Abstract
Surgical cure of glioblastomas is virtually impossible and their clinical course is mainly determined by the biologic behavior of the tumor cells and their response to radiation and chemotherapy. We investigated whether response to temozolomide (TMZ) chemotherapy differs in subsets of malignant glioblastomas defined by genetic lesions. Eighty patients with newly diagnosed glioblastoma were analyzed with comparative genomic hybridization and loss of heterozygosity. All patients underwent radical resection. Fifty patients received TMZ after radiotherapy (TMZ group) and 30 patients received radiotherapy alone (RT group). The most common aberrations detected were gains of parts of chromosome 7 and losses of 10q, 9p, or 13q. The spectrum of genetic aberrations did not differ between the TMZ and RT groups. Patients treated with TMZ showed significantly better survival than patients treated with radiotherapy alone (19.5 vs 9.3 months). Genomic deletions on chromosomes 9 and 10 are typical for glioblastoma and associated with poor prognosis. However, patients with these aberrations benefited significantly from TMZ in univariate analysis. In multivariate analysis, this effect was pronounced for 9p deletion and for elderly patients with 10q deletions, respectively. This study demonstrates that molecular genetic and cytogenetic analyses potentially predict responses to chemotherapy in patients with newly diagnosed glioblastomas.
Keywords: CGH, LOH, glioblastoma multiforme, survival, TMZ
Introduction
Treatment of diffuse gliomas still remains one of the most disappointing tasks in oncology. Surgical cure of these infiltrating brain tumors is virtually impossible. The clinical course is determined by the biologic behavior of the tumor, including growth rate and response to radiation and chemotherapy.
Neuropathologic diagnosis is the most valuable tool for the classification of human tumors. However, within neuropathologic entities, morphologic and immunohistochemical analyses usually cannot predict prognosis and response to therapy. However, molecular genetic analysis has succeeded in determining distinct genetic subgroups that may exhibit different biologic behavior [1–3].
Trials on the effects of systemic chemotherapy on survival and recurrence in adults with high-grade gliomas showed extended survival times, but there was no evidence that the effect of chemotherapy depended on age, sex, histology, Karnofsky performance status (KPS), or extent of resection [4]. However, patients of young age and with complete resection have a considerably better prognosis [5–7]. Temozolomide (TMZ), an orally administered second-generation imidazotetrazine, has been demonstrated to increase the survival time of glioma patients [8]. Phase II studies of TMZ (versus procarbazine) showed that TMZ has an acceptable safety profile and can improve the quality of life [8–10].
Numerous studies revealed that the most common somatic chromosomal changes in malignant gliomas are complete or partial loss of chromosome 10 and gain of chromosome 7. Various molecular genetic alterations have been identified, including the amplification of EGFR, CDK4, and MDM2, as well as the deletion of tumor-suppressor genes like TP53 (17p), RB (13q), CDKN2A (9p), CDKN2B (9p), PTEN (10q), and DMBT1 (10q) [2,11–15]. These tumor-suppressor genes play crucial roles in the regulation of cell proliferation and apoptosis. The TP53 gene product, p53, is involved in the regulation of cell repair, apoptosis, and cell cycle. Cyclin-dependent kinases (cdk), such as CDK4 and their inhibitors, p16 and p15, proteins from CDKN2A and CDKN2B, and pRB, are key regulators of the cell cycle. The gene products of CDKN2A/B locus on 9p also participate in the TP53 pathway through a protein encoded by an alternate reading frame, p14arf, which binds to the p53/MDM2 complex and inhibits MDM2-mediated degradation of p53. Therefore, homozygous deletion of the CDKN2A/B locus affects both Rb and TP53 pathways [16].
In recent years, studies have identified a correlation between alterations on chromosome 10q and shorter survival in patients with high-grade glioma. Tada et al. [3] reported significantly shorter survival rates of patients with glioblastoma multiforme (GBM) with loss of heterozygosity (LOH) on 10q containing the PTEN/MMAC1 gene, and in anaplastic astrocytoma patients with LOH on 10q in the region containing DMBT1. Several authors [3,17–19] detected poorer survival rates associated with LOH on 10q in glioblastomas, but showed that PTEN mutation is only marginally associated with survival [17,20].
A further candidate on chromosome arm 10q is MGMT. The MGMT gene encodes for the DNA repair enzyme O6-alkylguanine-DNA-alkyltransferase, which is responsible for protecting cells from alkylating agents. The enzyme efficiently removes methyl adducts at the O-6 position of guanine, which is an important target of alkylating agents [21]. It could been shown that the sensitivity to alkylating agents like BCNU correlates inversely to MGMT activity [22,23]. The responsiveness to BCNU is associated with an increase in overall survival rate [24]. Further on, the presence of aberrant promoter hypermethylation of MGMT was associated with loss of the MGMT protein, in contrast to retention of protein in the majority of tumors without hypermethylation [25]. Further clinical trials suggested that methylation of the MGMT promoter is predictive for better outcome in patients with malignant gliomas treated with alkylating agents such as TMZ [26–28].
Gains of chromosome 7 are known to be associated with shorter patient survival in anaplastic astrocytomas and low-grade astrocytomas [29,30], but, to our knowledge, no correlation between additional copies of chromosome 7 and survival in GBM has been found so far. However, EGFR amplification is considered to be an unfavorable marker for survival [31,32]. Further indicators of poor prognosis are LOH on 9p [17,33] and p16 mutations [34].
Chemosensitivity and prolonged overall survival of patients with anaplastic oligodendroglioma have recently been linked to specific genetic alterations, namely LOH on 1p or combined LOH on 1p and 19q, and the absence of homozygous deletion of the CDKN2A tumor-suppressor gene on 9p21 [19,35]. Apart from these data on the effect of genetic changes on the overall prognosis of gliomas, there is no information at the moment on the significance of further genetic changes on therapy response. Therefore, we analyzed a series of TMZ-treated patients in comparison to a retrospective, conventionally treated control group with newly diagnosed glioblastoma with respect to the abovementioned typical chromosomal alterations in glioblastomas.
The aim of this study was to determine whether specific genetic markers predict response to TMZ chemotherapy and may serve as parameters for the rational design of chemotherapy.
Materials and Methods
Patients
In total, 80 cases of newly diagnosed glioblastomas operated on during the period of 1997 to 2003 were studied (Table 1). The patients were treated in two centers: 48 patients in the Department of Neurosurgery of the Saarland University and 32 patients in the Department of Neurosurgery, Charité, University Berlin. Patients eligible for this nonrandomized study were 18 to 70 years of age, with a histologically proven GBM (World Health Organization [WHO] grade IV astrocytoma) [2] and a KPS of 70 or better. Patients with renal, hepatic, or bone marrow impairment; HIV infection; prior chemotherapy; or stereotactic biopsy were excluded. All patients underwent radical resection followed by radiotherapy within 4 weeks of surgery. Radiotherapy consisted of fractionated focal irradiation at a dose of 1.8 to 2 Gy per fraction, given once daily 5 days per week over a period of 6 weeks, for a total dose of 60 Gy. Radiotherapy was delivered to the gross tumor volume with a 2-cm margin volume for the clinical target volume on a preoperative magnetic resonance imaging (MRI).
Table 1.
Patient Characteristics.
| Patients | TMZ (n = 50) | Conventionally Treated (n = 30) |
| Age (years) | ||
| Median | 53 | 58 |
| Range | 26–72 | 24–77 |
| Still alive | 17 | 3 |
| Gender | ||
| Male | 33 | 20 |
| Female | 17 | 10 |
| KPS | ||
| 100 | 27 | 6 |
| 90 | 19 | 16 |
| 80 | 4 | 2 |
| 70 | 6 | |
Two groups of patients were defined. The first group included patients from March 1997 to April 1999, treated after radical resection with radiotherapy alone (control group). In the second group, patients from the time period from April 1999 to November 2003 were included. This group received, after radical resection, the abovementioned radiotherapy regime and an adjuvant chemotherapy consisting of TMZ (TMZ group). TMZ was administered as a test dosage of 150 mg/m2 body surface area per day (750 mg/m2 total dose per cycle) on days 1 to 5 in the first cycle because of its dose-limiting toxic effect resulting in thrombocytopenia. The following cycles were performed at a dosage of 200 mg/m2 per day (1000 mg/m2 total dose per cycle). Treatment cycles were repeated every 28 days. Altogether, eight cycles were carried out. Patients were seen at least every 3 months in the first 2 years. At each follow-up visit, clinical performance status, neurologic status, and MRI were recorded.
