Abstract
Taxotere showed antitumor activity against solid tumors including prostate cancer. However, the molecular mechanism(s) of action of Taxotere has not been fully elucidated. In order to establish such molecular mechanism(s) in both hormone-insensitive (PC3) and hormone-sensitive (LNCaP) prostate cancer cells, comprehensive gene expression profiles were obtained by Affymetrix Human Genome U133A Array. The RNA from the cells treated with 2 nM Taxotere was subjected to microarray analysis. We found that a total of 166, 365, and 1785 genes showed greater than twofold change in PC3 cells after 6, 36, and 72 hours of treatment, respectively compared to 57, 823, and 964 genes in LNCaP cells. The expression of tubulin was decreased, whereas the expression of microtubule-associated proteins was increased in Taxotere-treated prostate cancer cells, confirming the microtubule-targeting effect of Taxotere. Clustering analysis showed downregulation of some genes for cell proliferation and cell cycle. In contrast, Taxotere upregulated some genes that are related to induction of apoptosis and cell cycle arrest. From these results, we conclude that Taxotere caused alterations of a large number of genes, many of which may contribute to the molecular mechanism(s) by which Taxotere affects prostate cancer cells. Further molecular studies are needed in order to determine the cause and effect relationships between these genes altered by Taxotere. Nevertheless, our results could be further exploited for devising strategies to optimize therapeutic effects of Taxotere for the treatment of prostate cancer.
Keywords: Taxotere, gene expression, microarray, prostate cancer cells, apoptosis
Introduction
Taxotere is a semisynthetic anticancer agent derived from baccatin III of the needles of Taxus baccata. It has shown a wide spectrum of cytotoxicity for various solid tumors as well as clinical activity when used for the treatment of breast, lung, ovarian, and prostate cancers. In early-stage and metastatic breast, lung, and ovarian cancers, randomized trials have shown that Taxotere-containing therapies are superior to, or as effective as, established standard chemotherapeutic regimens and are often associated with an improved safety profile [1–3]. Recent clinical trials have found that weekly Taxotere in patients with metastatic hormone-refractory prostate cancer is associated with improvements in clinical benefit response and quality of life, and is well tolerated [4,5]. Thus, Taxotere is currently considered to be among the most important anticancer drugs in cancer chemotherapy. Its known basic mechanism of action is that it binds to tubulin and deranges the equilibrium between microtubule assembly and disassembly during mitosis [6]. Stabilization of microtubules by Taxotere impairs mitosis and exerts an anticancer effect in tumors [6]. Recent investigations suggest that Taxotere acts by additional mechanisms that are distinct from its effects on microtubules. It has been reported that Taxotere increases bcl-2 phosphorylation and downregulates bcl-xl, inhibiting the antiapoptotic function of bcl-2 family [7–9]. Avramis et al. [10] found increased p21WAF1 and p53 levels, and induced apoptosis following Taxotere treatment in human leukemia cells. Despite its ability to induce apoptosis, Taxotere also exhibits regulation of expression of cytokine genes [11], suggesting the pleiotropic effects of Taxotere on human cancers.
Prostate cancer is the most common nondermatological carcinoma in the United States with an estimated 220,900 new cases and 28,900 deaths in 2003 [12]. Up to 30% of men undergoing radical prostatectomy will relapse, often as a result of micrometastatic disease present at the time of surgery [13]. Metastatic androgen-independent prostate cancer is an advanced stage of prostate cancer and, unfortunately, conventional chemotherapeutic agents have not been, thus far, effective for metastatic prostate cancer. Therefore, there is a tremendous need for the development of mechanism-based and targeted strategies for the treatment of prostate cancer. Microarray permits rapid analysis of the expression of a large number of genes and, in turn, provides an opportunity for determining the effects of anticancer agents on cancer cells. The alterations of gene expression profiles by several anticancer agents have been reported [14,15]. The data from microarray are likely to contribute in devising therapeutic strategies more accurately, and will help to determine the molecular mechanism(s) of action of therapeutic agents.
Yoo et al. [16] have reported Taxotere-induced gene expression patterns in head and neck cancer cells by using cDNA microarray, which contains 1191 genes. They found that Taxotere regulated some genes that are related to cell cycle, apoptosis, angiogenesis, and tyrosine kinase signal transduction. However, little is known regarding the gene expression profiles of prostate cancer cells after Taxotere treatment. In addition, the precise molecular mechanism(s) by which Taxotere exerts its tumor-suppressive effects on prostate cancer is unclear. Hence, in this study, we utilized the high-throughput gene chip, which contains 22,215 known genes, to determine the alteration of gene expression profiles of hormone-insensitive (PC3) and hormone-sensitive (LNCaP) prostate cancer cells exposed to Taxotere.
Materials and Methods
Cell Culture and Growth Inhibition
PC3 (ATCC, Manassas, VA) and LNCaP (ATCC) human prostate cancer cells were cultured in RPMI-1640 media (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin in a 5% CO2 atmosphere at 37°C. Taxotere (Aventis Pharmaceuticals, Bridgewater, NJ) was dissolved in DMSO to make 4 mM stock solution. For growth inhibition, PC3 and LNCaP cells were seeded at a density of 5 x 104 per well in a six-well culture dish. After 24 hours, the cells were treated with 1, 2, and 4 nM Taxotere. Control PC3 and LNCaP cells received 0.01% DMSO for same time points. Cells treated with Taxotere or DMSO for 1 to 3 days were harvested by trypsinization, stained with 0.4% trypan blue, and counted using a hemocytometer. The concentrations of Taxotere used for our in vitro studies are easily achievable in humans, suggesting that our experimental results are relevant for human applications. The experiment was repeated three times and a t-test was performed to verify the significance of cell growth inhibition after treatment.
