Abstract
Early after infection, the retroviral RNA genome is reverse transcribed to generate a linear cDNA copy, then that copy is integrated into a chromosome of the host cell. We report that unintegrated viral cDNA is a substrate for the host cell non-homologous DNA end joining (NHEJ) pathway, which normally repairs cellular double-strand breaks by end ligation. NHEJ activity was found to be required for an end-ligation reaction that circularizes a portion of the unintegrated viral cDNA in infected cells. Consistent with this, the NHEJ proteins Ku70 and Ku80 were found to be bound to purified retroviral replication intermediates. Cells defective in NHEJ are known to undergo apoptosis in response to retroviral infection, a response that we show requires reverse transcription to form the cDNA genome but not subsequent integration. We propose that the double-strand ends present in unintegrated cDNA promote apoptosis, as is known to be the case for chromosomal double-strand breaks, and cDNA circularization removes the pro-apoptotic signal.
Keywords: DNA-PK/HIV/integrase/Ku/ligase IV
Introduction
Early steps of retroviral infection involve reverse transcription of the viral RNA to yield a linear cDNA copy, and integration of that cDNA into a chromosome of the host cell (Coffin et al., 1997; Hansen et al., 1998). The completion of reverse transcription produces a pre-integration complex (PIC), which contains the viral cDNA, the viral-encoded integrase protein and other components organized to carry out the integration reaction (Brown et al., 1987; Bowerman et al., 1989; Farnet and Haseltine, 1990; Gallay et al., 1995; Farnet and Bushman, 1997; Miller et al., 1997). Here we report that proteins of the cellular non-homologous DNA end joining (NHEJ) pathway also bind to PICs and modulate the course of infection.
The NHEJ pathway comprises the major double-strand break repair pathway detected during G0, G1 and early S phases of the cell cycle (Lieber, 1998; Smith and Jackson, 1999). Ku and the 469 kDa DNA-PKcs appear to be the initial components that bind to the DNA ends. Ku is a heterodimer of two subunits, Ku70 and Ku86 (also often called Ku80). The two DNA ends are typically modified by nucleases and polymerases in order to make them compatible for ligation. The final ligation of the double-strand break is carried out specifically by the XRCC4–DNA ligase IV complex. Cells defective for one of the subunits of Ku, DNA-PK, XRCC4 or DNA ligase IV are sensitive to ionizing radiation. Moreover, lymphocytes from animals with these NHEJ pathway mutations are able to carry out the initial double-strand cleavage steps of V(D)J recombination, but are unable to rejoin the resulting double-strand breaks (Pergola et al., 1993; Smider et al., 1994; Taccioli et al., 1993, 1994; Grawunder et al., 1998).
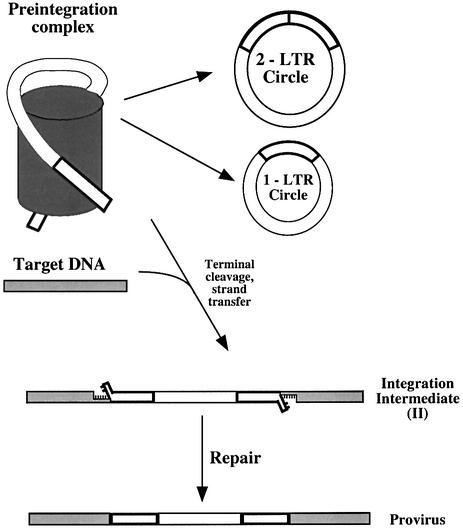
DNA double-strand ends are also generated by retroviral reverse transcription, which produces a linear cDNA product (Figure 1). Prior to integration, two nucleotides are removed from each cDNA 3′ end (Fujiwara and Mizuuchi, 1988; Brown et al., 1989; Roth et al., 1989). The recessed 3′ ends are then joined to protruding 5′ DNA ends in target DNA. Melting of the target DNA between the points of joining yields an integration intermediate with gaps at each host–virus DNA junction and overhanging dinucleotides derived from the 5′ cDNA ends (the integration intermediate is marked ‘II’ in Figure 1 and subsequent figures). These gaps are then filled in, the branching 5′ ends removed and the 5′ viral DNA strands joined. The terminal cleavage and first strand transfer steps can be modeled in vitro with purified integrase (Katzman et al., 1989; Bushman et al., 1990; Craigie et al., 1990; Katz et al., 1990; Sherman and Fyfe, 1990) and the gap-repair step modeled with purified DNA gap-repair proteins (Yoder and Bushman, 2000).
Fig. 1. Retroviral cDNA forms produced after infection. Retroviral genomes contained in preintegration complexes (PICs) can be integrated to generate an integration intermediate (II) which is then repaired to yield a provirus. Alternatively, the cDNA can be circularized to form 2-LTR or 1-LTR circles.
The retroviral cDNA can also be circularized rather than integrated (Figure 1). In one pathway, circularization takes place by ligation of the long terminal repeat (LTR) sequences at the cDNA ends, yielding a 2-LTR circle. Another pathway involves homologous recombination between LTRs, yielding a circle with a single LTR. A third pathway involves integration of the viral cDNA into itself, yielding internally rearranged circular forms. Circularization is believed to represent an unproductive pathway leading to loss of the viral genome (Coffin et al., 1997).
In earlier work, Daniel et al. (1999) reported that infections of cells mutant in components of the NHEJ pathway yielded reduced viral titers due to increased apoptosis of infected cells. This finding engendered considerable interest, in part because human immunodeficiency virus (HIV) infection is believed to kill cells in patients at least partly by an apoptotic mechanism (Corbeil and Richman, 1995). Daniel et al. (1999) reported that a virus mutant in integrase did not induce apoptosis, suggesting that the NHEJ activity might be required after the action of integrase to repair DNA gaps in integration intermediates. However, the NHEJ pathway proteins are not known to repair single-stranded gaps, only double-strand breaks (Smith and Jackson, 1999). Moreover, a recent paper by Baekelandt et al. (2000) reported that apoptosis was only observed after infections with high viral titers, leading them to conclude that the NHEJ pathway could not be required for gap repair after integration. These findings focus interest on other possible origins of the pro-apoptotic signal (Coffin and Rosenberg, 1999).
We investigated the role of the NHEJ pathway in retroviral replication, leading to a new proposal for its function. We found that the 2-LTR circular form of the viral cDNA was absent in infected cells mutant in the NHEJ pathway. This implicated the NHEJ pathway in cDNA circularization and focused attention on the role of unintegrated cDNA forms in inducing apoptosis. We also investigated whether NHEJ proteins could be found in PICs isolated from infected cells and found that the Ku heterodimer was indeed associated. The Ku proteins are not important for the cDNA integration reaction itself, since PICs isolated from wild-type cells or cells mutant in Ku80 were equally active for integration in vitro.