Specimens of resected tumors were immediately frozen and stored at -80°C, or fixed in formalin and embedded in paraffin. All patients gave written informed consent for the use of the tumor samples for genetic analysis.
Comparative Genomic Hybridization (CGH)
DNA was obtained using standard protocols. Reference DNA from the blood of a healthy donor and tumor DNA from frozen tumor tissues were labeled with biotin and digoxigenin by standard nick translation (Roche Diagnostics, Mannheim, Germany). Six hundred nanograms of each tumor and reference DNA was hybridized together with COT1 DNA (Roche Diagnostics) to normal chromosome metaphase spreads from peripheral blood lymphocytes prepared following standard procedures. After 3 to 4 days of hybridization at 37°C, posthybridization washes were performed at a stringency of 50% formamide/2x standard saline citrate (SSC), 2x SSC, and 0.1x SSC at 45°C. Tumor DNA was visualized with fluorescein isothiocyanate (Vector Laboratories, Burlingame, CA) and reference DNA with rhodamine (Roche Diagnostics). Fluorescence images were captured using a fluorescence microscope Olympus AX 70 (Olympus,Hamburg,Germany) with a cooled charged-coupled device camera. Image processing was performed by use of ISIS (MetaSystems, Altlussheim, Germany). Average ratio profiles were determined from analyses of 10 to 15 metaphases. The thresholds used for ratio profiles were 1.2 for gain and 0.8 for loss.
Because of suppression with COT1 DNA, the fluorescence intensities were not representative at chromosome regions with tandem repetitive DNA clusters (i.e., at the heterochromatic blocks on chromosomes 1, 9, 16, and Y, at the centromeric regions, and along the short arms of acrocentric chromosomes). These areas were excluded from evaluation. Chromosome 19 and the chromosomal segment 1p34-pter were also excluded from the analysis because results for 19 and 1p34-pter have been observed to be prone to artifacts in our and others' laboratories [36].
Microsatellite Analysis for LOH
DNA from frozen tumor tissues and, if not available, DNA from paraffin-embedded tumor sections (n = 24) was separated using previously published protocols [37,38].
The following regions were examined for allelic losses by nonradioactive microsatellite analysis: chromosomal arm 10p with markers D10S1172, D10S1159, D10S527, and D10S506; chromosomal arm 10q with markers D10S1419, D10S1171, D10S523, D10S1765, D10S1143, D10S 1173, D10S520, D10S521, D10S1141, D10S503, D10S1165, D10S1439, D10S505, D10S1134, and D10S1248; chromosomal arm 13q with markers D13S326, D13S887, D13S788, and D13S773; and chromosomal arm 9p with markers D9S759, D9S925, D9S1121, and D9S319. Nucleotide sequences and mapping information were retrieved from the Human Genome Database (www.gdb.org) or the Cooperative Human Linkage Center (www.chlc.org) database files. Polymerase chain reaction (PCR) was performed in a final volume of 10 µl containing 10 ng of DNA, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 200 µM deoxynucleotides, 0.1% gelatin, and 20 pmol of each primer. Taq polymerase (Gibco BRL/Life Technologies, Karlsruhe, Germany) was used and the MgCl2 concentration ranged from 1.0 to 2.0 mM, depending on the primer pair. Initial denaturation at 95°C for 3 minutes was followed by 29 cycles on a thermocycler (Biometra, Goettingen, Germany), denaturation at 95°C for 30 seconds, annealing at temperatures ranging from 52°C to 64°C for 40 seconds, and extension at 72°C for 30 seconds. A final extension step of 10 minutes at 72°C was added. PCR products were separated on 8% denaturating acrylamide gels and visualized by silver staining [39]. LOH was scored as previously described [38].
Statistical Analyses
Subgroups of patients were defined by genetic status, therapy, and age (old patients, > 53 years; young patients, ≤ 53 years). We chose this classification because the median age was 54 years. Comparisons between groups were performed by Kaplan-Meier curves and Cox regression analysis.
In the univariate Cox proportional hazards model, for a given genetic status, the main effect of therapy on survival was tested. In the first multivariate Cox model, for a given genetic status, main effects of therapy, age, gender, and KPS were estimated. In the second multivariate Cox model, main effects of therapy and age and an interaction effect between therapy and age were estimated. All effects were quantified by hazard ratio estimates with 95% confidence intervals (CIs). All P values were calculated with two-sided tests. Median survival rates were calculated using the Kaplan-Meier method. Association between dichotomous variables was tested by chi-square analysis.
LOH data were summarized per chromosomal arm. LOH was defined as present if at least one loss on the respective chromosomal arm was detected. For chromosome arm 10q with a large number of primer, LOH was defined as present if the majority of the considered regions were lost.
Results
Univariate and Multivariate Prognosis Analyses
Treatment, gender, and age In total, 60 patients had died and 20 were alive during the last follow-up (20 censored data). Median follow-up was 17.2 months and median age at diagnosis was 54 years.
Median overall survival for the TMZ group (n = 50; median age, 53 years) was 19.5 months and for the control group (n = 30; median age, 58 years) was 9.3 months. Univariate analysis showed that TMZ chemotherapy was significantly associated with longer survival rates (P = .00063; Figure 1).
Figure 1.
Kaplan-Meier estimates for patients with GBM treated with TMZ and radiotherapy (TMZ; solid lines) versus the untreated control group (radiotherapy alone; dashed lines) had significantly higher overall survival. Censored data (patients still alive) are plotted as hash marks.
Multivariate analysis indicated that the strongest factors associated with survival were chemotherapy and age. Younger patients survived longer than older patients (> 53 years of age) in the control group (P = .024). However, for older patients, a prolonged survival time under adjuvant chemotherapy treatment was detectable (P = .00014), but not for younger patients (P = .96). There was no significant difference in survival time by age in the TMZ-treated group (P = .83).
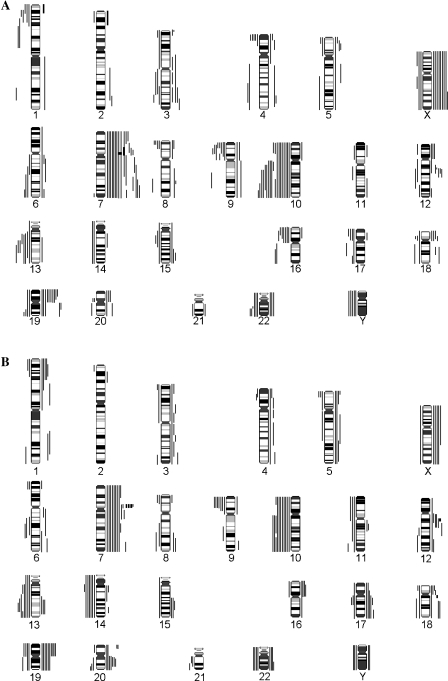
Genetic status, treatment, gender, and age Clinical data, CGH, and LOH results of patients in the TMZ and control groups are summarized in Tables 1–3. Both groups showed a similar genetic pattern. The most frequent genetic aberrations were complete or partial losses of chromosome 10 (63%) and complete or partial gains of chromosome 7 (58%). Further deletions were detectable on 9p (29%) and 13q (35%) (Figure 2).
Table 2.