Microarray Analysis for Gene Expression Profiles
PC3 and LNCaP cells were treated with 2 nM Taxotere for 6, 36, and 72 hours. Total RNA from each sample was isolated by Trizol (Invitrogen) and purified by RNeasy Mini Kit and RNase-free DNase Set (Qiagen, Valencia, CA) according to the manufacturer's protocols. cDNA for each sample was synthesized by using Superscript cDNA Synthesis Kit (Invitrogen) using the T7-(dT)24 primer instead of the oligo(dT) provided in the kit. Then, the biotin-labeled cRNA was transcripted in vitro (IVT) from cDNA by using BioArray High-Yield RNA Transcript Labeling Kit (Enzo Biochem, New York, NY), and purified by RNeasy Mini Kit. After fragmentation, the fragmented labeled cRNA was applied to Human Genome U133A Array (Affymetrix, Santa Clara, CA), which contains 22,215 human gene probes, and hybridized to the probes in the array. After washing and staining, the arrays were scanned. Two independent experiments were performed to verify the reproducibility of results. Correlation statistical analysis for the data obtained from the two experiments was accessed by using Pearson product moment correlation coefficient.
Microarray Data Normalization and Analysis
The gene expression levels of samples were normalized and analyzed by using Microarray Suite, MicroDB, and Data Mining Tool software (Affymetrix). The absolute call (present, marginal, and absent) and average difference of 22,215 gene expressions in a sample, and the absolute call difference, fold change, and average difference of gene expressions between two or several samples were also normalized and identified using these software. Statistical analysis of the mean expression average difference of genes, which show greater than two-fold change, was performed using a t-test between treated and untreated samples. Clustering and annotation of the gene expression were analyzed by using Cluster, TreeView [17], Onto-Express [18], and GenMAPP (www.genmapp.org). Genes that were not annotated or not easily classified were excluded from the functional clustering analysis.
Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis for Gene Expression
To verify the alterations of gene expression at the mRNA level, which appeared on the microarray, we chose 23 representative genes (Table 1) with varying expression profiles for real-time RT-PCR analysis. Two micrograms of total RNA from each sample was subjected to reverse transcription using the Superscript first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer's protocol. Real-time PCR reactions were then carried out in a total of 25 µl of reaction mixture (2 µl of cDNA, 12.5 µl of 2 x SYBR Green PCR Master Mix, 1.5 µl of each 5 µM forward and reverse primers, and 7.5 µl of H2O) in an ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). The PCR program was initiated by 10 minutes at 95°C before 40 thermal cycles, each of 15 seconds at 95°C and 1 minute at 60°C. Data were analyzed according to the comparative Ct method and were normalized by actin expression in each sample. Melting curves for each PCR reaction were generated to ensure the purity of the amplification product.
Table 1.
The Primers Used for Real-Time RT-PCR Analysis.
| Genes | Primer Sequence | PCR Product (bp) |
| α-Tubulin | gct ttg gtg ggg gaa ct | 104 |
| cgc cgg gta aat aga gaa ct | ||
| α1-Tubulin | cgg tct tca ggg ctt ctt gg | 139 |
| ctg ggg tgc tgg gta aat gg | ||
| β5-Tubulin | ccg cag aag agg agg agg att | 120 |
| gga gag gaa agg ggc agt tga | ||
| CHK1 | tag ata tga agc gtg ccg tag | 113 |
| gcc ttc tct cct gtg acc a | ||
| Cyclin A2 | aat cag ttt ctt acc caa tac | 127 |
| ctg atg gca aat act tga | ||
| Fas/Apo-1 | caa aag tgt taa tgc cca agt | 187 |
| gca gtc tgg ttc atc cc | ||
| FOXM1 | gcc aca ctt agc gag acc c | 189 |
| atc aca agc att tcc gag aca | ||
| GADD45 | cgc ctg tga gtg agt gc | 154 |
| ctt atc cat cct ttc ggt ctt | ||
| IGFBP2 | atg ggc gag ggc act t | 189 |
| cag ctc ctt cat acc cga ctt | ||
| IMPA2 | tga ccc tgc gac cct gaa | 197 |
| cgc ctg ctt ctc tga tga tga | ||
| Ki-67 | ccg ggc tcc atc atc t | 148 |
| ctc cag acg cca aaa taa gac | ||
| Microtubule-related | gga gat gtc tat tgc cga gta | 164 |
| protein | acc cca aac caa aca cc | |
| p21WAF1 | ctg gag act ctc agg gtc gaa | 98 |
| gga tta ggg ctt cct ctt gga | ||
| p27KIP1 | cgc tcg cca gtc cat t | 187 |
| aca aaa ccg aac aaa aca aag | ||
| PIR | cac tag ccc tcc atc ctc tac | 151 |
| ggg tct gcc aat gct tct | ||
| RANBP1 | ttc cga ttt gcc tct gag aac | 153 |
| cgg cgt gat gta gtg gtt g | ||
| STK6 | tca gcg ggt ctt gtg t | 162 |
| ctc ttt tgg gtg tta ttc agt | ||
| Survivin | cca ctg ccc cac tga gaa c | 118 |
| acc gga cga atg ctt ttt atg | ||
| TRIP13 | tct ggc agt gga caa gca gtt | 136 |
| tgg gag acg gct gtg tgg | ||
| TBP | cac gaa cca cgg cac tga tt | 96 |
| ttt tct tgc tgc cag tct gga c | ||
| β-Actin | cca cac tgt gcc cat cta cg | 99 |
| agg atc ttc atg agg tag tca gtc ag | ||
Western Blot Analysis
In order to verify whether the alterations of genes at the level of transcription ultimately result in the alterations at the level of translation, we conducted Western blot analysis for selected genes with varying expression profiles. The PC3 and LNCaP cells were treated with 1 and 2 nM Taxotere for 24, 48, and 72 hours. After treatment, the cells were lysed and protein concentration was measured using BCA protein assay (Pierce, Rockford, IL). The proteins were subjected to 10% or 14% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to nitrocellulose membrane. The membranes were incubated with antip21WAF1 (1:500; Upstate, Lake Placid, NY), anti-p27KIP1 (1:250; Novocastra, Newcastle upon Tyne, UK), anti-Bax (1:10000; Trevigen, Gaithersburg, MD), anti-survivin (1:200; Alpha Diagnostic, San Antonio, TX), anti-cell division cycle (CDC) 2 (1:200; Santa Cruz, Santa Cruz, CA), anti-cyclin A (1:250; NeoMarkers, Union City, CA), anti-cyclin E (1:250; NeoMarkers), and anti-β-actin (1:10000; Sigma, St. Louis, MO) primary antibodies, and subsequently incubated with secondary antibody conjugated with fluorescence dye. The signal was then detected and quantified by using Odyssey infrared imaging system (LI-COR, Lincoln, NE). The ratios of p21WAF1, p27KIP1, Bax, survivin, CDC2, cyclin A, or cyclin E against β-actin were calculated by standardizing the ratios of each control to the unit value.