As in previous studies, we found that retroviral infection of cells mutant in the NHEJ pathway was cytotoxic, particularly in high titer infections. Assays of caspase activity confirmed that cell death was by apoptosis. However, studies of an integrase-mutant virus revealed that it too was toxic, at odds with previous work, supporting a role for unintegrated viral cDNA in inducing apoptosis. Studies with the reverse transcriptase inhibitor nevirapine revealed that synthesis of the viral cDNA was nevertheless required for the pro-apoptotic signal. We propose that double-strand ends in the viral cDNA provide the pro-apoptotic signal, since they (i) are the normal substrate of the NHEJ pathway; (ii) are removed by cDNA circularization; and (iii) are known to induce apoptosis (Huang et al., 1996; Smith and Jackson, 1999). These findings extend an observation made by Howard Temin 20 years ago that the amount of unintegrated viral cDNA in infected cells correlates with the cytopathicity of retroviral infection (Temin et al., 1980).
Results
NHEJ activity is required for formation of 2-LTR circles
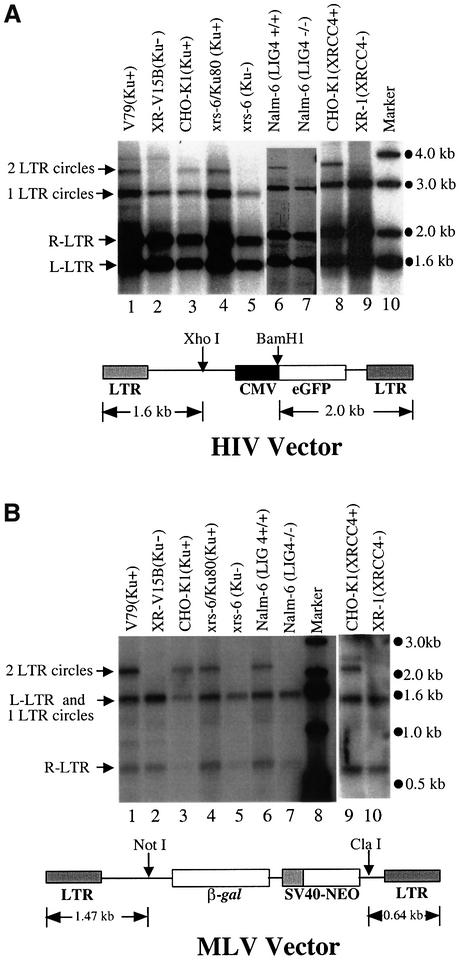
Since NHEJ proteins normally ligate double-strand DNA ends, we asked whether the production of circular viral cDNA forms was affected in cells lacking NHEJ activity. Cell lines mutant in Ku (xrs6 and XR-V15B), ligase IV (Nalm-6 LIG4–/–) and XRCC4 (XR-1) were studied together with wild-type controls. Cells were infected with HIV- or Moloney murine leukemia virus (MLV)-based vectors bearing the vesicular stomatitis virus (VSV)-G envelope, which permitted viral entry into the cell types studied. Extrachromosomal DNA fractions were prepared 24 h after infection. DNAs were cleaved with restriction enzymes that cut twice in the viral cDNA, thereby yielding discrete length fragments for the 2-LTR circle, the 1-LTR circle and the unintegrated linear DNA. Samples were analyzed by Southern blotting (Figure 2A and B).
Fig. 2. The NHEJ system is required for formation of 2-LTR circles. (A) Southern blot analysis of extrachromosomal HIV cDNA forms. (B) Southern blot analysis of extrachromosomal MLV cDNA forms. Unintegrated cDNA was isolated from the cell lines indicated above the autoradiograms after high titer infection (m.o.i. 5) using the Hirt procedure (Arad, 1998). Expected mobilities of DNAs from the unintegrated DNA forms are as indicated beside the gel (L-LTR is the left half of the linear genome, R-LTR is the right half). For the HIV-1 vector, the viral cDNA molecules were digested with XhoI and BamHI. For the MLV vector, the viral cDNAs were digested with NotI and ClaI. A 32P-labeled DNA matching each cognate LTR was used as probe.
The DNA fragment derived from 2-LTR circles was undetectable in the NHEJ-mutant cell lines but prominent in the matched wild-type controls for both HIV (Figure 2A) and MLV (Figure 2B). A derivative of the xrs6 cell line containing a wild-type copy of the human Ku80 gene also showed 2-LTR circle formation, indicating that the mutation in the Ku gene was responsible for the defect (Figure 2A and B, lanes 4). These data establish that the NHEJ pathway is required for formation of 2-LTR circles in the cells studied.
The abundance of 1-LTR circles was not affected, indicating that the NHEJ pathway is not involved in formation of this species. The 1-LTR circles can be formed either by recombination between LTRs or as stalled reverse transcription intermediates. It is not clear whether formation of 1-LTR circles provides another means for sequestering double-strand ends in the viral cDNA.
Ku is associated with PICs
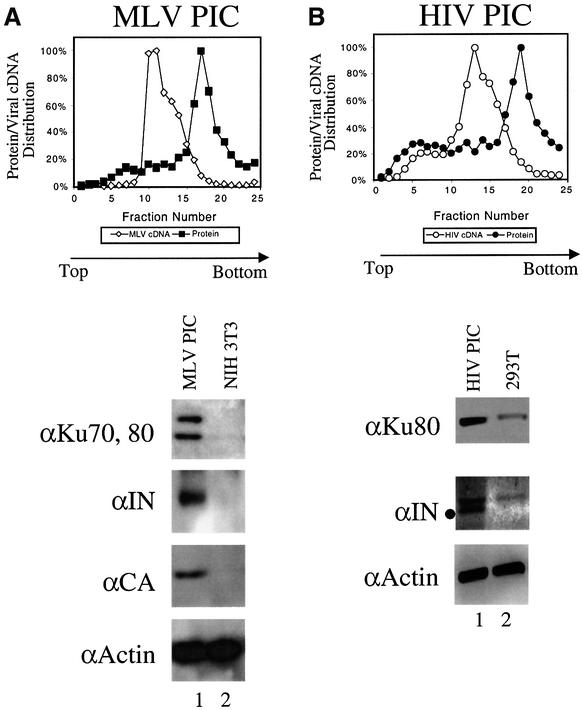
We next asked whether NHEJ proteins bound to retroviral PICs isolated from infected cells. PICs contain the viral cDNA, integrase, and other proteins organized in integration-competent particles (Brown et al., 1987; Ellison et al., 1990; Farnet and Haseltine, 1990). Cells were infected either with MLV- or HIV-based vectors, and PICs were harvested after the completion of reverse transcription. Cytoplasmic extracts from infected or mock-infected cells were purified by gel filtration, precipitation in the presence of lowered ionic strength, and Nycodenz density gradient fractionation. PIC proteins were then concentrated and analyzed by western blotting. Figure 3A shows that Ku70 and Ku80 were detected in the MLV PIC peak fraction but not control fractions from mock-infected cells. Analysis of protein composition across gradient fractions indicated that the peak of Ku protein corresponded to the peak of cDNA (data not shown). The viral integrase and capsid proteins were also detectable in the peak cDNA fractions. Similar amounts of actin were detectable in both the experimental and control fractions, documenting similar recovery of bulk protein. Studies of HIV PICs were complicated by the fact that human cells have much higher levels of Ku protein than do murine cells (Smith and Jackson, 1999), and with available methods it has not yet been possible to separate free Ku completely from HIV PICs. Figure 3B shows that Ku80 is enriched in fractions from the HIV cDNA peak, but lesser amounts of Ku80 are also present in the uninfected cell control. HIV integrase was also detected (lower band in the panel marked ‘anti-IN’).
