR- or hTERT-Cre at the lower ratios. We noted that cotransfection of the basic vector, which lacks a promoter, also resulted in significant induction of LPS at both the 2:1 and 8:1 ratios in these cells. Together, these data suggest that the Cre/Lox switch is extremely sensitive to activation by very low Cre doses in these cells, but may be less sensitive to differential Cre expression levels at higher doses in these cells.
In 5637 cells, neither hTR-Cre nor hTERT-Cre resulted in significant induction of CMV promoter activity. SV40-Cre cotransfection induced activity by 20-fold, but the maximum achievable induction was 388-fold in this cell line, suggesting that, overall, activation efficiency was extremely poor in these cells. Finally, in A549 cells, which had the weakest hTR and hTERT activities, the profile of switch activation by the SV40 promoter was higher than that observed for hTR or hTERT, although hTR and hTERT were themselves relatively similar across the concentration range. The hTR-Cre vector led to 24-, 11-, and 9-fold induction of LPS, whereas hTERT-Cre led to 17-, 11-, and 6-fold activation. In contrast, SV40 led to 72-, 38-, and 42-fold activation at the ratios tested. However, because the SV40 promoter is 22-fold stronger than the hTERT promoter in these cells, it would have been reasonable to expect a greater difference between activation levels induced by hTERT-Cre or SV40-Cre.
Sigmoid Dose–Response Characteristics of Cre/Lox Switch Activation
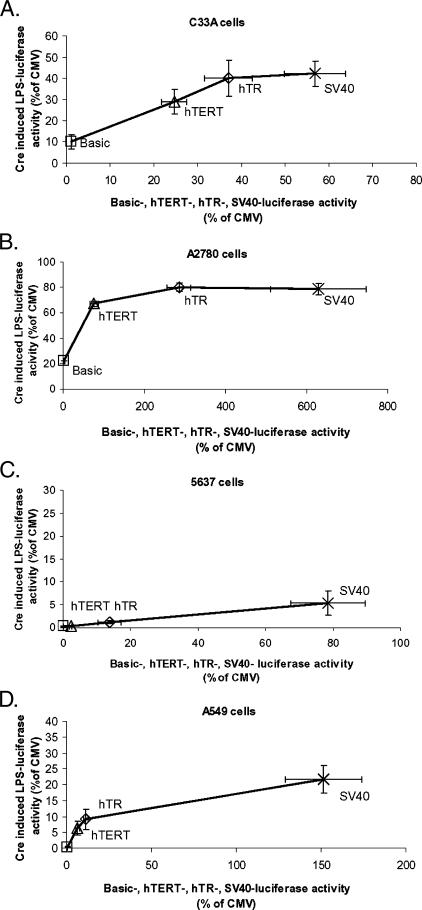
To determine whether induction of the Cre/Lox switch leads to enhancement of luciferase activity relative to direct expression through the hTR or hTERT promoters, we examined the relationship between the specific promoter activity of hTR-, hTERT-, SV40-, or basic-luciferase vectors (x-axis) and the level of switch activation achieved by hTR-, hTERT-, SV40-, or basic-Cre cotransfection in the switch system (y-axis) at the 2:1 ratio. As shown in Figure 4, A–D, both specific promoter activities and switch activation efficiency are represented as a percentage of the activity of the CMV promoter alone, which was included as a control in all experiments. Therefore, enhancement can be inferred, where y > x. The relationship between specific promoter strength and switch activation efficiency appeared roughly sigmoid for all cell lines.
Figure 4.
The Cre/Lox switch does not enhance telomerase-specific luciferase expression. Individual specific promoter activities measured for the transfection of 250 ng of hTR-, hTERT-, SV40-, and basic-luciferase expression vectors are represented on the x-axis as a percentage of CMV promoter activity in each cell line. Induced luciferase activity of the Cre/Lox switch after the cotransfection of hTR-, hTERT-, SV40-, and basic-Cre expression plasmids with 250 ng of vector LPS at a 2:1 ratio (LPS/Cre) is shown on the y-axis, also as a percentage of control CMV promoter activity (maximal activation efficiency) in each cell line. The criterion used to demonstrate enhancement is therefore y > x. Each data point represents means and standard errors from three pooled independent experiments performed in quadruplicate. Squares: basic-luciferase (x-axis), basic-Cre (y-axis); triangles: hTERT-luciferase (x-axis), hTERT-Cre (y-axis); diamonds: hTR-luciferase (x-axis), hTR-Cre (y-axis); crosses: SV40-luciferase (x-axis), SV40-Cre (y-axis). (A) C33A cells; (B) A2780 cells; (C) 5637 cells; (D) A549 cells.