Clinical Characteristics and CGH Results of TMZ-Treated and Conventionally Treated High-Grade Gliomas.
| Case Number | Tumor | Age/Sex | Chromosomal Imbalances by CGH | ||
| ST (months) | Gains | Losses | |||
| TMZ-treated high-grade gliomas | |||||
| 1782 | sGBM | 26/M | 24.2 | 19p, X | 16p13.1p13.3 |
| 265 | GBM | 46/F | 23.1 | None | 10q25.3qter, 16p12p13.3 |
| 1349 | GBM | 54/M | 33.8 | X | 16p, Y |
| 1099 | GBM | 49/M | 23.8 | 7, 12q13.2q13.3 | 1p31.3p32.3, 1q32.1q41, 10q |
| 369 | sGBM | 37/M | 14.3 | 1q, 5p15.1pter, 7q31.3qter, 8q21.1qter, 9p, 10p, 12p, 15q25qter, 18p11.2pter | 1p33p36.1, 4q21.1qter, 5q21qter, 10q21.3qter, 11p12p15.3, 13q12.1q31, 15q14q21.2, 19 |
| 1534 | GBM | 62/M | 18.4 | 7q11.2q21, 19p13.1p13.3, X | 4p16pter, 10q24.1qter, 13q21.3qter |
| 1106 | GBM | 70/M | 19.8 | 7q21.1qter, 12q13.1q14, 19p | 10q, 13q12.3q22, Y |
| 662 | GBM | 70/M | 13.4 | 3q24q26.3, 7p11.1p12, 7q31.1q34, 18q12.1q21.1, X | 4p16, 8p22pter, 10q, 16p11.2p13.2, Y |
| 1326 | GBM | 56/M | 19.2 | 7q34qter, 12p13.1pter | 10q21.3qter, 22q13.1q13.3 |
| 1515 | sGBM | 40/F | 11.2 | 19p | 2p23pter, 3p24.2p26, 21q21qter |
| 1707 | GBM | 47/M | 29.4 | 7, 12q13.3q14 | 10q21.1qter |
| 1691 | sGBM | 39/F | 17.4 | None | 1p35p36.1, 4q27q32, 9p21, 9q21.2q32, 14 |
| 497 | GBM | 58/F | 16.9 | 7p11.2p13, amp 1p36pter | 6q12q21, 9p23p24 |
| 1460 | GBM | 31/F | 11.9 | 2q34q37.1, 3p26pter, 3q23.2qter, 4p11p16, 4q11q13.3, 8p22pter, 9, 15q22.3qter, 18q12.3qter, 19q11q13.1, 20p, 22 | 3q11.1q25.2, 5p, 13q12.3qter, 16q, 20q11.2qter, Y |
| 896 | GBM | 51/M | 13.0 | 7 | 6q22.2qter, 9p21pter, 10p12.2pter, 10q23.1qter, 13q13qter, 14, 16q13qter, 22q12.2 |
| 1405 | GBM | 53/M | 7.4 | 4q31.3q33 | 8p23.1pter, 16p, 17, 19q13.2qter |
| 643 | GBM | 54/F | 15.3 | 4p16pter, 7p11.1p13, 7q32qter, 16p12pter, 19q, 22 | 10, 13q14.3q22.1 |
| 1795 | sGBM | 31/M | 6.9 | None | 5p15.1pter, 6q16.3qter, 8p23.2pter, 9p13pter 18q12.1qter |
| T4789 | GBM | 38/M | 44.1 | 8p, Xq23q28 | 3q13.1q25.3, 9p |
| T5958 | GBM | 63/F | 14.4 | 2q22q35, 3p11.1p14.1, 3p25pter, 3q13.1q21, 3q24q26.3, 4p14p15.3, 4q21.2q34, 5p14, 5q13.1q22, 6q14q23.1, 7, 8q22.3, 11p13p15.3, 11q14.1qter, 13q14.1q33, 14q12q22, 20p12pter, amp 7p11.1p11.2 | 1p32.3pter, 10, 12q23q24.3, 16p, 17p, 17q24qter, 18p11.2, 19, 22, Y |
| 784 | GBM | 52/M | 22.5 | 7p13p22, 7q11.1q22, 19p, X, amp 7p11.1p13 | 4p16, 8p22pter, 9p11p13, 9p23pter, 10p13pter, 10q11.2q21.2, 16p |
| T6002 | GBM | 53/M | 11.2 | 5, 6, 7, 12p, 12q11q14, 12q24.1qter, 15, 19, 20, amp 7p11.1p11.2 | 1p21p31.3, 3p, 3q23q26.3, 10, 13, 17p11.1p13, 18, 22 |
| 1536 | GBM | 53/M | 15.1 | 3p11.1p14.1, 3q11.1q26.3, 7, 9q, 21q21qter, X | 1p33p36.1, 4p15.1pter, 10, 12p12.1pter, 12q11q21.3, 15, 17q12q23, 18p11.1p11.2, 19q13.3qter, 20p11.1p11.2 |
| 1940 | GBM | 54/F | 17.3 | 5p13.3p15.2, 7p15.3pter, 18p | 8q22.1q24.1, 10, 12q14q23, Xp11.4pter, Xq13q21.3 |
| 6 | GBM | 41/M | 26.2 | 6q12q15, 6q25.1qter, 7, 12q14q21.1, X | 4p16, 9p11p23, 10, 11q12q14.3, 15q11.1q22.2, 20p11.1p11.2, Y |
| 596 | GBM | 32/M | 21.1 | 7, 12q13.3q21.1, 17p11.1p12, 19, Y | 3q26.1q36.3, 10, X |
| 947 | GBM | 46/F | 17.7 | 7, 15q24q26.1, 18p, 19, 20q | 9p21p23, 10q21.1q25.2 |
| XXL | GBM | 45/M | 8.6 | 7p15.1pter, 7q11.1q31.3, amp 2p22pter | 1p36pter, 10, 13q13q21.1, 17p12pter, 22q13.1qter, X |
| T6044 | GBM | 37/M | 7.6 | 18p11.1p11.2, 19p, X | 6q25.2qter, 7q36 |
| Conventionally treated high-grade gliomas | |||||
| 861 | GBM | 70/F | 22.7 | 2p21p23, 7p12, 12q14q21.3, 13q21.3q32 | 4p15.3pter, 8p22pter, 16p, 19p, 20q13.1qter |
| 1856 | GBM | 46/M | 65.8 | 13q21.1q32 | 1p34pter, 17q12q21.3, 19q, 22, Y |
| 1028 | GBM | 62/M | 5.6 | 6q11q14, 7, 19p, X, Y | 10, 14, 15, 18 |
| 2046 | GBM | 39/M | 39.8 | 1p13.3p31.1, 1q25q41, 3p24.1pter, 3p11.1p13, 3q11.2q13.3, 3q24q26.3, 4p11p15.2, 4q12q13.3, 4q22q28, 5p13.3pter, 5q12q23.3, 6q12q23.1, 7, 8p22pter, 9p, 14q11.2q24.1, 18p11.3pter, 20p13, 21q11.1q22.1 | 1p35pter, 10, 11, 12q22qter, 13q12.1q14.1, 13q32qter, 15q21.3qter, 17, 19, 20q11.2qter, 22 |
| T6025 | GBM | 73/M | 6.3 | 1p34.3p36.1, 1q31q43, 3p14.1p21.1, 7, 16p11.1p13.2, 17p11.1p12, 17q, 18p11.1p11.3, 19p, Y, amp 7p11.1p12 | 2p24pter, 4p15.1p15.3, 5p15.2pter, 8p21.3pter, 8q22.1q24.1, 10, 12q15q21.3, 13q14.3qter, 14, 15q21.3qter, 20p12pter |
| 393 | GBM | 65/M | 4.6 | 1p, 3, 7, 12q11q13.2, 12q23q24.3, 16p, 17q, 19, 20q, 22. amp 7p11.1p12 | 1q32.2qter, 9p, 10, 13q22q31, 21q11.1q21 |
| T5954 | GBM | 61/M | 3.3 | 1p24.1pter, 11q11q14.1, 16p, 18p11.1p11.2, 19, 20q, 22q12.3q13.3, Xp, Xq11.1q21.2 | 9p13pter, 13, 14q11.1q12, 18q12.1qter |
| 1819 | GBM | 76/F | 1 | 4q32qter, 5p14pter, 5q23.3q35.1, 6p23pter, 6q22.3qter, 7q33q36, 8q23q24.2, 13q21.3q33, 18q12.2qter | 17q11.2q21.3, 19p |