Results
Cell Growth Inhibition by Taxotere Treatment
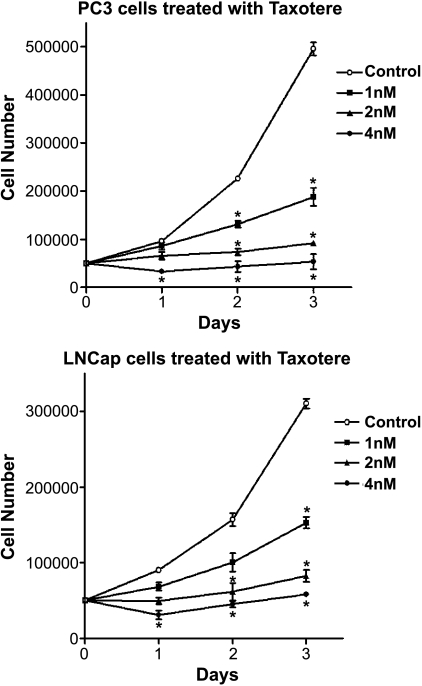
Cell growth inhibition assay showed that the treatment of PC3 and LNCaP prostate cancer cells with Taxotere resulted in a dose- and time-dependent inhibition of cell proliferation (Figure 1), demonstrating the growth-inhibitory effect of Taxotere on prostate cancer cells. Inhibition of cell proliferation observed by cell growth inhibition assay could be due to altered regulation of gene expression by Taxotere treatment. We further investigated the gene expression profiles of PC3 and LNCaP prostate cancer cells treated with Taxotere.
Figure 1.
Effects of Taxotere on the growth of PC3 and LNCaP cells. PC3 and LNCaP prostate cancer cells treated with Taxotere resulted in a doseand time-dependent inhibition of cell proliferation (*P < .05, n = 3).
Alteration of Global Gene Expression by Taxotere Treatment
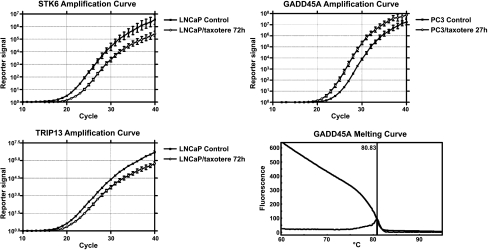
The alterations of gene expression were observed as early as 6 hours and were more evident with more genes altered after longer treatment (Figure 2). A total of 166, 365, and 1785 genes showed greater than two-fold change in PC3 cells after 6, 36, and 72 hours of Taxotere treatment, respectively compared to 57, 823, and 964 genes in LNCaP cells treated with Taxotere for the same time point. The results of real-time RT-PCR analysis for selected genes were in direct agreement with the microarray data (Figure 3; Table 2). The same alterations of gene expression were observed by real-time RT-PCR analysis, although the fold change in the expression level was not exactly similar between these two different analytical methods. furthermore, the results of western blot analysis also are in direct agreement with the microarray and RT-PCR data (Figure 4). These results suggest that Taxotere regulates the transcription and translation of genes that are involved in the processes of cell survival and physiological behaviors.
Figure 2.
Cluster analysis of genes showing alterations in mRNA expression after Taxotere treatment. The alterations of specific genes are shown.
Figure 3.
Real-time RT-PCR amplification curves showing the altered expression of specific genes from RNA of Taxotere-treated PC3 and LNCaP cells. Real-time RT-PCR melting curve showing that the PCR product is pure (only one peak).
Table 2.
Fold Changes of Specific Genes in PC3 and LNCaP Cells Treated with Taxotere.