In C33A cells, hTERT promoter activity alone was 24.6% of CMV promoter activity (± 2.9%). Cotransfection of a 2:1 LPS/hTERT-Cre ratio resulted in LPS induction to 28.9% of CMV activity (± 5.8%). In these cells, hTR promoter activity alone was 37.1% of CMV (± 5.5%), whereas when the hTR promoter was used to drive Cre expression, the activated switch activity was 39.1% of CMV (± 8.4%). As previously noted, the hTR promoter was actually stronger than CMV in A2780 cells and, therefore, there could have been no enhancement by activating the switch with this promoter. However, hTERT promoter activity in A2780 was 76.2% of CMV (± 10.8%). When hTERT was used to drive Cre expression, vector LPS was activated to only 67.4% of CMV activity (± 9.6%). Thus, although high-level switch activation efficiency was achieved in these cell lines, low differentials between hTR/hTERT and CMV promoter activities meant that no significant benefit was conferred by the Cre/Lox system relative to the expression of luciferase directly from either hTR or hTERT promoters.
In A549 cells, hTERT and hTR promoter activities were only 6.4% (± 1%) and 11.2% (± 1.3%) relative to CMV promoter activity, respectively, and it was therefore expected that the Cre/Lox system might enhance luciferase expression. Interestingly, activation of the switch resulted in relative activities of only 6.3% (± 2.1%) for hTERT-Cre expression and 9% (± 3.2%) for hTR-Cre expression. The lack of enhancement in these cells appeared to be due to a low peak in maximum efficiency of switch activation because the much stronger SV40 promoter (150% of CMV promoter strength) led to only 21.7% activation. Similarly, in 5637 cells, the hTR and hTERT promoter activities alone were 13.8% (± 3.4%) and 2.2% (± 0.3%) of CMV promoter activity, whereas on cotransfection of hTR-Cre and hTERT-Cre, the switch was activated only to 1.2% (± 0.4%) and 0.4% of the level of CMV promoter activity alone. Therefore, the Cre/Lox switch actually restricted the expression that could be achieved relative to using either promoter alone in this cell line, mainly because of poor overall activation.
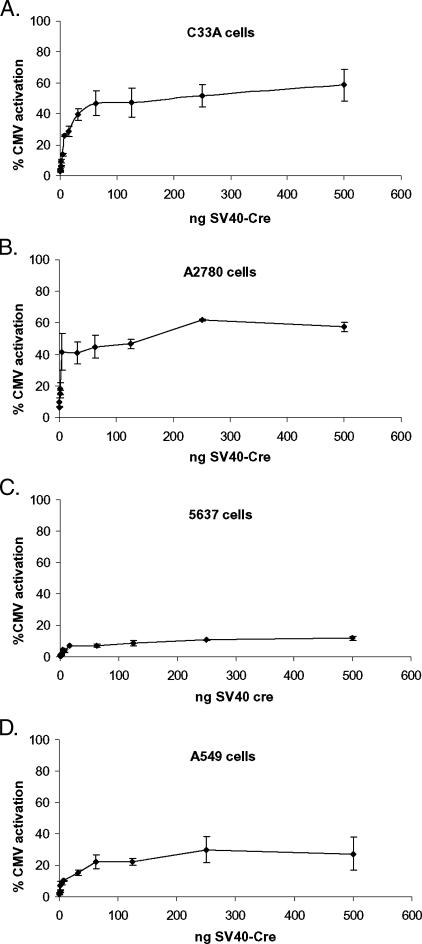
These data indicate that switch activation exhibits a broad plateau phase in all cell lines tested, at which increasing promoter activity—and hence Cre expression level—does not lead to equivalent linear increases in switch induction (“saturation efficiency”). Saturation efficiency differed significantly between the cell lines but was always significantly lower than 100%. Because we have shown that the activities of the hTR, hTERT, and SV40 promoters correlate well with Cre expression level in each cell line, these data also suggested that a relatively low “saturation dose” of Cre expression was required in each cell line to reach saturation efficiency. To test this relationship, we performed a titration of the SV40-Cre expression vector over a > 3-log concentration range using a fixed concentration of 250 ng of vector LPS (500–0.24 ng of SV40-Cre, 1:2 to 1000:1 ratios).