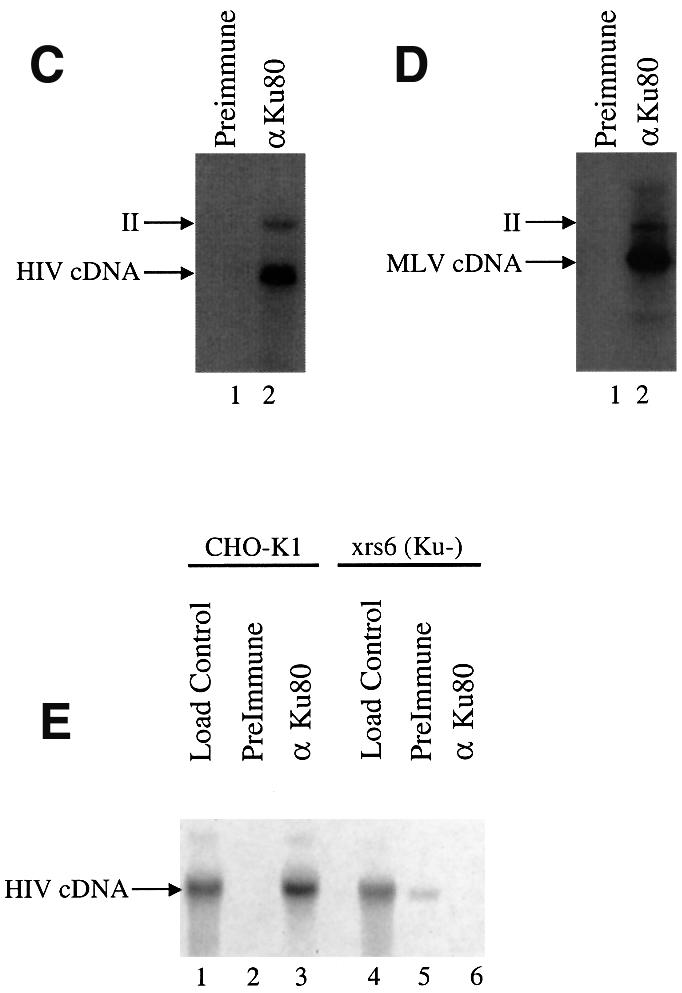
Fig. 3. Ku is associated with PICs. (A) Western blot analysis of proteins in purified MLV PIC fractions. Top: Nycodenz gradient analysis of viral genomes and total protein. Parallel fractions from mock-infected NIH 3T3 cells prepared identically are analyzed in the right lane in each panel. The anti-Ku70 and 80 lanes were probed with antibodies against both proteins. (B) Western blot analysis of proteins in the HIV PIC peak fractions. For the integrase blot the dot indicates HIV-integrase (the upper band is present in the control and represents a contaminating cellular protein). Only Ku80 was analyzed. (C) Immunoprecipitation (IP) of HIV vector PICs. (D) IP of MLV PICs. Lane 1, pre-immune serum; lane 2, anti-Ku80 antibody. PICs captured on protein A–agarose beads were exposed to target DNA, then the cDNA products were analyzed on Southern blots probed with the cognate LTR sequences labeled with 32P. ‘II’ indicates the integration intermediate generated by PIC integration, ‘cDNA’ indicates the unintegrated linear form. (E) Lack of IP of PICs with anti-Ku80 antibody from cells mutant in Ku80. PICs were prepared from CHO-K1 (lanes 1–3) or xrs6 cells (Ku-mutant; lanes 4–6). Load control, initial input PIC fraction; pre-immune, control IP with pre-immune serum; anti-Ku80, IP with anti-Ku80 antibody.
Binding of Ku to PICs of MLV and HIV was also documented by immunoprecipitation (IP; Figure 3C and D). Cytoplasmic extracts containing PICs were mixed with anti-Ku80 antibodies, then complexes were captured by addition of protein A–agarose beads. To test whether integration-competent complexes were present, the captured PICs were treated with target DNA while still bound to the beads. After incubation, reactions were deproteinized and cDNAs were visualized using Southern blots probed with 32P-labeled LTR sequences. IP with anti-Ku antibody resulted in the recovery of ∼30% of the total input cDNA (Figure 3C and D, lanes 2). A control pre-immune antibody displayed no detectable captured product (Figure 3C and D, lanes 1). Integration products were clearly detectable in the captured DNA species. The integration product is marked ‘II’ (integration intermediate), since the products of reactions in vitro are known to contain unrepaired DNA gaps (Fujiwara and Mizuuchi, 1988; Brown et al., 1989; Miller et al., 1995).
As a control, we repeated the IP experiment with PICs isolated from hamster cells mutant in Ku (Figure 3E). Parent CHO-K1 cells or Ku80-mutant xrs6 cells were infected with VSV-G pseudo-types of HIV-based vectors. PIC extracts were prepared as above and analyzed by IP with the anti-Ku80 antibody. PICs from CHO-K1 cells could be captured by IP (Figure 3E, lanes 1–3) but those from the Ku80-mutant cells could not (Figure 3E, lanes 4–6). These control experiments indicate that capture of PICs by IP with the anti-Ku80 antibody requires the presence of Ku80 in cells, documenting the specificity of the method. In summary, we conclude that Ku is associated with PICs of HIV and MLV.
Ku is not required for terminal cleavage or strand transfer
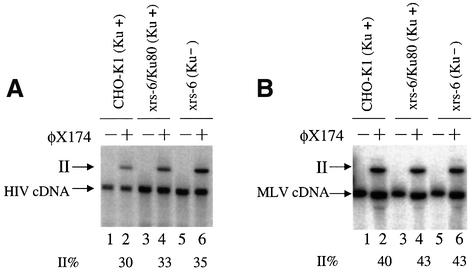
One possible function of Ku in PICs would be direct stimulation of the integration reaction itself. To test whether Ku was important for integration, PICs of HIV and MLV were produced in cells lacking Ku and the activity compared with PICs from wild-type cells (Figure 4A and B). For this study, hamster cells mutant in the Ku80 gene (xrs6) were compared with wild-type hamster cells (CHO-K1) or mutant cells complemented with an intact gene for Ku80 (xrs6/Ku). Each cell line was infected with MLV- or HIV-based vectors bearing the VSV-G envelope. PIC extracts were prepared and partially purified by gel filtration. PIC fractions were incubated with target DNA and the extent of integration compared on Southern blots. Reactions containing PICs from the mutant and wild-type cell lines yielded similar amounts of integration products, ranging from 30 to 35% for HIV (Figure 4A) and 40–43% for MLV (Figure 4B). These data indicate that Ku is not important for the terminal cleavage and first strand transfer reactions carried out by PICs.
Fig. 4. Lack of requirement for Ku80 during integration by PICs in vitro. The cell lines indicated above the autoradiograms were infected with HIV vectors (A) or MLV vectors (B) and PICs isolated. The PICs were mixed in vitro with target DNA (lanes 2, 4 and 6) or not (lanes 1, 3 and 5) and formation of the integration intermediate (labeled ‘II’, see Figure 1) quantitated by PhosphorImager.