| Genes | PC3 | LNCaP | ||||
| 6 hours | 36 hours | 72 hours | 6 hours | 36 hours | 72 hours | |
| Tubulin and microtubule-related | ||||||
| BC004188.1 tubulin, β2 | NC* | -1† | -1.3 | -1.4 | -3.2 | -3.5 |
| AF141349.1 β-tubulin mRNA | NC | -1 | -1.2 | -1.3 | -3.7 | -4.9 |
| BC004949.1 tubulin α6 | -1.1 | -0.8 | -1.5 | -1.3 | -2.6 | -3 |
| BC006379.1 tubulin α1 | -1.2 | -0.8 | -1.3 | -1.2 | -3.5 | -4 |
| BC005838.1 tubulin, β5 | NC | -0.9 | -1.2 | -1.3 | -4 | -4.3 |
| AL565074 tubulin, α1 | NC | -0.8 | -1.5 | -1.1 | -4.9 | -4.9 |
| BC001002.1 tubulin, β | -1.1 | -1.9 | -2 | -1 | -2.8 | -3.2 |
| AL581768 Tubulin, α | NC | -0.9 | -1.2 | -1.5 | -4 | -4.9 |
| AA515698 tubulin, β2 | NC | -0.9 | -1.5 | -1.3 | -3 | -3 |
| NM 012155.1 microtubule-associated protein like echin | -1.1 | 1.7‡ | 2.6 | 1.3 | 3.2 | 3.2 |
| AF183417.1 microtubule-associated proteins 1A1B | NC | 1.5 | 3 | -1.1 | 2 | 1.6 |
| Cell cycle | ||||||
| NM 006739.1 cell division cycle 46 | -1 | -0.9 | -2.6 | -1.4 | -45.3 | -36.8 |
| NM 021873.1 cell division cycle 25B (CDC25B) | -1.3 | -0.9 | -2 | -1.1 | -1.6 | -2.5 |
| NM 004526.1 mitotin mRNA | NC | -0.8 | -2.3 | -1.1 | -5.7 | -8 |
| NM 000075.1 cyclin-dependent kinase 4 (CDK4) | NC | -0.8 | -2.3 | -1.1 | -1.4 | -1.4 |
| NM 000389.1 cyclin-dependent kinase inhibitor p21 (Waf1) | 1.7 | 1.7 | 2.6 | 1.3 | 9.8 | 8.6 |
| NM 001255.1 CDC20 (CDC20) | NC | NC | -1.4 | -1.9 | -9.5 | -47 |
| NM 001786.1 cell division cycle 2 (CDC2) | NC | NC | -1.3 | -1.2 | -7.5 | -12.1 |
| NM 001237.1 cyclin A2 (CCNA2) | NC | -0.9 | -2 | -1.2 | -5.7 | -12.1 |
| NM 003503.2 CDC7 (cell division cycle 7) | -1.2 | -0.9 | -1.8 | NC | -2.1 | -2.3 |
| U17105.1 cyclin F mRNA | NC | -0.7 | -2.1 | -1.6 | -11.3 | -11.3 |
| NM 004702.1 cyclin E2 (CCNE2) | 1.8 | -0.9 | -2 | 1.6 | -5.3 | -3.2 |
| NM 006806.1 BTG family, member 3 (BTG3) | 1.3 | 1.2 | 1.6 | 1.1 | 1.6 | 2.1 |
| AI765445 BTG family, member 3 | 1.3 | 1.2 | 1.6 | 1.2 | 1.7 | 2.1 |
| BC001971.1 cyclin-dependent kinase inhibitor p27 (Kip1) | 1.1 | 1.6 | 2.3 | 1.2 | 1.1 | 1.2 |
| AI251890 CDC-like kinase 1 | 1.1 | 1.5 | 4.9 | NC | 1.7 | 2.1 |
| Apoptosis | ||||||
| NM 001168.1 survivin mRNA | -1.2 | -1 | -1.5 | -1.1 | -20.9 | -24.1 |
| NM 001924.2 GADD45A mRNA | 1.1 | 2.3 | 4.3 | 1.4 | 7.5 | 7.5 |
| NM 001455.1 forkhead box O3A (FOXO3A) | NC | 1 | 1.4 | 1.1 | 2 | 2.1 |
| AB028869.1 survivin-β | NC | NC | -1.3 | -1.1 | -1.3 | -4.9 |
| U72398 Bcl-xβ (bcl-x) gene | NC | NC | 1.5 | NC | 1.6 | 2.1 |
| X83493.1 Fas/Apo-1 | NC | 1.1 | 1.1 | 1.1 | 4.6 | 8 |
| AF082283.1 CARD-containing apoptotic signaling protein | NC | 2 | 2.3 | 1.1 | 1.3 | 2 |
| NM 001197.2 BCL2-interacting killer (apoptosis-inducing) | 1.4 | 1.2 | 1.7 | 1.1 | 2.6 | 2.6 |
| NM 004324.1 BCL2-associated X protein (BAX) | 1.1 | -0.8 | -1.4 | 1.3 | 3.2 | 3 |
| U19599.1BAX δ mRNA | 1.1 | NC | NC | 1.1 | 2.8 | 4.3 |
| Oncogenesis | ||||||
| NM 006342.1 acidic coiled coil containing protein 3 (TACC3) | -1.3 | NC | -1.6 | -1.2 | -11.3 | -45.3 |
| NM 005030.1 polo-like kinase (PLK) | NC | -0.9 | -1.6 | -1.4 | -5.3 | -5.3 |
| NM 014264.1 serine/threonine kinase 18 (STK18) | -1.3 | -0.9 | -1.9 | -1.4 | -8.6 | -16 |
| NM 001274.1 CHK1 | -1.1 | -0.8 | -2 | -1.1 | -4.3 | -7 |
| AU152107 Ki-67 | -1.2 | -0.8 | -3.3 | NC | -1.6 | -2.3 |
| NM 003600.1 serine/threonine kinase 15 (STK15) | -1.3 | NC | -1.5 | -1.2 | -3.7 | -8.6 |
| NM 003158.1 serine/threonine kinase 6 (STK6) | -1.2 | -1.1 | -1.7 | -1.7 | -11.3 | -9.8 |
| Transcription | ||||||
| NM 021953.1 forkhead box M1 (FOXM1) | NC | -0.8 | -2.1 | -1.2 | -13.9 | -32.2 |
| NM 004237.1 thyroid hormone receptor interactor 13 (TRIP13) | NC | NC | -1.7 | -1.1 | -4.9 | -3.5 |
| NM 007295.1 breast cancer 1, early onset (BRCA1) | -1.1 | NC | -1.4 | 1.2 | -1.7 | -24.3 |
| NM 005225.1 E2F transcription factor 1 (E2F1) | NC | -0.8 | -2.6 | NC | -9.2 | -13.9 |
| NM 002128.1 high-mobility group protein 1 (HMG1) | NC | -0.9 | -1.7 | -1.1 | -2 | -2.1 |
| NM 005342.1 high-mobility group protein 4 (HMG4) | -1.3 | -1.1 | -1.7 | -1.1 | -2.1 | -2.6 |
| NM 003662.1 Pirin (PIR) | -1.2 | 1.6 | 2.6 | NC | -2.5 | -3.5 |
| NM 006166.2 nuclear transcription factor Y, β (NFYB) | NC | NC | 1.5 | NC | -2.5 | -1.2 |
| Androgen receptor-related | ||||||
| NM 000597.1 insulin-like growth factor binding protein 2 | 1 | 1.1 | 1.3 | NC | 1.5 | 2.1 |
| NM 004114.1 fibroblast growth factor 13 (FGF13) | 1 | 1.6 | 1.9 | NC | 2.3 | 2.8 |
| BC003610.1 globule EGF factor 8 | NC | NC | 2.2 | 1.1 | 1.9 | 2.6 |
No change.