As shown in Figure 5, A–D, the titration experiment confirmed that activation of the Cre/Lox switch follows a sigmoid-like dose response in all cells tested here. Importantly, saturation efficiencies were very different between cell lines, with both A2780 and C33A cells reaching 50% to 60% maximal activation, whereas A549 cells reached only approximately 30% efficiency and 5637 cells reached only 10% activation. It is unclear how cell specific factors might influence the maximum activation efficiency in this system. One possibility is that cell-specific expression of CMV promoter-binding factors might block the access of Cre to its promoter-proximal site. Alternatively, cell-specific differentials in nuclear import or uncharacterized posttranslational modifications of Cre protein could play a role. Whatever the mechanistic basis for this finding, the data reported here have significant implications for the rational development of novel Cre/Lox switch systems for gene therapy.
Figure 5.
Sigmoid dose response of switch activation. Cells were transiently transfected with 250 ng of vector LPS together with a titration of SV40-Cre expression vector or with vector CMV alone. Forty-eight hours posttransfection, luciferase activities were determined. The concentrations of SV40-Cre ranged from 500 to 0.24 ng (LPS/SV40-Cre ratio ranged from 1:2 to 1000:1). Luciferase activities are represented as a percentage of the activity of vector CMV (maximal activation efficiency). Results are the pooled means and standard errors derived from three independent experiments in each cell line. (A) C33A cells; (B) A2780 cells; (C) 5637 cells; (D) A549 cells.
Discussion
Preclinical telomerase-directed gene therapy strategies show an extremely broad target cell range, coupled with a high degree of selectivity for tumor cells over normal cells [9]. However, for approaches such as enzyme/prodrug therapy, which may require high transgene expression levels, hTR, and particularly hTERT, promoter activities may occasionally be too low in some cancer cells to result in effective targeting [15,16]. Therefore, systems to enhance telomerase-specific transgene expression while maintaining selectivity may be advantageous in some cases. In this report, we tested a telomerase-specific Cre/Lox–dependent switch to determine whether coupling the specificity of the hTR and hTERT promoters with the activity of the CMV promoter could increase the likelihood of effectively targeting cancer cells with lower telomerase promoter activities in gene therapy applications. The switch was tested using a cell panel that exhibits a wide range of telomerase promoter activities, thus allowing us to examine different levels of Cre expression.
The hTR, hTERT, and SV40 promoters directed cell-specific levels of Cre expression that were well correlated with individual promoter strengths measured by luciferase assay. We also showed that incorporation of the SV40 polyadenylation signal in a LoxP-flanked stuffer fragment downstream of the CMV promoter effectively repressed CMV-dependent luciferase expression. However, in C33A cells, a significant level of luciferase activity was still detected even without Cre expression, indicating that the SV40 polyadenylation signal is not sufficient to completely block the activity of the CMV promoter in all cells. This configuration of the Cre/Lox switch may therefore display leakiness in some cells, which might give rise to off-target effects if the system was widely applied. The relative silencing of transgene expression could be improved by incorporation of additional or alternative silencing elements in the stuffer.
Cotransfection of Cre expression plasmids derepressed luciferase activity, indicating that the basic system was functional. However, the degree of switch activation was not well correlated with specific promoter activities and, thus, relative expression levels of Cre protein. Indeed, even cotransfection of the promoterless-Cre vector significantly induced the switch in A2780 and C33A cells. Titration of SV40-Cre revealed a sigmoid dose response characterized by a plateau of activation efficiency (saturation efficiency) at very low Cre doses (saturation dose).