Infection of cells defective in NHEJ is cytotoxic
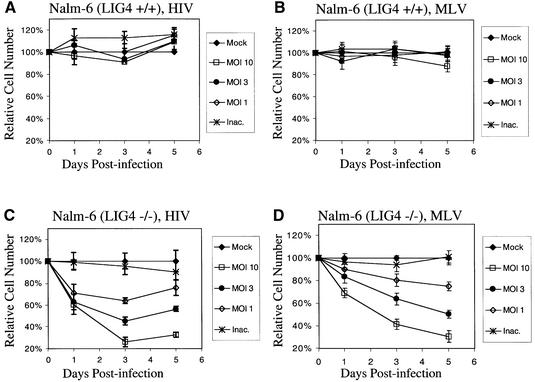
We next investigated the effect of the NHEJ pathway on the outcome of retroviral infection (Daniel et al., 1999; Baekelandt et al., 2000). Nalm-6 LIG4–/– cells have a particularly strong defect in NHEJ (Grawunder et al., 1998), and we found that Nalm-6 LIG4–/– displayed a robust cytotoxic response to infection, so we used this line as a model in subsequent studies. A cytotoxic response to infection was also seen with xrs6 (Ku-mutant) cells, although the extent of cell killing was less than for Nalm-6 LIG4–/– (data not shown).
Infection of Nalm-6 wild-type cells with the HIV (Figure 5A) or MLV vectors (Figure 5B) revealed little cytotoxicity. In contrast, infection of Nalm-6 LIG4–/– resulted in prominent cytotoxicity after infection with either virus (Figure 5C and D). Infection at a multiplicity of infection (m.o.i.) of 10 resulted in >70% reduction in cell number. In some very high titer infections (m.o.i. ∼30) some cytotoxicity was also seen for the Nalm-6 wild type, but always much less pronounced than in infections of Nalm-6 LIG4–/– (data not shown).
Fig. 5. Toxicity of retroviral infection in NHEJ-mutant (Nalm-6 LIG4–/–) cells. (A) Infection of Nalm-6 with the HIV-based vector. The m.o.i. of 10 corresponds to 60 ng of p24 capsid protein per 5 × 105 cells. (B) Infection of Nalm-6 with the MLV vector. (C) Infection of Nalm-6 LIG4–/– with the HIV vector. (D) Infection of Nalm-6 LIG4–/– with the MLV vector. Viable cell numbers were normalized to the mock-infected control. For the cytotoxicity experiments, an HIV vector containing the central polypurine tract was used to boost the titer (Follenzi et al., 2000). M.o.i. values reflect titers (gfp-transducing units per ml) on 293T cells. Each point represents the average of three separate cultures.
Induction of apoptosis requires reverse transcription but not integration
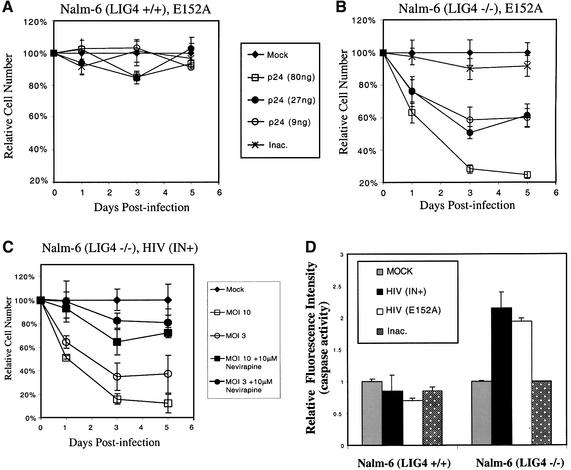
We next re-investigated the origin of the pro-apoptotic signal, since our data highlighted effects of NHEJ on unintegrated cDNA. To test the importance of integration, we carried out infections with HIV vector particles containing the well studied E152A substitution in the integrase active site (Drelich et al., 1992; Shin et al., 1994; Wiskerchen and Muesing, 1995; Engelman, 1999). HIV viruses containing the E152A integrase are capable of normal reverse transcription but are completely blocked for integration (Shin et al., 1994; Wiskerchen and Muesing, 1995). Purified integrase E152A is incapable of terminal cleavage and strand transfer in vitro (Drelich et al., 1992). Infections with the HIV E152A virus thus allow the consequences of cDNA synthesis in the absence of integration to be assessed.
We observed that infection with the HIV (E152A) integrase-minus vector was cytotoxic in Nalm-6 LIG4–/– cells (Figure 6A and B). Little cytotoxicity was seen in infections with media only (mock) or heat-inactivated virus. Infections of the Nalm-6 parent line similarly showed little cytotoxicity.
Fig. 6. Apoptosis of NHEJ-mutant cells (Nalm-6 LIG4–/–) after infection requires viral reverse transcription but not integration. (A) Infection of Nalm-6 cells with the HIV (E152A) vector. (B) Infection of Nalm-6 LIG4–/– with the HIV (E152A) vector. (C) Addition of 10 µM nevirapine inhibits the toxic effect of HIV (IN+) infection in Nalm-6 LIG4–/– cells. (D) Assay of caspase 3 activation after infection. Caspase 3 enzyme was captured by an immobilized anti-caspase 3 antibody, then a fluorogenic caspase 3 substrate added. Fluorescence intensity measured after incubation was normalized to the mock-infected control. Infections were initiated by adding 60 ng of p24 capsid antigen per 5 × 105 cells.
We next asked whether synthesis of the viral cDNA was required for the cytotoxic response. Addition of 10 µM nevirapine, which inhibits HIV-1 reverse transcription, to infections of Nalm-6 LIG4–/– with the HIV vector substantially reduced the toxic effect (Figure 6C). Assays by fluorescence-monitored PCR documented that addition of nevirapine at this concentration reduced cDNA synthesis at least 1000-fold (data not shown). This indicates that cDNA synthesis but not integration is required to generate the toxic signal.
We further compared the pro-apoptotic effects of the HIV vector or IN (E152A) by assaying activation of the pro-apoptotic protease caspase 3 (Figure 6D). Infection with HIV (IN+) or HIV (E152A) induced caspase 3 activity in the Nalm-6 LIG4–/– cells but not Nalm-6. Heat-inactivated virus did not induce caspase 3. These data indicate that the observed toxic effect involves induction of apoptosis.
The findings reported here for the integrase-mutant virus differed from those of Daniel et al. (1999). For this reason the phenotype of the E152A-mutant HIV vector was confirmed by detailed studies in the Nalm-6 cells (Figure 7). Figure 7A shows that infection of cells with an HIV vector transducing gfp yielded abundant fluorescent centers, while infection with HIV (E152A)-gfp yielded no infected centers. Analysis using fluorescence-monitored quantitative PCR (Butler et al., 2001) showed that reverse transcription took place efficiently after infection with both the E152A-mutant and integrase-wild-type particles, but integration was detected only in infections with integrase-wild-type particles (Figure 7B). Southern blot analysis also confirmed that abundant reverse transcription product was formed by both vectors 12 h after infection (Figure 7C, lanes 1 and 2) but integrated genomes were detected after 14 days of growth only in the integrase-wild-type culture (Figure 7C, lanes 3 and 4). These data confirm that the E152A-integrase-mutant particles were competent for reverse transcription but blocked for integration.