| 63 | GBM | 77/F | 10.9 | 7p11.1p12, 7q11.1q11.2 | 5p, 5q11.2q23.1, 6p23pter, 6q15q22.3, 9p23pter, 18p11.3pter |
| T6052 | GBM | 24/F | 4.8 | 5, 7, 12, 18, 19p, amp 12p13.1, 18q11.1q11.2 | 3p25pter, 3q26.3qter, 6p23pter, 8q22.1qter, 9p21pter, 11q23.3qter, 13q21.3q31, 20q13.2qter |
| 832 | GBM | 57/M | 8.9 | 4, 5, 7, 8q22.1q24.3, 14q11.1q12, 14q24.3q31, 17q24q25, 19, 20, 22, X | 3p, 9p13p24, 9q22.1q31, 10, 21q22 |
| Conventionally treated high-grade gliomas | |||||
| T4795 | GBM | 66/M | 0.2 | 4p15.1p16, 6q24q27, 7p15.1p22, 18q12.3q23, 20p13 | 10, 11p, 11q11q23.1, 13, 14, 19, 22, X, Y |
| 838 | GBM | 56/w | 1.3 | 7, 16, 17q12q21.3, 19, X, amp 7p11.2p12 | 9p21p24, 10, 13, 14, 22 |
| T4803 | GBM | 59/M | 9.4 | 1p32.1p36.3, 7, 9q32q34.2, 12q13.1q14, 15q22.1q24, 17, 20q11.1q13.1 | 10, 14 |
| H549 | GBM | 38/M | 5.5 | 3q21q23, 3q26.2qter, 7q21.3q31.1, amp 12q11q14 | 6q21q23.3, 10q, 11p, 11q11, 13q12.1qter, 14 |
| H321 | sGBM | 48/M | 18.2 | 19p, 20q11.2q13.1, X | 5p15.2pter, 9p21pter, 10q, Y |
| H147 | GBM | 48/M | 7.5 | 7q11.1q11.2, 19, 20q11.1q13.1 | 10p13pter, 10q11.2q21.3 |
| H281 | GBM | 56/M | 12.5 | 7p11.1p12 | None |
| N111 | sGBM | 24/M | 17.4 | 1p32pter, 11q13, 17, 19, 20, 22 | 6q16q23 |
sGBM, secondary glioblastoma multiforme; M, male; F, female; ST, survival time; amp, amplification.
Table 3.
LOH Results and Comparison with CGH Data of TMZ-Treated and Conventionally Treated High-Grade Gliomas.
| Tumor | ST (months) | CGH 10q* | PTEN LOH/Inf† | MGMT LOH/Inf† | CGH 13q* | RB LOH/Inf† | CGH 9p* | p16 LOH/Inf† |
| TMZ-treated high-grade gliomas | ||||||||
| 1795 | 6.9 | 0 | 2/2 | 0 | 0/3 | 1 | 3/3 | |
| 1534 | 40.9 | 1 | 1/4 | 2/4 | 0 | 0/2 | 0 | 0/2 |
| T4789 | 55.3 | 0 | 0/2 | 0 | 0/2 | 1 | 1/1 | |
| 1691 | 39.9 | 0 | 1/4 | 0 | 0/2 | 1 | 2/2 | |
| 1326 | 19.2 | 1 | 1/4 | 0/2 | 0 | 0/3 | 0 | 0/2 |
| 1460 | 11.9 | 0 | 0/5 | 1/5 | 1 | 2/2 | 0 | 0/2 |
| 643 | 35.3 | 1 | 3/4 | 1 | 0/3 | 0 | 3/3 | |
| 369 | 20.2 | 1 | 3/3 | 3/4 | 1 | 2/2 | 0 | 0/1 |
| 1515 | 11.2 | 0 | 1/1 | 0 | 0 | 1/1 | ||
| 1106 | 19.8 | 1 | 1/4 | 2/2 | 1 | 0/1 | 0 | 0/2 |
| 497 | 16.9 | 0 | 1/3 | 2/5 | 0 | 0/2 | 1 | |
| 662 | 13.4 | 1 | 2/2 | 2/6 | 0 | 0/3 | 0 | 0/3 |
| 1782 | 24.2 | 0 | 3/3 | 4/5 | 0 | 0/2 | 0 | 2/2 |
| 1349 | 56.8 | 0 | 0/5 | 0 | 0/3 | 0 | 0/2 | |
| 1099 | 23.8 | 1 | 4/4 | 4/6 | 0 | 0/1 | 0 | 1/2 |
| 1707 | 29.4 | 1 | 5/5 | 3/4 | 0 | 0/1 | 0 | 1/2 |
| 596 | 21.1 | 1 | 3/3 | 4/4 | 0 | 0/1 | 0 | 0/3 |
| 398 | 14.4 | 1/4 | 0/1 | 2/3 | ||||
| 1777 | 51.6 | 0/5 | 0/3 | 0/1 | ||||
| 1536 | 15.1 | 1 | 4/4 | 4/4 | 0 | 1/1 | 0 | 0/1 |
| 90 | 12.0 | 0/0 | 4/5 | 0/1 | ||||
| 947 | 17.7 | 1 | 5/5 | 0 | 0/3 | 1 | 2/3 | |
| XXL | 8.6 | 1 | 1/1 | 4/5 | 1 | 0 | 0/2 | |
| 784 | 22.4 | 1 | 1/1 | 0 | 0 | |||
| 1921 | 3.4 | 1 | 5/5 | 0 | 0/2 | 0 | 0/2 | |
| N529/02 | 21.4 | 0/5 | 0/2 | 0/1 | ||||
| N20/02 | 24.8 | 2/2 | 4/4 | 0/3 | 1/2 | |||
| N690/02 | 18.6 | 1/2 | 0/1 | 0/3 | ||||
| N1082/02 | 13.5 | 1/1 | 0/1 | |||||
| N1421/01 | 13.6 | 0/1 | 1/2 | 0/1 | ||||
| N934/02 | 24.5 | 1/1 | ||||||
| N1124/01 | 9.0 | 0/1 | 0/1 | |||||
| N1507/01 | 16.0 | 5/5 | 0/3 | 0/3 | ||||
| N363/02 | 18.9 | 0/4 | 0/4 | 0/1 | 1/3 | |||
| N449/03 | 8.2 | 2/3 | 3/3 | 2/4 | 2/3 | |||
| N1443/02 | 15.1 | 6/6 | 2/2 | |||||
| N528/03 | 6.7 | 4/4 | 0/4 | 1/2 | ||||
| N438/03 | 8.9 | 6/6 | 0/3 | 0/2 | ||||
| N202/03 | 10.8 | 4/4 | 2/2 | 0/2 | 1/2 | |||
| N412/03 | 9.0 | 0/5 | 0/3 | 0/2 | ||||
| N759/01 | 31.5 | 0/1 | 0/3 | 1/3 | 0/2 | |||
| N193/03 | 11.0 | 3/3 | 3/3 | 1/2 | 0/2 | |||
| N1618/02 | 10.3 | 1/2 | 1/2 | 0/1 | ||||
| Conventionally treated high-grade gliomas | ||||||||
| 1856 | 58.5 | 0 | 0/3 | 0 | 0/1 | 0 | 0/3 | |
| 1394 | 0.5 | 5/5 | 2/4 | 2/2 | 3/3 | |||
| 1819 | 1.0 | 0 | 0/4 | 0 | 0/3 | 0 | 0/2 | |
| 2046 | 39.8 | 1 | 4/4 | 1 | 0 | 0/1 | ||
| 63 | 10.9 | 0 | 0/6 | 2/5 | 0 | 1 | 0/1 | |
| T4803 | 9.4 | 1 | 3/3 | 0 | 0/3 | 0 | 0/3 | |
| N111 | 17.4 | 0 | 0/4 | 0 | 0/1 | 0 | 0/3 | |
| 2064 | 18.2 | 1 | 0/3 | 3/3 | 0 | 0/2 | 1 | 1/2 |
| 393 | 4.6 | 1 | 4/4 | 4/4 | 1 | 0/4 | 1 | 2/2 |
| 1028 | 5.6 | 1 | 0/5 | 0 | 0/3 | 0 | 0/2 | |
| N28/99 | 5.8 | 2/4 | 2/6 | 0/1 | 2/2 | |||
| N1106/00 | 39.0 | 1/3 | 3/4 | 1/2 | 0/2 | |||
| N113/99 | 9.5 | 3/4 | 0/2 | 2/3 | ||||
| N600/98 | 16.7 | 3/3 | 5/5 | 0/1 | 2/3 | |||
| N1047/00 | 0.7 | 4/4 | 1/1 | 1/1 | ||||
| N982/00 | 14.4 | 1/3 | 0/2 | 1/1 | ||||
| N823/01 | 14.4 | 2/2 | 0/1 | 1/1 | ||||
| N894/00 | 21.3 | 0/2 | 0/2 | 0/2 | ||||
| N860/01 | 21.3 | 0/3 | 1/2 | 0/3 | 0/3 | |||
| N1320/99 | 4.7 | 1/2 | ||||||
| N231/00 | 12.4 | 0/2 | 2/7 | 1/2 | 0/2 | |||
| N358/00 | 11.2 | 3/3 | 1/1 | |||||
1, deletion; 0, no deletion determined by CGH.