These results are consistent with the kinetics of Cre-mediated excision in vitro [38,39]. The behavior of Cre in vitro appears to be related to the stoichiometry and dissociation kinetics of the recombination complex and possibly to a time-dependent inactivation or sequestration mechanism. We show here that these characteristics are retained in the cellular milieu. Importantly, for gene therapy, our study indicates that dose response and saturation efficiency differ markedly between cell lines, with A2780 and C33A cells both reaching approximately 60% of full efficiency but with A549 and 5637 cells reaching only approximately 30% and 10%, respectively.
These data may explain the previous observation that effective enhancement of tissue-specific gene therapy by the Cre/Lox switch requires an extremely large differential between the activities of selective and constitutive promoters [33]. Our results indicate that optimal effects will be expected mainly for selective promoters with activity differing significantly from the saturation efficiency of the constitutive promoter in individual cell lines. Furthermore, there is no net benefit in terms of increased switch activation efficiency by increasing Cre expression levels above the saturation dose. Thus, the differential in promoter activities will also be optimal when the selective promoter is weak enough to direct Cre expression levels that lie close to the saturation dose. Because these parameters are likely to vary widely, it may prove difficult to make any general predictions regarding the fold difference required for enhancement.
As we show here, either a low differential between selective and constitutive promoter activities or a low saturation efficiency can limit the general applicability of this system. Therefore, our results do not support the development of the system in its current configuration for telomerase-specific enzyme–prodrug therapy because the telomerase promoters already show acceptable activity in a range of target cells.
Our results do not indicate, however, that the Cre/Lox system has no utility for telomerase gene therapy. Indeed, because hTERT promoter activity appeared to be close to the saturation dose in all cell lines, it is very likely to be low enough in some cells to result in enhancement given that the other variables are favorable. The use of a stronger constitutive promoter could significantly improve the chances of enhancement using an hTERT-specific switch for cancer therapy. Interestingly, the system might also be used to selectively target gene expression to normal stem cells of noncancer patients, which should be the main telomerase-positive populations. The current switch may also have utility if high expression levels can be sacrificed in return for selectivity, as may be the case for expression of siRNA.
Overall, only a subset of cancers will be good targets for this switch in its current configuration. Additionally, because of the dose–response profile of the Cre-mediated excision that we describe, off-target stuffer excision in nontarget cells with low telomerase could also result in transgene expression. However, an improvement could be made to limit cryptic activation in nontarget cells. Cre-mediated recombination of the mutant LoxAT sequence is inhibited at low Cre concentrations in vitro, and although the final saturation efficiency for LoxAT is equivalent with that of LoxP, more Cre is needed to maximally recombine LoxAT [39]. LoxAT thus has the potential to introduce a low-threshold Cre dose requirement for initial switch activation, which could reduce the potential for off-target effects.
Finally, we suggest a critical improvement for novel Cre/Lox switch systems to progress to successful clinical testing: because a large differential between selective and constitutive promoter activities is essential to predict enhancement by this system, it is also essential to use a constitutive promoter with activity that is estimable by standard techniques in patient biopsy samples. In this respect, viral promoters are a major disadvantage in this system because they have no acceptable biomarkers of activity. Our data show that, in A2780 cells, the hTR and hTERT promoters were stronger than CMV. Thus, viral promoters can restrict the efficacy of this system in an unpredictable way, rendering the approach extremely difficult to apply to hypothesis-led trials. A more predictable approach requires the use of a strong human housekeeping gene promoter. Thus, specific and constitutive target RNA levels could be judged in clinical samples, facilitating streamlined patient selection from clinical specimens.
In conclusion, telomerase-directed gene therapy continues to show promise in preclinical models. However, there is a hypothetical need to test strategies for overcoming low promoter activity in some target cells. Although we have not found significant benefits to the Cre/Lox system in the cells tested here, different configurations of this system might have greater utility. It is significant that telomerase-dependent expression of at least 12 different therapeutic transgenes has been reported, in addition to the development of telomerase-specific replicating adenoviral vectors [9]. Thus, tumor cells that are not susceptible to killing by any one approach may be targeted very efficiently by another. Furthermore, telomerase-specific gene therapies comprise the basic toolkit of both hTR and hTERT promoters, both of which show selective but widely different activities. Therefore, there is a suitably broad base of approaches with which to begin the clinical assessments that will ultimately define the efficacy of telomerase-specific targeting.