Fig. 7. Characterization of the integrase-mutant HIV (E152A)-gfp vector. (A) Fields of Nalm-6 cells infected with the HIV-gfp vector or HIV (E152A)-gfp vector analyzed 14 days after infection. Left column, phase contrast image of infected cells. Right column, fluorescent image of the same fields. (B) Quantitative PCR analysis of reverse transcription and integration by HIV (IN+) and HIV (E152A) in Nalm-6 cells. Filled symbols (‘RT’), quantitation of HIV genomes; open symbols (‘provirus’), quantitation of integrated proviruses by Alu-PCR. The approach to quantitating results of Alu-PCR is described in Butler et al. (2001). Circles, HIV (IN+); squares, HIV (E152A). (C) Southern blot analysis of DNA from Nalm-6 cells infected with the HIV-gfp vector or the HIV (E152A)-gfp vector. Unintegrated (Hirt) cDNA (lanes 1 and 2) was isolated 12 h after infection and analyzed by Southern blotting. Genomic DNA (lanes 3 and 4) was isolated 14 days after infection and cleaved with BamHI and EcoRI, which each cut once in the vector DNA. The blots were probed with a 32P-labeled sequence matching the internal vector sequences. Infections were initiated by adding 60 ng of p24 capsid antigen per 5 × 105 cells.
Discussion
We have investigated the role of the host cell NHEJ system in modulating the outcome of retroviral infection. We present data indicating that: (i) the Ku component of the NHEJ system binds to PICs; (ii) the NHEJ system is responsible for circularizing a portion of the viral cDNA to make 2-LTR circles; (iii) infection of cells mutant in the NHEJ system leads to apoptosis of infected cells [paralleling previous studies (Daniel et al., 1999; Baekelandt et al., 2000)]; and (iv) the unintegrated cDNA provides the pro-apoptotic signal. The latter observation echoes studies from Howard Temin and coworkers, who found that the amount of unintegrated retroviral cDNA in infected cells correlated with the extent of the cytopathic effect observed after infection (Temin et al., 1980).
Models to explain the role of NHEJ in suppressing apoptosis can be divided between those invoking a direct role for 2-LTR circle formation in suppressing apoptosis and those invoking an indirect effect. At present we favor the view that abnormal persistence of cDNA ends in NHEJ-mutant cells promotes apoptosis, and that circularization suppresses apoptosis directly by removing the free cDNA ends. Double-strand DNA breaks are known to be highly pro-apoptotic, with as little as one DNA double-strand break sufficient to arrest the cell cycle in G1 (Huang et al., 1996). Circular DNAs, in contrast, do not have this effect. This model also helps to explain the requirement for high titer infection for the cytopathic effect (Baekelandt et al., 2000), since increased m.o.i. results in increased accumulation of cDNA ends. The variable penetrance of the apoptotic effect is also accommodated by this model, since different cell types are known to have different predispositions to undergo apoptosis in response to double-strand breaks.
Models invoking indirect mechanisms are not, however, ruled out. For example, circularization of the viral cDNA may only be a marker for interaction of PICs with the NHEJ system, and not itself the mechanism that suppresses apoptosis. According to this idea, the unintegrated cDNA may be recognized by the NHEJ system, initiating signaling that a double-strand break is present, but circularization of the viral DNA is just a side product of this interaction. Consistent with this, the linear form of the viral cDNA is always more abundant in cells than the 2-LTR circles, calling into question the importance of circularizing a minor fraction of the total viral DNA.
To preserve the idea that circularization acts directly, one needs to propose that those cDNAs that form 2-LTR circles are especially active in initiating the pro-apoptotic signal. For example, the cDNAs that become circularized may be components of partially disassembled PICs in which the DNA ends are exposed and so particularly pro-apoptotic. According to this idea, the PIC is organized to facilitate circularization of the ends upon partial disassembly, potentially preserving the host cell and allowing replication of sibling viruses.
Many studies have implicated apoptosis as important in HIV pathogenesis (reviewed in Corbeil and Richman, 1995). Infection in vivo will rarely take place in cells lacking the NHEJ pathway, but we find that infection also promotes apoptosis in some wild-type cells, albeit at higher titer and with reduced efficiency compared with mutant cells. The effective titer of HIV in vivo has been proposed to be high at least in some tissues (Coffin et al., 1997), consistent with a potential role for induction of apoptosis by viral DNA in vivo.
Binding of Ku to PICs may also affect nuclear localization. HIV infects non-dividing cells, implying that integration complexes must cross the nuclear membrane. Extensive analysis of the mechanism of nuclear localization has yielded complicated and controversial results (Fouchier and Malim, 1999). Ku is reported to have a nuclear localization signal (Koike et al., 1999), thus providing a new candidate determinant for nuclear localization of PICs. Ku would be expected to bind to the ends of the linear cDNA, but the internally branched cDNA at the HIV central polypurine tract (cPPT) may also provide a high affinity site (Tuteja et al., 1994). The cPPT has been proposed to promote nuclear localization of HIV PICs (Follenzi et al., 2000; Zennou et al., 2000), suggesting a possible role for Ku at the cPPT.
It is unclear whether there is a role for the NHEJ proteins in repair after integration, as suggested by Daniel et al. (1999). Data presented here highlight the importance of unintegrated cDNA in generating the pro-apoptotic signal but do not rule out a contribution after integration as well.
Several recombination systems in addition to retroviral integrases are known to recruit the NHEJ pathway and thereby suppress toxicity of DNA double-strand breaks. Several of these are known or suspected to be members of the DDE superfamily of recombinases (Fayet et al., 1990; Rowland and Dyke, 1990; Kulkosky et al., 1992; Doak et al., 1994; Landree et al., 1999; Fugmann et al., 2000), which includes retroviral integrases, the recombination associated gene (RAG)-1 recombinase and the transposase of the Drosophila P-element transposon. Like retroviral infection, RAG recombination and P-element excision generate DNA double-strand ends. In the absence of the NHEJ system, these reactions are toxic to cells (Beall and Rio, 1996; Smith and Jackson, 1999). For P-elements and now for retroviruses, Ku is known to bind to recombination complexes, potentially positioning the NHEJ system to ligate double-strand DNA breaks. Ku has also been found to bind to integration complexes of the yeast retrotransposon Ty1, although it is not clear whether Ku ligates toxic double-strand breaks or serves some other purpose (Downs and Jackson, 1999). The NHEJ pathway has also been implicated in immunoglobulin heavy-chain switching (Rolnik et al., 1996; Casellas et al., 1998; Manis et al., 1998), which may also involve DNA double-strand breaks (Wuerffel et al., 1997). These parallels suggest that there may be more similarities in detailed conformation and function of these recombinase–NHEJ protein complexes than was previously suspected.