LOH at available informative markers.
Figure 2.
Overview of genetic imbalances of (A) TMZ-treated GBMs and (B) conventionally treated GBMs. Lines on the left represent losses, and lines on the right represent gains; amplifications are in bold.
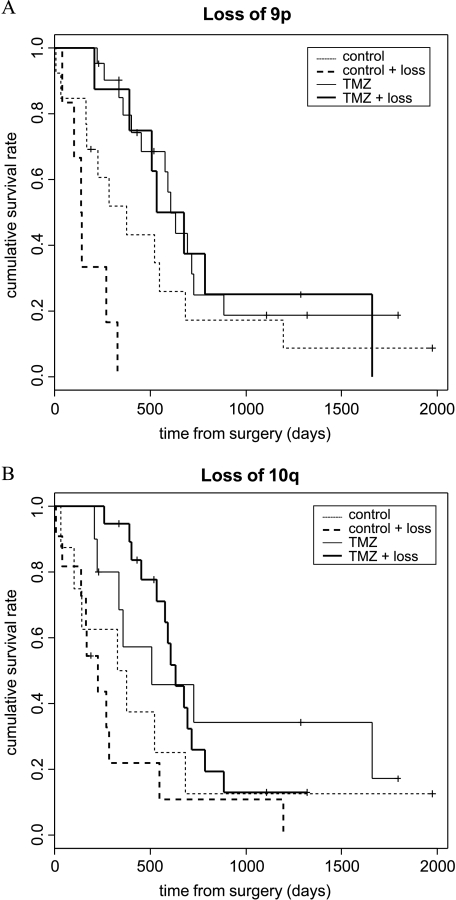
Subgroups of patients were defined by their genetic pattern. In the univariate model, patients with deletions of 9p and 10q, respectively, did associate with better prognosis under treatment (Figure 3; Table 4). No correlation with treatment was observed for gains of chromosomes 7p and 12, losses of chromosome 13, and gender. Due to the methodical limitations of LOH, only deletions could be detected. A positive effect on survival was observed for better KPS and younger age (Table 5).
Figure 3.
Kaplan-Meier estimates of overall survival, according to genetic alteration and assignment to TMZ and radiotherapy or radiotherapy alone. (A) Survival curves for patients with loss of 9p (thick lines) and without loss of 9p (thin lines). (B) Survival curves for patients with loss of 10q (thick lines) and without loss of 10q (thin lines), determined by CGH. The Kaplan-Meier estimates for overall survival indicate that the patients with losses of 9p and 10q had significantly longer overall survival rates under TMZ treatment.
Table 4.
Univariate Analysis of the TMZ Effect on Overall Survival.
| Alteration | Hazard Ratio CGH and LOH (95% CI) | P Value CGH and LOH (Two-Sided) |
| Loss 9p | 0.55 (0.0064–0.48) | .0085 |
| 0.16 (0.053–0.5) | .0015 | |
| Loss 10q | 0.31 (0.134–0.729) | .0071 |
| 0.63 (0.28–1.43) | .27 | |
Table 5.
Univariate Analysis of Effects of Predictors on Overall Survival.
| Predictor | Hazard (95% CI) | P Value (Two-Sided) |
| TMZ | 0.409 (0.245–0.683) | .00063 |
| Sex | 1.03 (0.589–1.79) | .93 |
| Age | 1.03 (1.00–1.05) | .017 |
| KPS | 0.955 (0.923–0.988) | .0074 |
In the multivariate Cox proportional hazards models, for the genetic subgroups, the main effects of TMZ treatment, gender, KPS, and age, as well as an interaction effect between treatment and age, were estimated. Patients with deletions of 9p showed a positive correlation between TMZ chemotherapy and survival also in multivariate analysis, whereas gender, age, and the interaction of age and treatment had no effect (Table 6). Patients with deletions on 10q and higher age had a poorer prognosis (P = .0076). However, the negative effect of the genetic status and age was compensated by TMZ treatment (P = .0042) (Table 7).
Table 6.
Subgroup Analysis of Differential TMZ, Age, Gender, and KPS Effects Depending on Genetic Aberration with Respect to Survival Assessed by Cox Proportional Hazard Regression.
| Alteration | Variable | Hazard Ratio CGH and LOH (95% CI) | P Value CGH and LOH (Two-Sided) |
| Loss 9p | TMZ | 0.065 (0.0043–0.98) | .048 |
| 0.12 (0.031–0.47) | .0024 | ||
| Age (> 53 years) | 1.63 (0.21–12.76) | .64 | |
| 1.04 (0.33–3.25) | .95 | ||
| Gender | 0.81 (0.23–2.87) | .75 | |
| 0.61 (0.16–2.34) | .47 | ||
| KPS (≥ 90) | 1.32 (0.21–8.30) | .76 | |
| 0.086 (0.017–0.43) | .003 | ||
| Loss 10q | TMZ | 0.32 (0.13–0.80) | .014 |
| 0.74 (0.30–1.85) | .52 | ||
| Age (> 53 years) | 1.20 (0.46–3.17) | .71 | |
| 2.39 (1.00–5.73) | .051 | ||
| Gender | 0.77 (0.21–2.87) | .77 | |
| 0.46 (0.13–1.61) | .22 | ||
| KPS (≥ 90) | 0.41 (0.075–2.28) | .31 | |
| 0.028 (0.0048–0.16) | .000068 | ||
In the multivariate Cox model, all predictors are included and only main effects are estimated.
Table 7.