Acknowledgments
We would like to thank Stephen Forrow (Beatson Institute for Cancer Research, Glasgow, UK) for the initial gift of pTurbo-Cre.
Footnotes
This work was supported by grants from Cancer Research UK and the University of Glasgow.
References
- 1.Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc Natl Acad Sci USA. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 3.Bachand F, Autexier C. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA–protein interactions. Mol Cell Biol. 2001;21:1888–1897. doi: 10.1128/MCB.21.5.1888-1897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascolo E, Wenz C, Lingner J, Hauel N, Priepke H, Kauffmann I, Garin-Chesa P, Rettig WJ, Damm K, Schnapp A. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J Biol Chem. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- 5.El-Daly H, Kull M, Zimmermann S, Pantic M, Waller CF, Martens UM. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood. 2005;105:1742–1749. doi: 10.1182/blood-2003-12-4322. [DOI] [PubMed] [Google Scholar]
- 6.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 7.Naasani I, Seimiya H, Yamori T, Tsuruo T. FJ5002: a potent telomerase inhibitor identified by exploiting the disease-oriented screening program with COMPARE analysis. Cancer Res. 1999;59:4004–4011. [PubMed] [Google Scholar]
- 8.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith WN, Bilsland A, Hardie M, Evans TRJ. Cancer cell immortality—telomerase as a target for novel cancer gene therapies. Nat Clin Pract Oncol. 2004;1:88–96. doi: 10.1038/ncponc0044. [DOI] [PubMed] [Google Scholar]
- 10.Keith WN, Bilsland A, Evans TR, Glasspool RM. Telomerase-directed molecular therapeutics. Expert Rev Mol Med. 2002;2002:1–25. doi: 10.1017/S1462399402004507. [DOI] [PubMed] [Google Scholar]
- 11.Lavelle F, Riou JF, Laoui A, Mailliet P. Telomerase: a therapeutic target for the third millennium? Crit Rev Oncol Hematol. 2000;34:111–126. doi: 10.1016/s1040-8428(00)00057-3. [DOI] [PubMed] [Google Scholar]
- 12.White LK, Wright WE, Shay JW. Telomerase inhibitors. Trends Biotechnol. 2001;19:114–120. doi: 10.1016/s0167-7799(00)01541-9. [DOI] [PubMed] [Google Scholar]
- 13.Majumdar AS, Hughes DE, Lichtsteiner SP, Wang Z, Lebkowski JS, Vasserot AP. The telomerase reverse transcriptase promoter drives efficacious tumor suicide gene therapy while preventing hepatotoxicity encountered with constitutive promoters. Gene Ther. 2001;8:568–578. doi: 10.1038/sj.gt.3301421. [DOI] [PubMed] [Google Scholar]
- 14.Takeda T, Inaba H, Yamazaki M, Kyo S, Miyamoto T, Suzuki S, Ehara T, Kakizawa T, Hara M, DeGroot LJ, et al. Tumor-specific gene therapy for undifferentiated thyroid carcinoma utilizing the telomerase reverse transcriptase promoter. J Clin Endocrinol Metab. 2003;88:3531–3538. doi: 10.1210/jc.2002-021856. [DOI] [PubMed] [Google Scholar]
- 15.Plumb JA, Bilsland A, Kakani R, Zhao J, Glasspool RM, Knox RJ, Evans TR, Keith WN. Telomerase-specific suicide gene therapy vectors expressing bacterial nitroreductase sensitize human cancer cells to the pro-drug CB1954. Oncogene. 2001;20:7797–7803. doi: 10.1038/sj.onc.1204954. [DOI] [PubMed] [Google Scholar]