Materials and methods
Materials
NIH 3T3, 293T, MoMLV-K producer cells (Li et al., 1998), xrs6, xrs6/Ku, XR-V15B and XR-1 (Zdzienicka, 1995) were cultured in Dulbecco’s modified Eagle’s medium with high glucose (BioWhittaker) plus 10% heat-inactivated fetal calf serum (FCS) (Biowhittaker) and 1× Pen/strep (Gibco-BRL). V79 was maintained in the same media with 5% FCS. CHO-K1 cells were grown in F-12K medium with 10% FCS. The HIV-1 vector producer line SODKICG2 was cultured essentially as described (Hansen et al., 1999). Nalm-6 and Nalm-6 LIG4–/– cells (Grawunder et al., 1998) were grown in RPMI with 10% FCS, 10 mM HEPES, 55 µM β-mercaptoethanol, non-essential amino acids, sodium pyruvate, penicillin, streptomycin and l-glutamine (included at the manufacturer’s recommended concentrations).
Antibodies against Ku were obtained from Serotec, against actin from Boehringer Mannheim Biochemicals, against MLV CA from Quality Biotech, and secondary antibodies were obtained from Jackson ImmunoResearch Laboratories Inc. Antibodies against MLV integrase and HIV integrase were raised in-house in White New Zealand Rabbits.
Production of vectors derived from MLV and HIV-1
Large-scale production of VSV-G pseudotyped HIV-1-based vector (used in the experiments in Figures 2–4), a derivative of the SODKICG2-SM2-inducible producer cell line that yields vector particles transducing gfp (Hansen et al., 1999), was carried out using a Corning Cell Cube apparatus, which permitted feed-back control of pH, dissolved oxygen and CO2. Glucose consumption was measured manually and adjusted during the run. Cells were expanded, then introduced into the Cell Cube and cultured for 3 days. Expression of the HIV vector was induced upon introduction of cells into the Cell Cube by removal of doxycycline (Sigma). Culturing under these conditions yielded 3 l of vector stock at a titer of 6 × 106 gfp-transducing particles per ml.
For the experiments in Figures 5–7, an HIV vector containing the cPPT (p156RRLsinPPTCMVGFPPRE) was used (Follenzi et al., 2000) in order to boost the transduction efficiency. To make the integrase-deficient vector, a DNA fragment encoding the E152A mutant in the HXB2 background (Shin et al., 1994) was transferred to the gag-pol expression plasmid delta-R9 (Naldini et al., 1996) as a BclI–SalI restriction fragment. As a control, the wild-type HXB2 fragment was transferred in parallel. HIV-based vector supernatants were then prepared by three-plasmid cotransfection into 293T cells with pVSVG, the modified gag-pol constructs, and p156RRLsinPPTCMVGFPPRE.
For the generation of VSV-G pseudotyped MLV vector, three plasmids, pVSVG (Stratagene), pCgp (encoding MLV gag-pol) and pCnBg (Han et al., 1997), were co-transfected into 293T cells using the calcium phosphate method (experiments in Figures 2, 4 and 5). Sixteen hours later, the media was replaced with media containing 10 mM sodium butyrate. After 12 h, the media was then replaced with fresh media, and the vector supernatant was harvested 16–24 h later, yielding vector particles with a titer of ∼2 × 106 infectious units/ml.
Production of PICs and analysis of their activity
MLV-K PICs (wild-type MLV, used in the experiments in Figure 3) were prepared as described (Brown et al., 1987; Li et al., 1998). PICs derived from VSV-G pseudotyped MLV-based vectors were generated by cell-free infections (Figure 4). Cells (3 × 106 CHO-K1, XRS or XRS/Ku) were plated in 10 cm dishes (Falcon) 12–16 h prior to transduction. Eight milliliters of vector supernatant (m.o.i. ∼5) were used to transduce these cells in the presence of 10 µg/ml DEAE–dextran. Two hours after the initial transduction, the vector supernatant were removed and replaced with fresh culture media. Eight hours after transduction, cells were trypsinized and PICs were harvested as described (Brown et al., 1987; Li et al., 1998). The preparation of HIV vector PICs followed the protocol described in Hansen et al. (1999) except that m.o.i. 5 was used to initiate the infection (Figures 3 and 4).
For Southern blot analysis of integration activity, PICs (100 µl) were mixed with 10 µg/ml linearized φX174 RFI and incubated for 30 min at 37°C. For detection of integration products, the DNAs were purified by treatment with SDS and proteinase K followed by extraction with phenol–chloroform, ethanol precipitated, separated on 0.8% native agarose gels, transferred to a nylon membrane and hybridized with a 32P-labeled LTR sequence as probe.
Purification of PICs and analysis of protein content
Purification of MLV and HIV PICs for western blot analysis was carried out as follows. Nine milliliters of PIC-containing crude extract were partially purified by gel filtration using Sepharose Cl-4B spin columns, then the flow through was adjusted to 20 mM HEPES pH 7.5, 75 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT) and pelleted at 150 000 g for 1 h. The pelleted PICs were suspended in 1.5 ml of buffer A (20 mM HEPES pH 7.5, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 20 U/ml aprotinin), 10 mM ammonium sulfate, 10% glycerol, and layered on top of 10–60% Nycodenz gradients, spun at 150 000 g at 4°C overnight and collected as 24 fractions. Protein concentration was determined using the Bradford Assay (Bio-Rad) and the cDNA copy number assayed by Southern blotting (MoMLV) or quantitative PCR (HIV, described below). The mock-infected cell extracts were made by the same procedure from the same number of cells as in the PIC preparation. After analyzing cDNA content, the PIC peak fractions were pooled and the corresponding fractions from uninfected cell extracts pooled separately. The proteins from the pooled fractions were purified, concentrated by methanol– chloroform extraction (Wessel and Flugge, 1984) and separated by 8–12% SDS–PAGE. Proteins were transferred to a PVDF membrane (Millipore) and analyzed on western blots developed with the Pierce SuperSignal West Femto Maximum Sensitivity Substrate.
Immunoprecipitation of PICs using anti-Ku antibody
Fifty microliters of MoMLV-K PICs (∼5 × 107 cDNA copies) harvested from NIH 3T3 cells or 100 µl of HIV-1 vector PICs (∼3 × 107 cDNA copies) harvested from 293T cells were mixed with 4 µl of pre-immune serum or rabbit anti-Ku80 antibodies. The reactions were incubated on ice for 1 h and then 25 µl of protein A–agarose beads (Sigma) were added. The mixtures were rocked at 4°C for 1 h. The beads were washed four times using buffer A and resuspended in 100 µl of buffer A with 30% glycerol. Integration reactions were carried out with PICs still bound to the beads by the addition of 1 µg of linearized φX174 RFI and incubated at 37°C for 1 h. The DNAs were then purified, ethanol precipitated and analyzed by Southern blotting as above.
Analysis of unintegrated viral cDNA forms
The unintegrated viral cDNA was isolated after high titer infection (m.o.i. 5) using a modified Hirt extraction procedure (Arad, 1998). Samples were harvested 24 h after infection. For Southern blot analysis, isolated cDNA was ethanol precipitated in the presence of glycogen and suspended in restriction enzyme digestion buffer. For the HIV-1 vector, the viral cDNA molecules were digested with XhoI and BamHI. For the MLV vector, the viral cDNAs were digested with NotI and ClaI. DNAs were separated by electrophoresis on 0.8% agarose gel and analyzed by Southern blotting probed with 32P-labeled LTR sequences.