Subgroup Analysis of Differential TMZ and Age Effects Depending on Genetic Aberration with Respect to Survival Assessed by Cox Proportional Hazard Regression.
| Alteration/Normal | Variable | Hazard Ratio CGH and LOH (95% CI) | P Value CGH and LOH (Two-Sided) |
| Loss 9p | TMZ | 0.037 (0.0017–0.78) | .034 |
| 0.49 (0.054–4.51) | .53 | ||
| Age (> 53 years) | 0.81 (0.081–8.09) | .86 | |
| 4.31 (0.47–39.36) | .20 | ||
| TMZ x age | 3.57 (0.13–99.15) | .45 | |
| (> 53 years) | 0.20 (0.015–2.71) | .23 | |
| Loss 10q | TMZ | 0.85 (0.25–2.91) | .79 |
| 5.64 (0.71–44.57) | .10 | ||
| Age (> 53 years) | 10.47 (1.87–58.65) | .0076 | |
| 33.56 (3.67–307.35) | .0019 | ||
| TMZ x age | 0.040 (0.0044–0.36) | .0042 | |
| (> 53 years) | 0.017 (0.0014–0.21) | .0015 | |
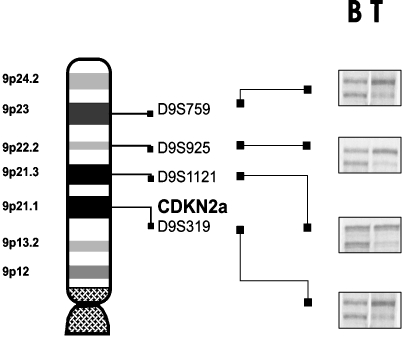
LOH analysis of the CDKN2A region, as well as LOH on chromosome arm 10q and the MGMT region, respectively, produced results similar to that of CGH (Tables 6 and 7; Figures 4 and 5). The only difference was the association of high KPS and better prognosis in patients with losses of 9p and 10q, in contrast to the corresponding CGH data.
Figure 4.
Microsatellite analysis for LOH on chromosome arm 9p.
Figure 5.
Microsatellite analysis for LOH on chromosome 10.
Discussion
Today, the aim of chemotherapy in patients with glioblastomas is palliation and improvement of health-related quality of life (HQL), as well as prevention of neurologic deterioration. Multimodal therapy approaches had limited success in the treatment of patients with malignant glioma [40–42]. The present studies demonstrate that TMZ chemotherapy after prior radiotherapy significantly prolongs survival among patients with newly diagnosed glioblastomas. Their median overall survival time was 19.5 months, compared to 9.3 months of patients with radiotherapy alone. Due to the retrospective nature of our study, patient selection cannot be excluded. However, the outcome of patients with radiotherapy alone in our study compares to the outcome in the literature. These patients had an overall survival of 7.7 and 12.1 months, respectively [43,44], whereas patients with TMZ chemotherapy and radiotherapy had an overall survival of 13.4 to 16 months [23,43,44]. The survival time of our TMZ-treated group is better than that reported in the papers before, but is in line with the study of Hegi et al. [28]. They reported a median survival of 21.7 months for patients treated with radiation and chemotherapy when defining subgroups according to the MGMT promoter methylation status.
Several clinical and histopathologic features are known to be of prognostic significance for survival in patients with glioblastomas. Favorable features include young age and good KPS [2,7,19]. For lymphomas, it was already suggested that increased perioperative and postoperative morbidity and mortality, as well as diminished tolerance to therapy of elderly patients, may be caused by the presence of concomitant diseases [45]. For gliomas, to our knowledge, there are no appropriate results concerning this matter.
In our present study, young age also correlated with better prognosis, but interestingly, the negative impact of age on survival was compensated by the TMZ treatment.
Recent studies suggest that there are genetic subtypes of diffuse gliomas associated with survival time [3,29], and that their clinical course as well as their response to radiation and chemotherapy are primarily determined by the biologic behavior of the tumor cells. Attention has already been drawn to gliomas with LOH on 1p and 19q. These alterations are typical for oligodendroglial tumors [46–49] and have been proposed to be a powerful independent factor for prolonged survival time and favorable response to chemotherapy in WHO grade III anaplastic oligodendrogliomas [35,50]. These genomic alterations are also observed in GBM, however, at lower frequencies, with approximately 10% for LOH 1p and up to 30% for LOH 19q [19,51], but indicating better survival [19].
Uniform numerical and/or structural alterations affecting chromosomes 7, 9, 10, 12, and 13 represent the most common genetic abnormalities in high-grade gliomas [11–13,51] despite genetic heterogeneity [52,53]. Several investigators have noted that LOH on 9p and 10q is associated with shorter survival time in these tumors [3,17–19]. The candidate genes CDKN2A and CDKN2B on the short arm of chromosome 9, and PTEN and DMBT1 on 10q are discussed to influence survival time. LOH around PTEN has been associated with shorter survival time [3,18]; however, mutations of PTEN did not affect prognosis for survival [20,34].
Esteller et al. [25] were able to show that aberrant MGMT promoter hypermethylation was associated with loss of MGMT protein, in contrast to retention of protein in the majority of tumors without aberrant hypermethylation. Recent studies suggested that methylation of the MGMT promoter is predictive for good outcome in patients with glioblastomas treated with alkylating agents [24,26–28]. Our data support the results mentioned above because MGMT is included in the deleted regions on 10q. The results for our control group showed that deletions on chromosome 10 indicate a trend to shorter survival. The same results are observed for LOH of MGMT, whereas LOH of PTEN showed no association with survival as reported before [17,20]. Interestingly, patients with deletion on 10q benefited significantly from adjuvant chemotherapy in univariate analysis.
Therefore, deletion as well as inactivation by hypermethylation of MGMT will predict responses to alkylating chemotherapy, probably in a dose-related manner. Further on, multivariate analysis showed that patients benefited significantly from the therapy especially if they belonged to the older age group (Table 7). Hence, our data suggest that the negative effect of age was compensated by TMZ treatment.
Homozygous deletion of CDKN2A gene mapping on chromosome 9p21 has been reported as an indicator of poor prognosis and resistance to chemotherapy for patients with anaplastic oligodendroglioma. However, homozygous CDKN2A losses were observed exclusively in tumors without LOH on 1p [35]. In glioblastomas with oligodendroglial components, similar results were found [54,55].
An association of tumor necrosis and microvascular proliferation with 9p deletion and CDKN2A alterations was observed in oligodendrogliomas. The higher vascularization of tumors harboring 9p deletion may explain the reasons for better response to chemotherapy [56]. This may also hold true for glioblastomas. However, this effect has never been proven in trials.
To our knowledge, this is the first report demonstrating an effect of 9p deletion on chemotherapy response in glioblastomas. The unfavorable prognostic factor of this alteration [15,17,33,34,57] was confirmed by CGH and LOH analyses in our control group. Further on, the effectivity of TMZ is enhanced by deletion of 9p by CGH and LOH of CDKN2A, respectively, indicating also a crucial role of CDKN2A in chemotherapy response to alkylating agents.
Further, our present data indicate that gains of chromosomes 7 and 12 have no influence on response and survival in TMZ treatment. Therefore, our results suggest that there is no association between frequently amplified regions on these chromosomes (EGFR, CDK4, and MDM2) and TMZ response. Deletions on chromosome arm 13q did not correlate with TMZ chemotherapy either.
In conclusion, we demonstrate a positive effect of TMZ treatment on survival in patients with newly diagnosed glioblastoma. Although deletions on 9p and 10q indicate poorer survival in patients without adjuvant therapy, patients with these molecular alterations benefit from TMZ treatment. This effect was pronounced also in elderly patients with 10q deletion having a very poor prognosis with conventional treatments. Thus, a controlled prospective study should be performed to confirm that TMZ chemotherapy is effective in patients with major factors for poor prognosis, deletions on 9p and 10, and older age.
Acknowledgements
We would like to thank S. Keller, U. Lass, U. Bechtel, U. Lindemann, and D. Fries for expert technical assistance.
Abbreviations
- CGH
comparative genomic hybridization
- GBM
glioblastoma multiforme
- KPS
Karnofsky performance status
- LOH
loss of heterozygosity
- MGMT
methylguanine methyltransferase
- MRI
magnetic resonance imaging
- sGBM
secondary glioblastoma multiforme
- SSC
sodium saline citrate
- ST
survival time
- TMZ
temozolomide
- WHO
World Health Organization
Footnotes
This work was supported by the Hedwig-Stalter Foundation (S. Wemmert), research advancement training program HOMFOR A/2003/25 (R. Ketter), and BMBF grant 01GR0453 (J. Rahnenführer). The work at the Max-Planck Institute for Informatics has been performed in the context of the BioSapiens Network of Excellence (EU contract no. LSHG-CT-2003-503265).