- 16.Bilsland AE, Anderson CJ, Fletcher-Monaghan AJ, McGregor F, Jeffry Evans TR, Ganly I, Knox RJ, Plumb JA, Nicol Keith W. Selective ablation of human cancer cells by telomerase-specific adenoviral suicide gene therapy vectors expressing bacterial nitroreductase. Oncogene. 2003;22:370–380. doi: 10.1038/sj.onc.1206168. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Zou WG, Lang MF, Luo J, Sun LY, Wang XN, Qian QJ, Liu XY. Cancer-specific killing by the CD suicide gene using the human telomerase reverse transcriptase promoter. Int J Oncol. 2002;21:661–666. [PubMed] [Google Scholar]
- 18.Komata T, Koga S, Hirohata S, Takakura M, Germano IM, Inoue M, Kyo S, Kondo S, Kondo Y. A novel treatment of human malignant gliomas in vitro and in vivo: FADD gene transfer under the control of the human telomerase reverse transcriptase gene promoter. Int J Oncol. 2001;19:1015–1020. doi: 10.3892/ijo.19.5.1015. [DOI] [PubMed] [Google Scholar]
- 19.Komata T, Kondo Y, Kanzawa T, Hirohata S, Koga S, Sumiyoshi H, Srinivasula SM, Barna BP, Germano IM, Takakura M, et al. Treatment of malignant glioma cells with the transfer of constitutively active caspase-6 using the human telomerase catalytic subunit (human telomerase reverse transcriptase) gene promoter. Cancer Res. 2001;61:5796–5802. [PubMed] [Google Scholar]
- 20.Koga S, Hirohata S, Kondo Y, Komata T, Takakura M, Inoue M, Kyo S, Kondo S. A novel telomerase-specific gene therapy: gene transfer of caspase-8 utilizing the human telomerase catalytic subunit gene promoter. Hum Gene Ther. 2000;11:1397–1406. doi: 10.1089/10430340050057477. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Zhang L, Huang X, Lin T, Yin M, Xu K, Ji L, Roth JA, Fang B. A novel single tetracycline-regulative adenoviral vector for tumor-specific Bax gene expression and cell killing in vitro and in vivo. Oncogene. 2002;21:4757–4764. doi: 10.1038/sj.onc.1205582. [DOI] [PubMed] [Google Scholar]
- 22.Groot-Wassink T, Aboagye EO, Glaser M, Lemoine NR, Vassaux G. Adenovirus biodistribution and noninvasive imaging of gene expression in vivo by positron emission tomography using human sodium/iodide symporter as reporter gene. Hum Gene Ther. 2002;13:1723–1735. doi: 10.1089/104303402760293565. [DOI] [PubMed] [Google Scholar]
- 23.Lin T, Huang X, Gu J, Zhang L, Roth JA, Xiong M, Curley SA, Yu Y, Hunt KK, Fang B. Long-term tumor-free survival from treatment with the GFP–TRAIL fusion gene expressed from the hTERT promoter in breast cancer cells. Oncogene. 2002;21:8020–8028. doi: 10.1038/sj.onc.1205926. [DOI] [PubMed] [Google Scholar]
- 24.Kim E, Kim JH, Shin HY, Lee H, Yang JM, Kim J, Sohn JH, Kim H, Yun CO. Ad-mTERT-Delta19, a conditional replication-competent adenovirus driven by the human telomerase promoter, selectively replicates in and elicits cytopathic effect in a cancer cell–specific manner. Hum Gene Ther. 2003;14:1415–1428. doi: 10.1089/104303403769211637. [DOI] [PubMed] [Google Scholar]
- 25.Boyd M, Mairs RJ, Keith WN, Ross SC, Welsh P, Akabani G, Owens J, Vaidyanathan G, Carruthers R, Dorrens J, et al. An efficient targeted radiotherapy/gene therapy strategy utilising human telomerase promoters and radioastatine and harnessing radiation-mediated by stander effects. J Gene Med. 2004;6:937–947. doi: 10.1002/jgm.578. [DOI] [PubMed] [Google Scholar]
- 26.Fullerton NE, Boyd M, Mairs RJ, Keith WN, Alderwish O, Brown MM, Livingstone A, Kirk D. Combining a targeted radiotherapy and gene therapy approach for adenocarcinoma of prostate. Prostate Cancer Prostatic Dis. 2004;7:355–363. doi: 10.1038/sj.pcan.4500760. [DOI] [PubMed] [Google Scholar]