Negative value, decrease.
Positive value, increase.
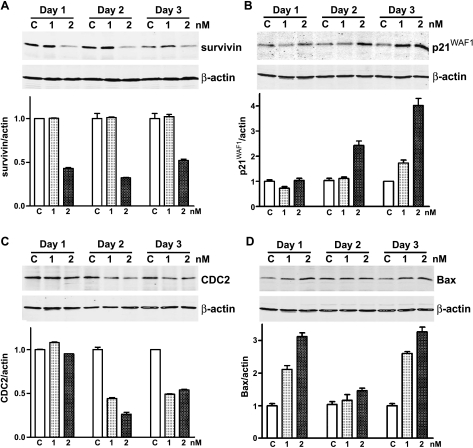
Figure 4.
Western blot analysis of selected gene expression in Taxotere-treated LNCaP cells at the protein level. (A) The expression of survivin was downregulated with Taxotere treatment. (B) The expression of p21WAF1 was upregulated with Taxotere treatment. (C) The expression of CDC2 was downregulated with Taxotere treatment. (D) The expression of Bax was upregulated with Taxotere treatment.
The altered genes were subjected to cluster analysis according to the location and cellular components. The genes showing altered expression were mostly located on chromosomes 1, 2, 6, 12, 17, and 19, and were mainly responsible for the transcription and translation of components of the nucleus, cytoplasm, and integral plasma membrane proteins.
Taxotere exerted differential effects on gene expression profiles between PC3 and LNCaP cells. However, we found no effect of Taxotere on the expression of androgen receptor (AR), although upregulation of several genes involved in steroid-independent AR activation [insulin-like growth factor binding protein (IGFBP) 2, fibroblast growth factor (FGF) 13, and epidermal growth factor (EGF) 8] was observed in LNCaP cells (Table 2). This interesting observation needs further in-depth investigation and is currently ongoing in our laboratory.
Regulation of Microtubule-Related Genes by Taxotere Treatment
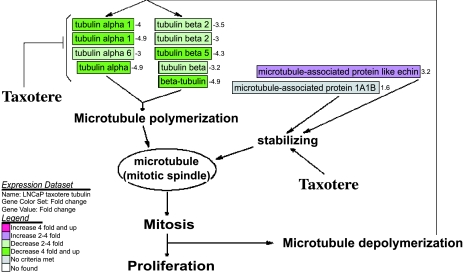
By using microarray, GenMAPP computerized analysis, and RT-PCR, we found that Taxotere downregulated the expression of tubulins including tubulin α1, tubulin α6, tubulin β2, and tubulin β5 (Table 2; Figures 2 and 5), suggesting the inhibitory effect of Taxotere on microtubule. We also found that Taxotere upregulated the expression of microtubule-associated proteins (Table 2; Figures 2 and 5), suggesting the effects of Taxotere on microtubule assembly.
Figure 5.
Effect of Taxotere on microtubule-related gene expression analyzed and visualized by GenMAPP software integrated with cDNA microarray data (positive value: increase in fold change; negative value: decrease in fold change).
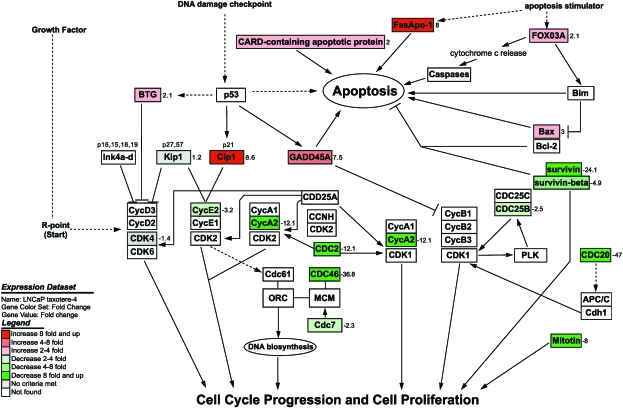
Regulation of Genes Involved in Cell Cycle and Apoptotic Process
After clustering based on biological function using Onto- Express and GenMAPP computerized analysis, we found that in both PC3 and LNCaP cells, Taxotere downregulated the expression of some genes, which are critically involved in the regulation of cell proliferation and cell cycle (Table 2; Figures 2 and 6). In contrast, Taxotere upregulated the expression of some genes that are related to induction of apoptosis and cell cycle arrest (Table 2; Figures 2 and 6). Western blot analysis showed that the protein levels of p21WAF1 and Bax in LNCaP cells treated with Taxotere were upregulated, whereas survivin, CDC2, cyclin A, and cyclin E were downregulated (Figure 4). In Taxotere-treated PC3 cells, the protein levels of p21WAF1 and p27KIP1 were upregulated, and survivin, cyclin A, and cyclin E were downregulated. These results are novel and have not been reported previously. However, further mechanistic studies are needed in order to establish cause-and-effect relationships between these genes with apoptosis-inducing effect of Taxotere in prostate cancer cells.
Figure 6.
Effects of Taxotere on cell cycle- and apoptosis pathway-related gene expression analyzed and visualized by GenMAPP software integrated with cDNA microarray data (positive value: increase in fold change; negative value: decrease in fold change).