S. Wemmert and R. Ketter contributed equally to this work.
References
- 1.von Deimling A, von Ammon K, Schoenfeld D, Wiestler OD, Seizinger BR, Louis DN. Subsets of glioblastoma multiforme defined by molecular genetic analysis. Brain Pathol. 1993;3:19–26. doi: 10.1111/j.1750-3639.1993.tb00721.x. [DOI] [PubMed] [Google Scholar]
- 2.Kleihues P, Cavenee WK. WHO Classification, Tumors of the Central Nervous System. Lyon, France: IARC Press; 2000. [Google Scholar]
- 3.Tada K, Shiraishi S, Kamiryo T, Nakamura H, Hirano H, Kuratsu JI, Kochi M, Saya H, Ushio Y. Analysis of loss of heterozygosity on chromosome 10 in patients with malignant astrocytic tumors: correlation with patient age and survival. J Neurosurg. 2001;95:651–659. doi: 10.3171/jns.2001.95.4.0651. [DOI] [PubMed] [Google Scholar]
- 4.Stewart LA. Chemotherapy in adult high-grade glioma: a systematic review and meta-analysis of individual patient data from 12 randomised trials. Lancet. 2002;359:1011–1018. doi: 10.1016/s0140-6736(02)08091-1. [DOI] [PubMed] [Google Scholar]
- 5.Devaux BC, O'Fallon JR, Kelly PJ. Resection, biopsy, and survival in malignant glial neoplasms. A retrospective study of clinical parameters, therapy, and outcome. J Neurosurg. 1993;78:767–775. doi: 10.3171/jns.1993.78.5.0767. [DOI] [PubMed] [Google Scholar]
- 6.Albert FK, Forsting M, Sartor K, Adams HP, Kunze S. Early postoperative magnetic resonance imaging after resection of malignant glioma: objective evaluation of residual tumor and its influence on regrowth and prognosis. Neurosurgery. 1994;34:45–60. doi: 10.1097/00006123-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Burger PC, Green SB. Patient age, histologic features, and length of survival in patients with glioblastoma multiforme. Cancer. 1987;59:1617–1625. doi: 10.1002/1097-0142(19870501)59:9<1617::aid-cncr2820590916>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 8.Yung WKA, Albright RE, Olson J, Fredericks R, Fink K, Prados MD, Brada M, Spence A, Hohl RJ, Shapiro W, et al. A phase II study of temozolomide vs procarbazine in patients with glioblastoma multiforme at first relapse. Br J Cancer. 2000;83:588–593. doi: 10.1054/bjoc.2000.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osoba D, Brada M, Yung WKA, Prados M. Health-related quality of life in patients treated with temozolomide versus procarbazine for recurrent glioblastoma multiforme. J Clin Oncol. 2000;18:1481–1491. doi: 10.1200/JCO.2000.18.7.1481. [DOI] [PubMed] [Google Scholar]
- 10.Brada M, Hoang-Xuan K, Rampling R, Dietrich PY, Dirix LY, Macdonald D, Heimans JJ, Zonnenberg BA, Bravo-Marques JM, Henriksson R, et al. Multicenter phase II trial of temozolomide in patients with glioblastoma multiforme at first relapse. Ann Oncol. 2001;12:259–266. doi: 10.1023/a:1008382516636. [DOI] [PubMed] [Google Scholar]
- 11.Bigner SH, Mark J, Burger PC, Mahaley MS, Jr, Bullard DE, Muhlbaier LH, Bigner DD. Specific chromosomal abnormalities in malignant human gliomas. Cancer Res. 1988;48:405–411. [PubMed] [Google Scholar]
- 12.Thiel G, Losanowa T, Kintzel D, Nisch G, Martin H, Vorpahl K, Witkowski R. Karyotypes in 90 human gliomas. Cancer Genet Cytogenet. 1992;58:109–120. doi: 10.1016/0165-4608(92)90095-p. [DOI] [PubMed] [Google Scholar]
- 13.Louis DN, Gusella JF. A tiger behind many doors—multiple genetic pathways to malignant glioma. Trends Genet. 1995;11:412–415. doi: 10.1016/s0168-9525(00)89125-8. [DOI] [PubMed] [Google Scholar]
- 14.Fults D, Brockmeyer D, Tullous MW, Pedone CA, Cawthon RM. p53 mutation and loss of heterozygosity on chromosomes 17 and 10 during human astrocytoma progression. Cancer Res. 1992;52:674–679. [PubMed] [Google Scholar]
- 15.Ueki K, Ono Y, Hensen JW, Efird JT, von Deimling A, Louis DN. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–153. [PubMed] [Google Scholar]
- 16.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 17.Rasheed A, Herndon JE, Stenzel TT, Raetz JG, Kendelhardt J, Friedman HS, Friedman AH, Bigner DD, Bigner SH, McLendon RE. Molecular markers of prognosis in astrocytic tumors. Cancer. 2002;94:2688–2697. doi: 10.1002/cncr.10544. [DOI] [PubMed] [Google Scholar]
- 18.Terada K, Tamiya T, Daido S, Kambara H, Tanaka H, Ono Y, Matsumoto K, Ito S, Ouchida M, Ohmoto T, et al. Prognostic value of loss of heterozygosity around three candidate tumor suppressor genes on chromosome 10q in astrocytomas. J Neuro-Oncol. 2002;58:107–114. doi: 10.1023/a:1016017711033. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt MC, Antweiler S, Urban N, Mueller W, Kuklik A, Meyer-Puttlitz B, Wiestler OD, Louis DN, Fimmers R, von Deimling A. Impact of genotype and morphology on the prognosis of glioblastoma. J Neuropathol Exp Neurol. 2002;61:321–328. doi: 10.1093/jnen/61.4.321. [DOI] [PubMed] [Google Scholar]
- 20.Kraus JA, Glesmann N, Beck M, Krex D, Klockgether T, Schackert G, Schlegel U. Molecular analysis of the PTEN, TP53 and CDKN2A tumor suppressor genes in long-term survivors of glioblastoma multiforme. J Neuro-Oncol. 2000;48:89–94. doi: 10.1023/a:1006402614838. [DOI] [PubMed] [Google Scholar]
- 21.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 22.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002;20:2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 23.Stupp R, Gander M, Leyvraz S, Newlands E. Current and future developments in the use of temozolomide for the treatment of brain tumours. Lancet Oncol. 2001;2:552–560. doi: 10.1016/S1470-2045(01)00489-2. [DOI] [PubMed] [Google Scholar]
- 24.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 25.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 26.Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Guirre-Cruz L, Garcia-Lopez JL, Piquer J, Safont MJ, Balana C, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 27.Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van MG, de TN, Stupp R. Clinical trial substantiates the predictive value of O6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 28.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de TN, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 29.Kunwar S, Mohapatra G, Bollen A, Lamborn KR, Prados M, Feuerstein BG. Genetic subgroups of anaplastic astrocytomas correlate with patient age and survival. Cancer Res. 2001;61:7683–7688. [PubMed] [Google Scholar]
- 30.Wessels PH, Twijnstra A, Kessels AG, Krijne-Kubat B, Theunissen PH, Ummelen MI, Ramaekers FC, Hopman AH. Gain of chromosome 7, as detected by in situ hybridization, strongly correlates with shorter survival in astrocytoma grade 2. Genes Chromosomes Cancer. 2002;33:279–284. doi: 10.1002/gcc.10029. [DOI] [PubMed] [Google Scholar]
- 31.Huncharek M, Kupelnick B. Epidermal growth factor receptor gene amplification as a prognostic marker in glioblastoma multiforme: results of a meta-analysis. Oncol Res. 2000;12:107–112. doi: 10.3727/096504001108747576. [DOI] [PubMed] [Google Scholar]