Quantitative PCR analysis of HIV cDNA
HIV cDNA was analyzed using fluorescent-monitored quantitative PCR (Taqman) with an ABI Prism 7700 sequence detection system (PE-Applied Biosystems) essentially as described (Butler et al., 2001). Total DNA was isolated from infected cells using the Qiagen DNeasy kit, quantitated by OD260, and brought to the same concentration before use in quantitative PCR. The primer sets used to detect each sequence (from GenSet or Integrated DNA Technologies) were as follows: total viral cDNA (late RT), forward, MH531(5′-TGTGTGCCCGTCTGTTGTGT-3′); reverse, MH532 (5′-GAGTCCTGCGTCGAGAGAGC-3′); probe, LRT-P [5′-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3′]. For proviral cDNA: Alu forward, MH535 (5′-AACTAGGGAACCCACTGCTTAAG-3′); Alu reverse, SB704 (5′-TGCTGGGATTACAGGCGTGAG-3′); Alu probe, MH603 [5′-(FAM)-ACACTACTTGAAGCACTCAAGGCAAGCTTT-(TAMRA)-3′]. For late RT (HIV genome) sequences, a plasmid encoding the p156RRLsinPPTCMVGFPPRE vector was used as a copy number standard. The Alu copy number standard DNA was generated by quantitation of proviruses integrated into genomic DNA in infected cells as described (Butler et al., 2001). PCRs contained 1× Taqman universal master mix (Perkin-Elmer), 300 nM forward primer, 300 nM reverse primer, 100 nM probe primer and 250 ng of template DNA. All samples were measured in triplicate.
Cell viability assay
For the cell viability assay, the retroviral vector was first purified by pelleting at 23 000 r.p.m. (SW28 rotor) for 2 h then resuspending in fresh RPMI medium. For infections, 5 × 105 Nalm-6 or Nalm-6 LIG4–/– cells were plated in each well of a 24-well plate. Mock (media only), virus supernatant or heat-inactivated virus were spin inoculated (O’Doherty et al., 2000) onto cells at 2500 r.p.m. for 1 h in the presence of 4 µg/ml polybrene in a final volume of 1 ml in each well. Spinoculation increases transduction efficiency 5- to 10-fold (L.Li, J.M.Olvera and F.D.Bushman, unpublished data). The heat-inactivated control virus was incubated at 60°C for 30 min. After spinoculation, an additional 1 ml of fresh media was added per well. The number of viable cells were counted after trypan blue staining. The m.o.i. was determined by infection of 293T cells (for HIV-gfp). Stocks of HIV-gfp and HIV (E152A)-gfp were normalized by measuring p24 capsid antigen. Apoptosis was analyzed using the caspase 3 detection kit from Boehringer Mannheim. Infections in the presence of nevirapine (10 µM) or with HIV (E152A)-gfp did not yield gfp-positive infected centers.
Acknowledgments
Acknowledgements
We thank Dr Leslie Orgel, Dr Fred Alt and members of the Bushman laboratory and the Salk Institute Infectious Disease Laboratory for helpful discussions, and H.T.Lin for pCgp. We also thank Lynn Artale for artwork and help preparing the manuscript. L.L. was supported in part by the Rau Foundation. This work was supported by NIH grants GM56553 and AI34786 to F.D.B., the James B.Pendleton Charitable Trust, the Berger Foundation and Cornelia Mackey. F.D.B. is a Scholar and S.L.B. a fellow of the Leukemia and Lymphoma Society of America. S.L.M. was supported by NSF MCB9806033.
References
- Arad U. (1998) Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques, 24, 760–762. [DOI] [PubMed] [Google Scholar]
- Baekelandt V., Claeys,A., Cherepanov,P., De Clercq,E., Strooper,B.D., Nuttin,B. and Debyser,Z. (2000) DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol., 74, 11278–11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall E.L. and Rio,D.C. (1996) Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev., 10, 921–933. [DOI] [PubMed] [Google Scholar]
- Bowerman B., Brown,P.O., Bishop,J.M. and Varmus,H.E. (1989) A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev., 3, 469–478. [DOI] [PubMed] [Google Scholar]
- Brown P.O., Bowerman,B., Varmus,H.E. and Bishop,J.M. (1987) Correct integration of retroviral DNA in vitro. Cell, 49, 347–356. [DOI] [PubMed] [Google Scholar]
- Brown P.O., Bowerman,B., Varmus,H.E. and Bishop,J.M. (1989) Retroviral integration: structure of the initial covalent complex and its precursor, and a role for the viral IN protein. Proc. Natl Acad. Sci. USA, 86, 2525–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F.D., Fujiwara,T. and Craigie,R. (1990) Retroviral DNA integration directed by HIV integration protein in vitro. Science, 249, 1555–1558. [DOI] [PubMed] [Google Scholar]
- Butler S., Hansen,M. and Bushman,F.D. (2001) A quantitative assay for HIV cDNA integration in vivo. Nature Med., 7, 631–634. [DOI] [PubMed] [Google Scholar]
- Casellas R., Nussenzweig,A., Wuerffel,R., Pelanda,R., Rajewsky,K. and Nussenzweig,M. (1998) Ku80 is required for immunoglobulin isotype switching. EMBO J., 17, 2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J.M. and Rosenberg,N. (1999) Retroviruses. Closing the joint. Nature, 399, 413–416. [DOI] [PubMed] [Google Scholar]
- Coffin J.M., Hughes,S.H. and Varmus,H.E. (1997) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Corbeil J. and Richman,D.D. (1995) The role of surface CD4 in HIV-induced apoptosis. Adv. Exp. Med. Biol., 374, 91–99. [DOI] [PubMed] [Google Scholar]
- Craigie R., Fujiwara,T. and Bushman,F. (1990) The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell, 62, 829–837. [DOI] [PubMed] [Google Scholar]
- Daniel R., Katz,R.A. and Skalka,A.M. (1999) A role for DNA-PK in retroviral DNA integration. Science, 284, 644–647. [DOI] [PubMed] [Google Scholar]
- Doak T.G., Doerder,F.P., Jahn,C.L. and Herrick,G. (1994) A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common ‘D35E’ motif. Proc. Natl Acad. Sci. USA, 91, 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J.A. and Jackson,S.P. (1999) Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol. Cell. Biol., 19, 6260–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drelich M.R., Wilhelm,R. and Mous,J. (1992) Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 in protein in vitro. Virology, 188, 459–468. [DOI] [PubMed] [Google Scholar]
- Ellison V.H., Abrams,H., Roe,T., Lifson,J. and Brown,P.O. (1990) Human immunodeficiency virus integration in a cell-free system. J. Virol., 64, 2711–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A. (1999) In vivo analysis of retroviral integrase structure and function. In Maramorosch,K., Murphy,F.A. and Shatkin,A.J. (eds), Advances in Virus Research. Academic Press, San Diego, CA, pp. 411–426. [DOI] [PubMed]
- Farnet C. and Bushman,F.D. (1997) HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell, 88, 1–20. [DOI] [PubMed] [Google Scholar]
- Farnet C.M. and Haseltine,W.A. (1990) Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl Acad. Sci. USA, 87, 4164–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O., Ramond,P., Polard,P., Prere,M.F. and Chandler,M. (1990) Functional similarities between the IS3 family of bacterial insertion elements? Mol. Microbiol., 4, 1771–1777. [DOI] [PubMed] [Google Scholar]