In addition to the effects of Taxotere on microtubule, cell cycle, and apoptosis, Taxotere also showed the regulation of genes related to signal transduction, transcription factor, oncogenesis, and tumor suppression (Table 2; Figure 2).
Discussion
Microtubules are formed from α-tubulin and β-tubulin, and their polymerization and depolymerization are complex and interesting processes with important biological roles [19]. The mitotic spindle, one of the microtubules, forms after the cytoplasmic microtubules disassemble during mitosis. It has been known that Taxotere binds tightly to microtubules and stabilizes them, causing mitotic arrest in cancer cells [6]. Taxotere also induces apoptosis through inhibition of the depolymerization of microtubules [20]. However, the precise molecular mechanism is still unknown. In the present study, we found that Taxotere inhibited the expression of α-tubulin and β-tubulin, in addition to stabilizing microtubules. In contrast, the expression of microtubuleassociated proteins was upregulated after Taxotere treatment. Microtubule-associated proteins are a family of proteins, which are involved in the regulation of microtubule assembly and used as a marker of differentiation [21]. Upregulation of microtubule-associated proteins by Taxotere treatment will promote stabilization of microtubules and differentiation of cells, whereas downregulation of tubulin will lead to insufficiency of material for mitotic spindle formation, resulting in the cell cycle arrest of cancer cells (Figure 6). This may be another molecular mechanism by which Taxotere exerts its growth-inhibitory effect on cancer cells.
It has been found that CDCs regulate the molecules related to the cell cycle, and they are essential and important for the initiation and progression of successive phases of the cell cycle [22,23]. It has been well known that cyclins associate with cyclin-dependent protein kinases (CDKs) and CDCs to control the process of cell cycle [24–27]. The CDK inhibitors including p21WAF1 and p27KIP1 have been demonstrated to arrest the cell cycle and inhibit the growth of cancer cells [28,29]. Our results showed that cyclins (cyclin A2, cyclin E2, and cyclin F), CDK4, and CDCs (CDC2, CDC 7, CDC20, and CDC25B) were downregulated in Taxotere-treated prostate cancer cells, whereas CDK inhibitors (p21WAF1 and p27KIP1) were upregulated, suggesting that Taxotere inhibited the growth of both hormone-insensitive and hormone-sensitive prostate cancer cells through the arrest of the cell cycle and inhibition of proliferation (Figure 6). These observations are novel in Taxotere-treated prostate cancer cells. Similar effects on cell cycle were also observed in Taxotere-treated head and neck cancer cells [16], suggesting some common mode of action of Taxotere in cancer cells. However, most of the altered genes by Taxotere in head and neck cancer cells are different from our data, reflecting the different cellular context; hence, the mode of action of Taxotere in prostate cancer cells is unique and has not been reported previously.
Inhibition of cell growth by Taxotere could also be due to the induction of apoptosis in addition to cell cycle arrest. It has been reported that Taxotere is able to induce apoptosis through caspase-3-dependent or caspase-3-independent cell death mechanism [30]. We found a decreased level of survivin and increased level of growth arrest and DNA damage gene 45A (GADD45A), Fas/Apo-1, and forkhead transcription factor FKHR-L1 (FOXO3A), all of which are related to apoptotic processes, and these effects of Taxotere have not been previously reported. Survivin is overexpressed in a variety of cancer cells. Xia et al. [31] reported that antisurvivin oligonucleotides induced apoptosis in mesothelioma cells, suggesting that downregulation of survivin appears to be an effective therapy for mesothelioma. It has been known that GADD45 promotes apoptosis and regulates G2/M arrest [32]. Fas/Apo-1 and FOXO3A also induce apoptosis [33,34]. Induction of apoptosis by FOXO3A has been correlated with the disruption of mitochondrial membrane integrity, cytochrome c release, and upregulation of Bim [35]. Taxotere regulated the expression of survivin, GADD45A, Fas/Apo-1, and FOXO3A in prostate cancer cells, suggesting its effect on induction of apoptosis (Figure 6). The induction of apoptosis mediated by these molecules could be additional molecular mechanism(s) by which Taxotere exerts its growth-inhibitory effects on prostate cancer cells.
We found that Taxotere also inhibited the expression of transcription factors [high-mobility group protein (HMG) 1, HMG4, nuclear factor-Y (NF-Y) B, and pirin], serine/threonine kinase 15 (STK15), checkpoint kinase 1 (Chk1), polo-like kinase (PLK), and transforming acidic coiled coil containing protein 3 (TACC3), which have been related to oncogenesis [36-39]. In contrast, it upregulated the expression of tumor-suppressor genes including p21WAF1, p27KIP1, and N-myc downregulated gene 1 (NDRG1). These results are novel, and suggest that Taxotere may inhibit cancer cell growth and survival through inhibition of transcription and oncogenesis.
Taxotere showed differential effects on gene expression profiles between androgen-insensitive PC3 cells and androgen- sensitive LNCaP cells. We observed significant alteration in the expression of CDC46, survivin, TACC3, and forkhead box transcription factor M1 (FOXM1) in Taxoteretreated LNCaP cells compared to PC3 cells, suggesting that LNCaP cell is more sensitive to Taxotere-induced apoptosis. These results are consistent with published data from other investigators showing that different Taxotere-induced apoptotic pathways exist in LNCaP and PC3 prostate cancer cell lines [40]. Because Taxotere showed no effect on AR in LNCaP cells, it is likely that Taxotere may exert its growthinhibitory effects on prostate cancer through AR-independent pathways. Further in-depth studies are needed to address this issue, particularly the gene regulation by Taxotere in hormone-responsive and nonresponsive prostate cancers. Nevertheless, Taxotere inhibited cell growth in both androgen-insensitive PC3 cells and androgen-sensitive LNCaP cells through regulation of gene expression related to control of cell proliferation, cell cycle, apoptosis, transcription factor, and oncogenesis, suggesting that Taxotere may be used for the treatment of prostate cancer including advanced hormone-insensitive prostate cancer.