- 32.Muracciole X, Romain S, Dufour H, Palmari J, Chinot O, Ouafik L, Grisoli F, Branger DF, Martin PM. PAI-1 and EGFR expression in adult glioma tumors: toward a molecular prognostic classification. Int J Radiat Oncol Biol Phys. 2002;52:592–598. doi: 10.1016/s0360-3016(01)02699-2. [DOI] [PubMed] [Google Scholar]
- 33.De Souza NP, Maciel CM, Kawamura MT, Oliveira JA, Teixeira A, Carvalho MG, Alves G. Molecular analysis of CDKN2 (p16) in gliomas associated with clinical data. Oncol Rep. 2001;8:1039–1043. doi: 10.3892/or.8.5.1039. [DOI] [PubMed] [Google Scholar]
- 34.James CD, Galanis E, Frederick L, Kimmel DW, Cunningham JM, Atherton-Skaff PJ, O'Fallon JR, Jenkins RB, Buckner JC, Hunter SB, et al. Tumor suppressor gene alterations in malignant gliomas: histopathological associations and prognostic evaluation. Int J Oncol. 1999;15:547–553. doi: 10.3892/ijo.15.3.547. [DOI] [PubMed] [Google Scholar]
- 35.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 36.Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA-sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 37.Louis DN, von Deimling A, Seizinger BR. A (CA)n dinucleotide repeat assay for evaluating loss of allelic heterozygosity in small and archival human brain tumor specimens. Am J Pathol. 1992;141:777–782. [PMC free article] [PubMed] [Google Scholar]
- 38.Louis DN, Rubio MP, Correa KM, Gusella JF, von Deimling A. Molecular genetics of pediatric brain stem gliomas. Application of PCR techniques to small and archival brain tumor specimens. J Neuropathol Exp Neurol. 1993;52:507–515. doi: 10.1097/00005072-199309000-00009. [DOI] [PubMed] [Google Scholar]
- 39.Budowle B, Koons BW, Errera JD. Multiplex amplification and typing procedure for the loci D1S80 and amelogenin. J Forensic Sci. 1996;41:660–663. [PubMed] [Google Scholar]
- 40.Shapiro WR, Green SB, Burger PC, Mahaley MS, Jr, Selker RG, Van Gilder JC, Robertson JT, Ransohoff J, Mealey J, Jr, Strike TA, et al. Randomized trial of three chemotherapy regimens and two radiotherapy regimens and two radiotherapy regimens in postoperative treatment of malignant glioma. Brain Tumor Cooperative Group Trial 8001. J Neurosurg. 1989;71:1–9. doi: 10.3171/jns.1989.71.1.0001. [DOI] [PubMed] [Google Scholar]
- 41.Selker RG, Shapiro WR, Burger P, Blackwood MS, Arena VC, Gilder JC, Malkin MG, Mealey JJ, Jr, Neal JH, Olson J, et al. The Brain Tumor Cooperative Group NIH Trial 87-01: a randomized comparison of surgery, external radiotherapy, and carmustine versus surgery, interstitial radiotherapy boost, external radiation therapy, and carmustine. Neurosurgery. 2002;51:343–355. [PubMed] [Google Scholar]
- 42.Deutsch M, Green SB, Strike TA, Burger PC, Robertson JT, Selker RG, Shapiro WR, Mealey J, Jr, Ransohoff J, Paoletti P, et al. Results of a randomized trial comparing BCNU plus radiotherapy, streptozotocin plus radiotherapy, BCNU plus hyperfractionated radiotherapy, and BCNU following misonidazole plus radiotherapy in the postoperative treatment of malignant glioma. Int J Radiat Oncol Biol Phys. 1989;16:1389–1396. doi: 10.1016/0360-3016(89)90939-5. [DOI] [PubMed] [Google Scholar]
- 43.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 44.Athanassiou H, Synodinou M, Maragoudakis E, Paraskevaidis M, Verigos C, Misailidou D, Antonadou D, Saris G, Beroukas K, Karageorgis P. Randomized phase II study of temozolomide and radiotherapy compared with radiotherapy alone in newly diagnosed glioblastoma multiforme. J Clin Oncol. 2005;23:2372–2377. doi: 10.1200/JCO.2005.00.331. [DOI] [PubMed] [Google Scholar]
- 45.Corry J, Smith JG, Wirth A, Quong G, Liew KH. Primary central nervous system lymphoma: age and performance status are more important than treatment modality. Int J Radiat Oncol Biol Phys. 1998;41:615–620. doi: 10.1016/s0360-3016(97)00571-3. [DOI] [PubMed] [Google Scholar]
- 46.Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B, Schlegel U, Schramm J, Wiestler OD, Reifenberger G. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–130. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Deimling A, Louis DN, von Ammon K, Petersen I, Wiestler OD, Seizinger BR. Evidence for a tumor suppressor gene on chromosome 19q associated with human astrocytomas, oligodendrogliomas, and mixed gliomas. Cancer Res. 1992;52:4277–4279. [PubMed] [Google Scholar]
- 48.Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 49.Kraus JA, Koopmann J, Kaskel P, Maintz D, Brandner S, Schramm J, Louis DN, Wiestler OD, von Deimling A. Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol. 1995;54:91–95. doi: 10.1097/00005072-199501000-00011. [DOI] [PubMed] [Google Scholar]
- 50.Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 51.von Deimling A, Fimmers R, Schmidt MC, Bender B, Fassbender F, Nagel J, Jahnke R, Kaskel P, Duerr EM, Koopmann J, et al. Comprehensive allelotype and genetic analysis of 466 human nervous system tumors. J Neuropathol Exp Neurol. 2000;59:544–558. doi: 10.1093/jnen/59.6.544. [DOI] [PubMed] [Google Scholar]
- 52.Loeper S, Romeike BFM, Heckmann N, Jung V, Henn W, Feiden W, Zang KD, Urbschat S. Frequent mitotic errors in tumor cells of genetically micro-heterogeneous glioblastomas. Cytogenet Cell Genet. 2001;94:1–8. doi: 10.1159/000048773. [DOI] [PubMed] [Google Scholar]
- 53.Jung V, Romeike BF, Henn W, Feiden W, Moringlane JR, Zang KD, Urbschat S. Evidence of focal genetic microheterogeneity in glioblastoma multiforme by area-specific CGH on microdissected tumor cells. J Neuropathol Exp Neurol. 1999;58:993–999. doi: 10.1097/00005072-199909000-00009. [DOI] [PubMed] [Google Scholar]
- 54.Kraus JA, Lamszus K, Glesmann N, Beck M, Wolter M, Sabel M, Krex D, Klockgether T, Reifenberger G, Schlegel U. Molecular genetic alterations in glioblastomas with oligodendroglial component. Acta Neuropathol (Berl) 2001;101:311–320. doi: 10.1007/s004010000258. [DOI] [PubMed] [Google Scholar]
- 55.Fuller CE, Schmidt RE, Roth KA, Burger PC, Scheithauer BW, Banerjee R, Trinkaus K, Lytle R, Perry A. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J Neuropathol Exp Neurol. 2003;62:1118–1128. doi: 10.1093/jnen/62.11.1118. [DOI] [PubMed] [Google Scholar]
- 56.Godfraind C, Rousseau E, Ruchoux MM, Scaravilli F, Vikkula M. Tumour necrosis and microvascular proliferation are associated with 9p deletion and CDKN2A alterations in 1p/19q-deleted oligodendrogliomas. Neuropathol Appl Neurobiol. 2003;29:462–471. doi: 10.1046/j.1365-2990.2003.00484.x. [DOI] [PubMed] [Google Scholar]
- 57.Puduvalli VK, Kyritsis AP, Hess KR, Bondy ML, Fuller GN, Kouraklis GP, Levin VA, Bruner JM. Patterns of expression of Rb and p16 in astrocytic gliomas, and correlation with survival. Int J Oncol. 2000;17:963–969. doi: 10.3892/ijo.17.5.963. [DOI] [PubMed] [Google Scholar]