- Follenzi A., Ailes,L.E., Bakovic,S., Gueuna,M. and Naldini,L. (2000) Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nature Genet., 25, 217–222. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A. and Malim,M.H. (1999) Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv. Virus Res., 52, 275–299. [DOI] [PubMed] [Google Scholar]
- Fugmann S., Villey,I., Ptaszek,L. and Schatz,D. (2000) Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell, 5, 97–107. [DOI] [PubMed] [Google Scholar]
- Fujiwara T. and Mizuuchi,K. (1988) Retroviral DNA integration: structure of an integration intermediate. Cell, 54, 497–504. [DOI] [PubMed] [Google Scholar]
- Gallay P., Swingler,S., Song,J., Bushman,F. and Trono,D. (1995) HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell, 83, 569–576. [DOI] [PubMed] [Google Scholar]
- Grawunder U., Zimmer,D., Fugmann,S., Schwarz,K. and Lieber,M.R. (1998) DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell, 2, 477–484. [DOI] [PubMed] [Google Scholar]
- Han J.Y., Cannon,P.M., Lai,K.M., Zhao,Y., Eiden,M.V. and Anderson,W.F. (1997) Identification of envelope protein residues required for the expanded host range of 10A1 murine leukemia virus. J. Virol., 71, 8103–8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M.S.T., Carteau,S., Hoffmann,C., Li,L. and Bushman,F. (1998) Retroviral cDNA integration: mechanism, applications and inhibition. In Setlow,J.K. (ed.), Genetic Engineering. Principles and Methods, Vol. 20. Plenum Press, New York, NY, pp. 41–62. [DOI] [PubMed]
- Hansen M.S.T., Smith,G.J.I., Kafri,T., Molteni,V., Siegel,J.S. and Bushman,F.D. (1999) Integration complexes derived from HIV vectors for rapid assays in vitro. Nature Biotechnol., 17, 578–582. [DOI] [PubMed] [Google Scholar]
- Huang L.-C., Clarkin,K.C. and Wahl,G.M. (1996) Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc. Natl Acad. Sci. USA, 93, 4827–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R.A., Merkel,G., Kulkosky,J., Leis,J. and Skalka,A.M. (1990) The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell, 63, 87–95. [DOI] [PubMed] [Google Scholar]
- Katzman M., Katz,R.A., Skalka,A.M. and Leis,J. (1989) The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol., 63, 5319–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M., Ikuta,T., Miyasaka,T. and Shiomi,T. (1999) Ku80 can translocate to the nucleus independent of the translocation of Ku70 using its own nuclear localization signal. Oncogene, 18, 7495–7505. [DOI] [PubMed] [Google Scholar]
- Kulkosky J., Jones,K.S., Katz,R.A., Mack,J.P.G. and Skalka,A.M. (1992) Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol., 12, 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landree M., Wibbenmeyer,J. and Roth,D. (1999) Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev., 13, 3059–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Farnet,C.M., Anderson,W.F. and Bushman,F.D. (1998) Modulation of activity of MoMuLV preintegration complexes by several host factors in vitro. J. Virol., 72, 2125–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. (1998) Pathological and physiological double-strand breaks. Am. J. Pathol., 153, 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Gu,Y.S., Lansford,R., Sonoda,E., Ferrini,R., Davidson,L., Rajewsky,K. and Alt,F.W. (1998) Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med., 187, 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.D., Wang,B. and Bushman,F.D. (1995) HIV-1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J. Virol., 69, 3938–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.D., Farnet,C.M. and Bushman,F.D. (1997) Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol., 71, 5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blomer,U., Gallay,P., Ory,D., Mulligan,R., Gage,F.H., Verma,I.M. and Trono,D. (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science, 272, 263–267. [DOI] [PubMed] [Google Scholar]
- O’Doherty U., Swiggard,W.J. and Malim,M.H. (2000) Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol., 74, 10074–10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergola F., Zdzienicka,M.Z. and Lieber,M.R. (1993) V(D)J recombination in mammalian cell mutants defective in DNA double-strand break repair. Mol. Cell. Biol., 13, 3464–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolnik A., Melchers,F. and Andersson,J. (1996) The SCID but not the RAG-2 gene product is required for Sµ–Sε heavy chain class switching. Immunity, 5, 319–330. [DOI] [PubMed] [Google Scholar]
- Roth M.J., Schwartzberg,P.L. and Goff,S.P. (1989) Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence of IN function and terminal DNA sequence. Cell, 58, 47–54. [DOI] [PubMed] [Google Scholar]
- Rowland S.-J. and Dyke,K.G.H. (1990) Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol., 4, 961–975. [DOI] [PubMed] [Google Scholar]
- Sherman P.A. and Fyfe,J.A. (1990) Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl Acad. Sci. USA, 87, 5119–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C.-G., Taddeo,B., Haseltine,W.A. and Farnet,C.M. (1994) Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol., 68, 1633–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smider V., Rathmell,W.K., Lieber,M.R. and Chu,G. (1994) Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science, 266, 288–290. [DOI] [PubMed] [Google Scholar]
- Smith G.C.M. and Jackson,S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- Taccioli G.E., Rathbun,G., Oltz,E., Stamato,T., Jeggo,P.A. and Alt,F.W. (1993) Impairment of V(D)J recombination in double-strand break repair mutants. Science, 260, 207–210. [DOI] [PubMed] [Google Scholar]
- Taccioli G.E. et al. (1994) Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science, 265, 1442–1445. [DOI] [PubMed] [Google Scholar]
- Temin H.M., Keshet,E. and Weller,S.K. (1980) Correlation of transient accumulation of linear unintegrated viral DNA and transient cell killing by avian leukosis and reticuloendotheliosis viruses. Cold Spring Harb. Symp. Quant. Biol., 44, 773–778. [DOI] [PubMed] [Google Scholar]
- Tuteja N. et al. (1994) Human DNA helicase II: a novel DNA unwinding enzyme identified as the Ku autoantigen. EMBO J., 13, 4991–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D. and Flugge,U.I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem., 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Wiskerchen M. and Muesing,M.A. (1995) Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol., 69, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R., Du,J., Thompson,R. and Kenter,A. (1997) Ig Sλ3 DNA-specific double-strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J. Immunol., 159, 4139–4144. [PubMed] [Google Scholar]
- Yoder K. and Bushman,F.D. (2000) Repair of gaps in retroviral DNA integration intermediates. J. Virol., 74, 11191–11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzienicka M.Z. (1995) Mammalian cell mutants defective in the response to ionizing radiation-induced DNA damage. Mutat. Res., 336, 203–213. [DOI] [PubMed] [Google Scholar]
- Zennou V., Petit,C., Guetard,D., Nerbass,U., Montagnier,L. and Charneau,P. (2000) HIV-1 genome nuclear import is mediated by a central DNA flap. Cell, 101, 173–185. [DOI] [PubMed] [Google Scholar]