- 27.Fullerton NE, Mairs RJ, Kirk D, Keith WN, Carruthers R, McCluskey AG, Brown M, Wilson L, Boyd M. Application of targeted radiotherapy/gene therapy to bladder cancer cell lines. Eur Urol. 2005;47:250–256. doi: 10.1016/j.eururo.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Yao M, Zhang Z, Gu J, Zhang Y, Li B, Sun L, Liu X. Enhanced suicide gene therapy by chimeric tumor-specific promoter based on HSF1 transcriptional regulation. FEBS Lett. 2003;546:315–320. doi: 10.1016/s0014-5793(03)00606-9. [DOI] [PubMed] [Google Scholar]
- 29.Cui W, Gusterson B, Clark AJ. Nitroreductase-mediated cell ablation is very rapid and mediated by a p53-independent apoptotic pathway. Gene Ther. 1999;6:764–770. doi: 10.1038/sj.gt.3300873. [DOI] [PubMed] [Google Scholar]
- 30.Anlezark GM, Melton RG, Sherwood RF, Coles B, Friedlos F, Knox RJ. The bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954): I. Purification and properties of a nitroreductase enzyme from Escherichia coli—a potential enzyme for antibody-directed enzyme prodrug therapy (ADEPT) Biochem Pharmacol. 1992;44:2289–2295. doi: 10.1016/0006-2952(92)90671-5. [DOI] [PubMed] [Google Scholar]
- 31.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh K, Van Duyne GD. Cre–loxP biochemistry. Methods. 2002;28:374–383. doi: 10.1016/s1046-2023(02)00244-x. [DOI] [PubMed] [Google Scholar]
- 33.Lin X, Fischer AH, Ryu KY, Cho JY, Sferra TJ, Kloos RT, Mazzaferri EL, Jhiang SM. Application of the Cre/loxP system to enhance thyroid-targeted expression of sodium/iodide symporter. J Clin Endocrinol Metab. 2004;89:2344–2350. doi: 10.1210/jc.2003-031963. [DOI] [PubMed] [Google Scholar]
- 34.Nagayama Y, Nishihara E, Iitaka M, Namba H, Yamashita S, Niwa M. Enhanced efficacy of transcriptionally targeted suicide gene/prodrug therapy for thyroid carcinoma with the Cre–loxP system. Cancer Res. 1999;59:3049–3052. [PubMed] [Google Scholar]
- 35.Yoshimura I, Ikegami S, Suzuki S, Tadakuma T, Hayakawa M. Adenovirus mediated prostate specific enzyme prodrug gene therapy using prostate specific antigen promoter enhanced by the Cre–loxP system. J Urol. 2002;168:2659–2664. doi: 10.1016/S0022-5347(05)64239-5. [DOI] [PubMed] [Google Scholar]
- 36.Ueda K, Iwahashi M, Nakamori M, Nakamura M, Matsuura I, Yamaue H, Tanimura H. Carcinoembryonic antigen-specific suicide gene therapy of cytosine deaminase/5-fluorocytosine enhanced by the cre/loxP system in the orthotopic gastric carcinoma model. Cancer Res. 2001;61:6158–6162. [PubMed] [Google Scholar]
- 37.Takikawa H, Mafune K, Hamada H, Nettelbeck DM, Muller R, Makuuchi M, Kaminishi M. An advanced strategy of enhanced specific gene expression for hepatocellular carcinoma. Int J Oncol. 2003;22:1051–1056. [PubMed] [Google Scholar]
- 38.Ringrose L, Lounnas V, Ehrlich L, Buchholz F, Wade R, Stewart AF. Comparative kinetic analysis of FLP and cre recombinases: mathematical models for DNA binding and recombination. J Mol Biol. 1998;284:363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]
- 39.Martin SS, Chu VC, Baldwin E. Modulation of the active complex assembly and turnover rate by protein–DNA interactions in Cre–LoxP recombination. Biochemistry. 2003;42:6814–6826. doi: 10.1021/bi0272306. [DOI] [PMC free article] [PubMed] [Google Scholar]