In conclusion, Taxotere directly and indirectly caused changes in the expression of a large number of genes that are critically involved in the control of cell survival, cell physiological behaviors, and oncogenesis. This information may provide novel additional molecular mechanism(s) by which Taxotere exerts its pleiotropic effects on prostate cancer cells. These results could be important in devising mechanism-based and targeted therapeutic strategies for prostate cancer, especially in devising combination therapy with Taxotere and other chemotherapeutic agents based on the molecular mechanism(s). However, further in-depth investigation is needed in order to establish cause-and-effect relationships between these altered genes with apoptosisinducing effect of Taxotere in prostate cancer cells.
Abbreviations
- CDC
cell division cycle
- CDKs
cyclin-dependent protein kinases
- Chk1
checkpoint kinase 1
- EGF
epidermal growth factor
- FGF
fibroblast growth factor
- FOXM1
forkhead box transcription factor M1
- FOXO3A
forkhead transcription factor FKHR-L1
- GADD45A
growth arrest and DNA damage gene 45A
- HMG
high-mobility group protein
- IGFBP
insulin-like growth factor binding protein
- NDRG1
N-myc downregulated gene 1
- NF-Y
nuclear factor-Y
- PLK
polo-like kinase
- STK15
serine/threonine kinase 15
- TACC3
transforming acidic coiled coil containing protein 3
- TBP
TATA box binding protein
Footnotes
This work was partly funded by a grant from Aventis Pharmaceuticals (awarded to F.H.S.).
References
- 1.Hong WK. The current status of docetaxel in solid tumors. An M. D. Anderson Cancer Center review. Oncology (Huntington) 2002;16:9–15. [PubMed] [Google Scholar]
- 2.Friedland D, Cohen J, Miller R, Voloshin M, Gluckman R, Lembersky B, Zidar B, Keating M, Reilly N, Dimitt B. A phase II trial of docetaxel (Taxotere) in hormone-refractory prostate cancer: correlation of antitumor effect to phosphorylation of Bcl-2. Semin Oncol. 1999;26:19–23. [PubMed] [Google Scholar]
- 3.Oh WK, Kantoff PW. Docetaxel (Taxotere)-based chemotherapy for hormone-refractory and locally advanced prostate cancer. Semin Oncol. 1999;26:49–54. [PubMed] [Google Scholar]
- 4.Gravis G, Bladou F, Salem N, Macquart-Moulin G, Serment G, Camerlo J, Genre D, Bardou VJ, Maraninchi D, Viens P. Weekly administration of docetaxel for symptomatic metastatic hormone-refractory prostate carcinoma. Cancer. 2003;98:1627–1634. doi: 10.1002/cncr.11687. [DOI] [PubMed] [Google Scholar]
- 5.Petrioli R, Pozzessere D, Messinese S, Sabatino M, Di Palma T, Marsili S, Correale P, Manganelli A, Salvestrini F, Francini G. Weekly low-dose docetaxel in advanced hormone-resistant prostate cancer patients previously exposed to chemotherapy. Oncology. 2003;64:300–305. doi: 10.1159/000070285. [DOI] [PubMed] [Google Scholar]
- 6.Fulton B, Spencer CM. Docetaxel. A review of its pharmacodynamic and pharmacokinetic properties and therapeutic efficacy in the management of metastatic breast cancer. Drugs. 1996;51:1075–1092. doi: 10.2165/00003495-199651060-00011. [DOI] [PubMed] [Google Scholar]
- 7.Stein CA. Mechanisms of action of taxanes in prostate cancer. Semin Oncol. 1999;26:3–7. [PubMed] [Google Scholar]
- 8.udny V, Nakano S. Src tyrosine kinase augments Taxotere-induced apoptosis through enhanced expression and phosphorylation of Bcl-2. Br J Cancer. 2002;86:463–469. doi: 10.1038/sj.bjc.6600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haldar S, Basu A, Croce CM. Bcl2 is the guardian of microtubule integrity. Cancer Res. 1997;57:229–233. [PubMed] [Google Scholar]
- 10.Avramis VI, Nandy P, Kwock R, Solorzano MM, Mukherjee SK, Danenberg P, Cohen LJ. Increased p21/WAF-1 and p53 protein levels following sequential three drug combination regimen of fludarabine, cytarabine and docetaxel induces apoptosis in human leukemia cells. Anticancer Res. 1998;18:2327–2338. [PubMed] [Google Scholar]
- 11.Chan OT, Yang LX. The immunological effects of taxanes. Cancer Immunol Immunother. 2000;49:181–185. doi: 10.1007/s002620000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American Cancer Society, author. Cancer Facts and Figures 2002. Atlanta, GA: American Cancer Society, Inc.; 2002. p. 6. [Google Scholar]
- 13.Gopalkrishnan RV, Kang DC, Fisher PB. Molecular markers and determinants of prostate cancer metastasis. J Cell Physiol. 2001;189:245–256. doi: 10.1002/jcp.10023. [DOI] [PubMed] [Google Scholar]
- 14.Kudoh K, Ramanna M, Ravatn R, Elkahloun AG, Bittner ML, Meltzer PS, Trent JM, Dalton WS, Chin KV. Monitoring the expression profiles of doxorubicin-induced and doxorubicin-resistant cancer cells by cDNA microarray. Cancer Res. 2000;60:4161–4166. [PubMed] [Google Scholar]
- 15.Zembutsu H, Ohnishi Y, Tsunoda T, Furukawa Y, Katagiri T, Ueyama Y, Tamaoki N, Nomura T, Kitahara O, Yanagawa R, Hirata K, Nakamura Y. Genome-wide cDNA microarray screening to correlate gene expression profiles with sensitivity of 85 human cancer xenografts to anticancer drugs. Cancer Res. 2002;62:518–527. [PubMed] [Google Scholar]
- 16.Yoo GH, Piechocki MP, Ensley JF, Nguyen T, Oliver J, Meng H, Kewson D, Shibuya TY, Lonardo F, Tainsky MA. Docetaxel induced gene expression patterns in head and neck squamous cell carcinoma using cDNA microarray and PowerBlot. Clin Cancer Res. 2002;8:3910–3921. [PubMed] [Google Scholar]
- 17.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using Onto-Express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 19.Downing KH, Nogales E. Tubulin and microtubule structure. Curr Opin Cell Biol. 1998;10:16–22. doi: 10.1016/s0955-0674(98)80082-3. [DOI] [PubMed] [Google Scholar]
- 20.Miller ML, Ojima I. Chemistry and chemical biology of taxane anticancer agents. Chem Rec. 2001;1:195–211. doi: 10.1002/tcr.1008. [DOI] [PubMed] [Google Scholar]
- 21.Fabre-Jonca N, Allaman JM, Radlgruber G, Meda P, Kiss JZ, French LE, Masson D. The distribution of murine 115-kDa epithelial microtubule-associated protein (E-MAP-115) during embryogenesis and in adult organs suggests a role in epithelial polarization and differentiation. Differentiation. 1998;63:169–180. doi: 10.1111/j.1432-0436.1998.00169.x. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson I, Hoffmann I. Cell cycle regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res. 2000;4:107–114. doi: 10.1007/978-1-4615-4253-7_10. [DOI] [PubMed] [Google Scholar]
- 23.Poggioli GJ, Dermody TS, Tyler KL. Reovirus-induced sigma1s- dependent G(2)/M phase cell cycle arrest is associated with inhibition of p34(cdc2) J Virol. 2001;75:7429–7434. doi: 10.1128/JVI.75.16.7429-7434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obaya AJ, Sedivy JM. Regulation of cyclin-Cdk activity in mammalian cells. Cell Mol Life Sci. 2002;59:126–142. doi: 10.1007/s00018-002-8410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erlanson M, Landberg G. Prognostic implications of p27 and cyclin E protein contents in malignant lymphomas. Leuk Lymphoma. 2001;40:461–470. doi: 10.3109/10428190109097645. [DOI] [PubMed] [Google Scholar]
- 26.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 27.Brechot C. Oncogenic activation of cyclin A. Curr Opin Genet Dev. 1993;3:11–18. doi: 10.1016/s0959-437x(05)80335-1. [DOI] [PubMed] [Google Scholar]
- 28.el Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, Hill DE, Healy E, Rees JL, Hamilton SR. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- 29.Kawamata N, Morosetti R, Miller CW, Park D, Spirin KS, Nakamaki T, Takeuchi S, Hatta Y, Simpson J, Wilcyznski S. Molecular analysis of the cyclin-dependent kinase inhibitor gene p27/Kip1 in human malignancies. Cancer Res. 1995;55:2266–2269. [PubMed] [Google Scholar]
- 30.Kolfschoten GM, Hulscher TM, Duyndam MC, Pinedo HM, Boven E. Variation in the kinetics of caspase-3 activation, Bcl-2 phosphorylation and apoptotic morphology in unselected human ovarian cancer cell lines as a response to docetaxel. Biochem Pharmacol. 2002;63:733–743. doi: 10.1016/s0006-2952(01)00895-4. [DOI] [PubMed] [Google Scholar]
- 31.Xia C, Xu Z, Yuan X, Uematsu K, You L, Li K, Li L, McCormick F, Jablons DM. Induction of apoptosis in mesothelioma cells by antisurvivin oligonucleotides. Mol Cancer Ther. 2002;1:687–694. [PubMed] [Google Scholar]
- 32.Maeda T, Hanna AN, Sim AB, Chua PP, Chong MT, Tron VA. GADD45 regulates G2/M arrest, DNA repair, and cell death in keratinocytes following ultraviolet exposure. J Invest Dermatol. 2002;119:22–26. doi: 10.1046/j.1523-1747.2002.01781.x. [DOI] [PubMed] [Google Scholar]
- 33.Yakirevich E, Maroun L, Cohen O, Izhak OB, Rennert G, Resnick MB. Apoptosis, proliferation, and Fas (APO-1, CD95)/Fas ligand expression in medullary carcinoma of the breast. J Pathol. 2000;192:166–173. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH689>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt M, van der HA, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 36.Miyoshi Y, Iwao K, Egawa C, Noguchi S. Association of centrosomal kinase STK15/BTAK mRNA expression with chromosomal instability in human breast cancers. Int J Cancer. 2001;92:370–373. doi: 10.1002/ijc.1200. [DOI] [PubMed] [Google Scholar]
- 37.Luo Y, Rockow-Magnone SK, Kroeger PE, Frost L, Chen Z, Han EK, Ng SC, Simmer RL, Giranda VL. Blocking Chk1 expression induces apoptosis and abrogates the G2 checkpoint mechanism. Neoplasia. 2001;3:411–419. doi: 10.1038/sj.neo.7900175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spankuch-Schmitt B, Wolf G, Solbach C, Loibl S, Knecht R, Stegmuller M, von Minckwitz G, Kaufmann M, Strebhardt K. Downregulation of human polo-like kinase activity by antisense oligonucleotides induces growth inhibition in cancer cells. Oncogene. 2002;21:3162–3171. doi: 10.1038/sj.onc.1205412. [DOI] [PubMed] [Google Scholar]
- 39.Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58:165–170. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- 40.Muenchen HJ, Poncza PJ, Pienta KJ. Different docetaxelinduced apoptotic pathways are present in prostate cancer cell lines LNCaP and PC-3. Urology. 2001;57:366–370. doi: 10.1016/s0090-4295(00)00935-3. [DOI] [PubMed] [Google Scholar]